MDR1 Inhibition Reverses Doxorubicin-Resistance in Six Doxorubicin-Resistant Canine Prostate and Bladder Cancer Cell Lines
Abstract
:1. Introduction
2. Results
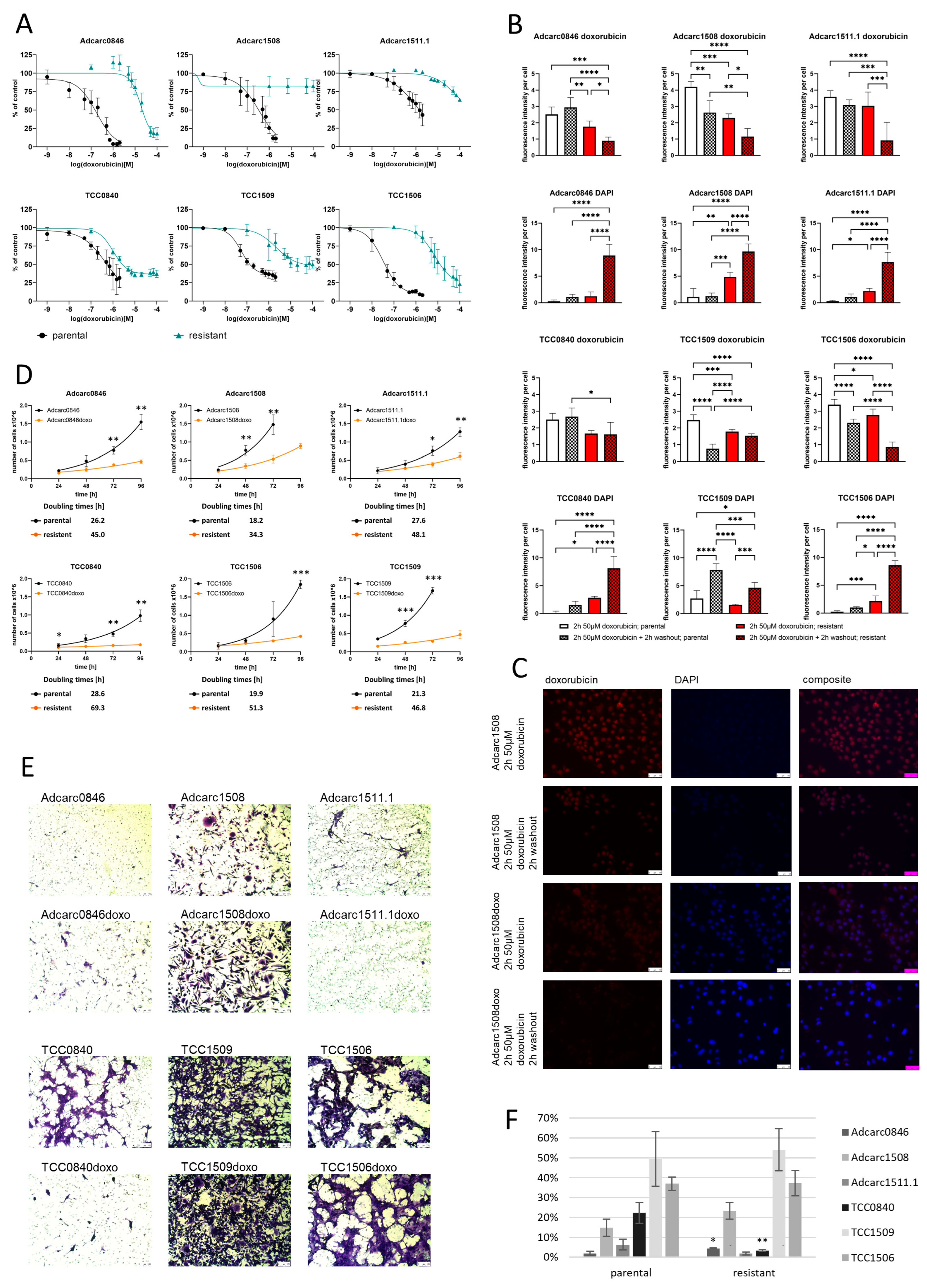
2.1. Six out of Nine Cell Lines Became Resistant to 2 µM Doxorubicin
2.2. Resistance Increased Drastically to Doxorubicin, without Establishing Cross-Resistance to Carboplatin
2.3. The Doxorubicin Resistance Is Based on Drug Efflux
2.4. Resistant Sublines Proliferate Slower Than Parental Cell Lines
2.5. TCC1509doxo and TCC1506doxo Remained Highly Invasive, while TCC0840doxo Lost Its Invasive Potential
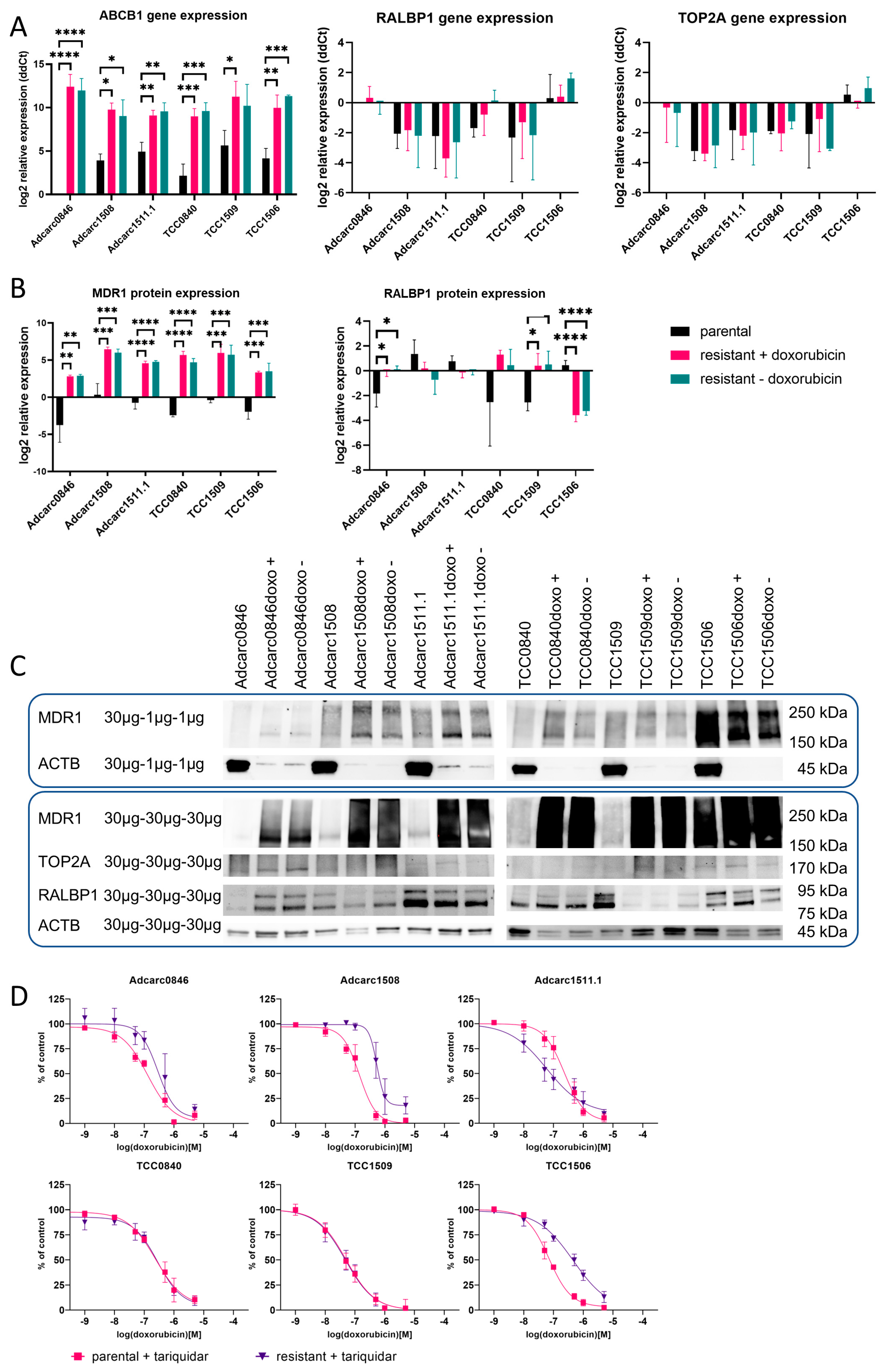
2.6. At the mRNA Level, ABCB1 Was Upregulated and Highly Expressed in All Resistant Cell Lines
2.7. At the Protein Level, ABCB1 Was Upregulated and Highly Expressed in All Resistant Cell Lines
2.8. MDR1 Inhibition Reversed the Doxorubicin Resistance in Varying Degrees
3. Discussion
4. Materials and Methods
4.1. Generation of Doxorubicin-Resistant Cells
4.2. Visualization of Doxorubicin Efflux by Fluorescence Microscopy
4.3. Quantification of Chemoresistance and Cross-Resistance to Carboplatin
4.4. Chemoresistance under MDR1 Inhibition
4.5. Calculation of Population Doubling Times
4.6. Estimation of Invasive Potential
4.7. Expression of Resistance-Associated Genes
4.8. Expression of Resistance-Associated Proteins
4.9. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Palmieri, C.; Foster, R.; Grieco, V.; Fonseca-Alves, C.; Wood, G.; Culp, W.; Escobar, H.M.; De Marzo, A.; Laufer-Amorim, R. Histopathological Terminology Standards for the Reporting of Prostatic Epithelial Lesions in Dogs. J. Comp. Pathol. 2019, 171, 30–37. [Google Scholar] [CrossRef]
- Fonseca-Alves, C.E.; Vicente, I.; Calazans, S.G.; Amorim, R.L. Canine Prostate Cancer: Would the Dog be an Important Model for the Study of New Drugs? Am. J. Drug Discov. Dev. 2013, 3, 220–224. [Google Scholar] [CrossRef]
- Leroy, B.R.; Northrup, N. Prostate cancer in dogs: Comparative and clinical aspects. Vet. J. 2009, 180, 149–162. [Google Scholar] [CrossRef] [PubMed]
- Ryman-Tubb, T.; Lothion-Roy, J.H.; Metzler, V.M.; Harris, A.E.; Robinson, B.D.; Rizvanov, A.A.; Jeyapalan, J.N.; James, V.H.; England, G.; Rutland, C.S.; et al. Comparative pathology of dog and human prostate cancer. Vet. Med. Sci. 2021, 8, 110–120. [Google Scholar] [CrossRef] [PubMed]
- Thiemeyer, H.; Taher, L.; Schille, J.T.; Packeiser, E.-M.; Harder, L.K.; Hewicker-Trautwein, M.; Brenig, B.; Schütz, E.; Beck, J.; Nolte, I.; et al. An RNA-Seq-Based Framework for Characterizing Canine Prostate Cancer and Prioritizing Clinically Relevant Biomarker Candidate Genes. Int. J. Mol. Sci. 2021, 22, 11481. [Google Scholar] [CrossRef]
- Wu, Q.; Yang, Z.; Nie, Y.; Shi, Y.; Fan, D. Multi-drug resistance in cancer chemotherapeutics: Mechanisms and lab approaches. Cancer Lett. 2014, 347, 159–166. [Google Scholar] [CrossRef]
- Klopfleisch, R.; Kohn, B.; Gruber, A. Mechanisms of tumour resistance against chemotherapeutic agents in veterinary oncology. Vet. J. 2016, 207, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Zandvliet, M.; Teske, E. Mechanisms of Drug Resistance in Veterinary Oncology—A Review with an Emphasis on Canine Lymphoma. Vet. Sci. 2015, 2, 150–184. [Google Scholar] [CrossRef]
- Wittenburg, L.A.; Weishaar, K.; Ramirez, D.; Gustafson, D.L. Doxorubicin area under the curve is an important predictor of neutropenia in dogs with naturally occurring cancers. Vet. Comp. Oncol. 2019, 17, 147–154. [Google Scholar] [CrossRef]
- Thorn, C.F.; Oshiro, C.; Marsh, S.; Hernandez-Boussard, T.; McLeod, H.; Klein, T.E.; Altman, R.B. Doxorubicin Pathways. Pharm. Genom. 2011, 21, 440–446. [Google Scholar] [CrossRef]
- Wyles, J.P.; Wu, Z.; Mirski, S.E.; Cole, S.P. Nuclear interactions of topoisomerase II and with phospholipid scramblase 1. Nucleic Acids Res. 2007, 35, 4076–4085. [Google Scholar] [CrossRef]
- Mealey, K.; Fidel, J. P-Glycoprotein Mediated Drug Interactions in Animals and Humans with Cancer. J. Vet. Intern. Med. 2015, 29, 1–6. [Google Scholar] [CrossRef]
- Zandvliet, M.; Teske, E.; Schrickx, J.; Mol, J. A longitudinal study of ABC transporter expression in canine multicentric lymphoma. Vet. J. 2015, 205, 263–271. [Google Scholar] [CrossRef]
- Uozurmi, K.; Nakaichi, M.; Yamamoto, Y.; Une, S.; Taura, Y. Development of multidrug resistance in a canine lymphoma cell line. Res. Vet. Sci. 2005, 78, 217–224. [Google Scholar] [CrossRef]
- Sahabi, K.; Selvarajah, G.T.; Mokrish, A.; Rasedee, A.; Kqueen, C.Y. Development and molecular characterization of doxorubicin-resistant canine mammary gland tumour cells. J. Appl. Anim. Res. 2022, 50, 125–145. [Google Scholar] [CrossRef]
- Zandvliet, M.; Teske, E.; Schrickx, J. Multi-drug resistance in a canine lymphoid cell line due to increased P-glycoprotein expression, a potential model for drug-resistant canine lymphoma. Toxicol. Vitr. 2014, 28, 1498–1506. [Google Scholar] [CrossRef]
- Mollberg, N.; Steinert, G.; Aigner, M.; Hamm, A.; Lin, F.-J.; Elbers, H.; Reissfelder, C.; Weitz, J.; Buchler, M.W.; Koch, M. Overexpression of RalBP1 in colorectal cancer is an independent predictor of poor survival and early tumor relapse. Cancer Biol. Ther. 2012, 13, 694–700. [Google Scholar] [CrossRef]
- Singhal, P.; Vatsyayan, R.; Chaudhary, P.; Lelsani, P.C.R.; Awasthi, Y.C.; Awasthi, S. Role of RLIP76 in doxorubicin resistance in lung cancer (Review). Int. J. Oncol. 2009, 34, 1505–1511. [Google Scholar] [CrossRef] [PubMed]
- Jullien-Flores, V.; Dorseuil, O.; Romero, F.; Letourneur, F.; Saragosti, S.; Berger, R.; Tavitian, A.; Gacon, G.; Camonis, J.H. Bridging Ral GTPase to Rho Pathways. J. Biol. Chem. 1995, 270, 22473–22477. [Google Scholar] [CrossRef] [PubMed]
- Rossé, C.; L’Hoste, S.; Offner, N.; Picard, A.; Camonis, J. RLIP, an Effector of the Ral GTPases, Is a Platform for Cdk1 to Phosphorylate Epsin during the Switch Off of Endocytosis in Mitosis. J. Biol. Chem. 2003, 278, 30597–30604. [Google Scholar] [CrossRef] [PubMed]
- McDermott, M.; Eustace, A.J.; Busschots, S.; Breen, L.; Crown, J.; Clynes, M.; O’Donovan, N.; Stordal, B.; O’Donovan, N. In vitro Development of Chemotherapy and Targeted Therapy Drug-Resistant Cancer Cell Lines: A Practical Guide with Case Studies. Front. Oncol. 2014, 4, 40. [Google Scholar] [CrossRef]
- ExPASy—Cellosaurus. Available online: http://web.expasy.org/cellosaurus/ (accessed on 10 January 2023).
- Mealey, K.L.; Barhoumi, R.; Rogers, K.; Kochevar, D.T. Doxorubicin induced expression of P-glycoprotein in a canine osteosarcoma cell line. Cancer Lett. 1998, 126, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Replogle-Schwab, T.S.; Schwab, E.D.; Pienta, K. Development of doxorubicin resistant rat prostate cancer cell lines. Anticancer Res. 1998, 17, 4535–4538. [Google Scholar]
- Packeiser, E.-M.; Hewicker-Trautwein, M.; Thiemeyer, H.; Mohr, A.; Junginger, J.; Schille, J.T.; Escobar, H.M.; Nolte, I. Characterization of six canine prostate adenocarcinoma and three transitional cell carcinoma cell lines derived from primary tumor tissues as well as metastasis. PLoS ONE 2020, 15, e0230272. [Google Scholar] [CrossRef]
- Packeiser, E.-M.; Taher, L.; Kong, W.; Ernst, M.; Beck, J.; Hewicker-Trautwein, M.; Brenig, B.; Schütz, E.; Escobar, H.M.; Nolte, I. RNA-seq of nine canine prostate cancer cell lines reveals diverse therapeutic target signatures. Cancer Cell Int. 2022, 22, 54. [Google Scholar] [CrossRef] [PubMed]
- Tegze, B.; Szallasi, Z.; Haltrich, I.; Pénzváltó, Z.; Tóth, Z.; Likó, I.; Gyorffy, B. Parallel Evolution under Chemotherapy Pressure in 29 Breast Cancer Cell Lines Results in Dissimilar Mechanisms of Resistance. PLoS ONE 2012, 7, e30804. [Google Scholar] [CrossRef]
- Vail, D.M.; Thamm, D.H.; Liptak, J.M. Hematopoietic Tumors. In Withrow and MacEwen’s Small Animal Clinical Oncology; Elsevier: St. Louis, MO, USA, 2019; pp. 688–772. [Google Scholar] [CrossRef]
- Valdivia, G.; Alonso-Diez, Á.; Pérez-Alenza, D.; Peña, L. From Conventional to Precision Therapy in Canine Mammary Cancer: A Comprehensive Review. Front. Vet. Sci. 2021, 8, 623800. [Google Scholar] [CrossRef]
- Fulkerson, C.M.; Knapp, D.W. Management of transitional cell carcinoma of the urinary bladder in dogs: A review. Vet. J. 2015, 205, 217–225. [Google Scholar] [CrossRef]
- Pérez-Blanco, J.S.; Gatta, M.D.M.F.D.; Hernández-Rivas, J.M.; Sánchez, M.J.G.; Marinero, M.L.S.; López, F.G. Validation and clinical evaluation of a UHPLC method with fluorescence detector for plasma quantification of doxorubicin and doxorubicinol in haematological patients. J. Chromatogr. B 2014, 955-956, 93–97. [Google Scholar] [CrossRef]
- Gustafson, D.; Thamm, D. Pharmacokinetic Modeling of Doxorubicin Pharmacokinetics in Dogs Deficient in ABCB1 Drug Transporters. J. Vet. Intern. Med. 2010, 24, 579–586. [Google Scholar] [CrossRef]
- Yang, L.-Y.; Trujillo, J.M.; Siciliano, M.J.; Kido, Y.; Siddik, Z.H.; Su, Y.-Z. Distinct P-glycoprotein expression in two subclones simultaneously selected from a human colon carcinoma cell line by cis-diamminedichloroplatinum (II). Int. J. Cancer 1993, 53, 478–485. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, C.; Grieco, V. Proposal of Gleason-like grading system of canine prostate carcinoma in veterinary pathology practice. Res. Vet. Sci. 2015, 103, 11–15. [Google Scholar] [CrossRef]
- Creemers, S.G.; Van Koetsveld, P.M.; De Herder, W.W.; Dogan, F.; Franssen, G.J.H.; A Feelders, R.; Hofland, L.J. MDR1 inhibition increases sensitivity to doxorubicin and etoposide in adrenocortical cancer. Endocr. Relat. Cancer 2019, 26, 367–378. [Google Scholar] [CrossRef]
- Kelly, R.J.; Draper, D.; Chen, C.C.; Robey, R.W.; Figg, W.D.; Piekarz, R.L.; Chen, X.; Gardner, E.R.; Balis, F.M.; Venkatesan, A.M.; et al. A Pharmacodynamic Study of Docetaxel in Combination with the P-glycoprotein Antagonist Tariquidar (XR9576) in Patients with Lung, Ovarian, and Cervical Cancer. Clin. Cancer Res. 2011, 17, 569–580. [Google Scholar] [CrossRef]
- Zandvliet, M.; Teske, E.; Chapuis, T.; Fink-Gremmels, J.; Schrickx, J.A. Masitinib reverses doxorubicin resistance in canine lymphoid cells by inhibiting the function of P-glycoprotein. J. Vet. Pharmacol. Ther. 2013, 36, 583–587. [Google Scholar] [CrossRef] [PubMed]
- Shao, M.; Shi, R.; Gao, Z.-X.; Gao, S.-S.; Li, J.-F.; Li, H.; Cui, S.-Z.; Hu, W.-M.; Chen, T.-Y.; Wu, G.-R.; et al. Crizotinib and Doxorubicin Cooperatively Reduces Drug Resistance by Mitigating MDR1 to Increase Hepatocellular Carcinoma Cells Death. Front. Oncol. 2021, 11. [Google Scholar] [CrossRef]
- Klose, K.; Packeiser, E.; Granados-Soler, J.; Hewicker-Trautwein, M.; Escobar, H.M.; Nolte, I. Evaluation of the therapeutic potential of masitinib and expression of its specific targets c-Kit, PDGFR -α, PDGFR -β, and Lyn in canine prostate cancer cell lines. Vet. Comp. Oncol. 2022, 20, 641–652. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.L.; Zhu, K.P.; Zhang, C.L. Identification of Gene Expression Profiles Associated with Doxorubicin Resistance in Paired Doxorubicin-Resistant and Doxorubicin-Sensitive Osteosarcoma Cell Lines. Int. J. Clin. Exp. Pathol. 2017, 10, 6254–6267. [Google Scholar]
- Hamaguchi, K.; Godwin, A.K.; Yakushiji, M.; O’Dwyer, P.J.; Ozols, R.F.; Hamilton, T.C. Cross-resistance to diverse drugs is associated with primary cisplatin resistance in ovarian cancer cell lines. Cancer Res. 1993, 53. [Google Scholar]
- Awasthi, S.; Singhal, S.S.; Sharma, R.; Zimniak, P.; Awasthi, Y.C. Transport of glutathione conjugates and chemotherapeutic drugs by RLIP76 (RALBP1): A novel link between G-protein and tyrosine kinase signaling and drug resistance. Int. J. Cancer 2003, 106, 635–646. [Google Scholar] [CrossRef]
- Chen, Z.; Shi, T.; Zhang, L.; Zhu, P.; Deng, M.; Huang, C.; Hu, T.; Jiang, L.; Li, J. Mammalian drug efflux transporters of the ATP binding cassette (ABC) family in multidrug resistance: A review of the past decade. Cancer Lett. 2016, 370, 153–164. [Google Scholar] [CrossRef]
- Silverton, L.; Dean, M.; Moitra, K. Variation and evolution of the ABC transporter genes ABCB1, ABCC1, ABCG2, ABCG5 and ABCG8: Implication for pharmacogenetics and disease. Drug Metab. Drug Interact. 2011, 26, 169–179. [Google Scholar] [CrossRef]
- Kimchi-Sarfaty, C.; Oh, J.M.; Kim, I.-W.; Sauna, Z.E.; Calcagno, A.M.; Ambudkar, S.V.; Gottesman, M.M. A “Silent” Polymorphism in the MDR1 Gene Changes Substrate Specificity. Science 2007, 315, 525–528. [Google Scholar] [CrossRef]
- Mao, Y.; Yu, C.; Hsieh, T.-S.; Nitiss, J.L.; Liu, A.A.; Wang, H.; Liu, L.F. Mutations of Human Topoisomerase IIα Affecting Multidrug Resistance and Sensitivity. Biochemistry 1999, 38, 10793–10800. [Google Scholar] [CrossRef]
- Harbottle, A.; Daly, A.; Atherton, K.; Campbell, F.C. Role of glutathioneS-transferase P1, P-glycoprotein and multidrug resistance-associated protein 1 in acquired doxorubicin resistance. Int. J. Cancer 2001, 92, 777–783. [Google Scholar] [CrossRef] [PubMed]
- Tsou, S.-H.; Chen, T.-M.; Hsiao, H.-T.; Chen, Y.-H. A Critical Dose of Doxorubicin Is Required to Alter the Gene Expression Profiles in MCF-7 Cells Acquiring Multidrug Resistance. PLoS ONE 2015, 10, e0116747. [Google Scholar] [CrossRef] [PubMed]
- Calcagno, A.M.; Salcido, C.D.; Gillet, J.-P.; Wu, C.-P.; Fostel, J.M.; Mumau, M.D.; Gottesman, M.M.; Varticovski, L.; Ambudkar, S.V. Prolonged Drug Selection of Breast Cancer Cells and Enrichment of Cancer Stem Cell Characteristics. Gynecol. Oncol. 2010, 102, 1637–1652. [Google Scholar] [CrossRef] [PubMed]
- Greife, A.; Tukova, J.; Steinhoff, C.; Scott, S.D.; Schulz, W.A.; Hatina, J. Establishment and characterization of a bladder cancer cell line with enhanced doxorubicin resistance by mevalonate pathway activation. Tumor Biol. 2015, 36, 3293–3300. [Google Scholar] [CrossRef] [PubMed]
- Amack, J.D. Cellular dynamics of EMT: Lessons from live in vivo imaging of embryonic development. Cell Commun. Signal. 2021, 19, 1–16. [Google Scholar] [CrossRef]
- Tam, S.; Gross, N.D. Cutaneous Squamous Cell Carcinoma in Immunosuppressed Patients. Curr. Oncol. Rep. 2019, 21, 82. [Google Scholar] [CrossRef]
- Boffetta, P.; Kaldor, J.M. Secondary Malignancies Following Cancer Chemotherapy. Acta Oncol. 1994, 33, 591–598. [Google Scholar] [CrossRef]
- Gustafson, D.L.; Rastatter, J.C.; Colombo, T.; Long, M.E. Doxorubicin pharmacokinetics: Macromolecule binding, metabolism, and excretion in the context of a physiologic model. J. Pharm. Sci. 2002, 91, 1488–1501. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Hellemans, J.; Mortier, G.; De Paepe, A.; Speleman, F.; Vandesompele, J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007, 8, R19. [Google Scholar] [CrossRef] [PubMed]
- Rempel, L.M.; Lillevang, K.T.A.; Straten, A.-K.T.; Friðriksdóttir, S.B.; Körber, H.; Wehrend, A.; Kowalewski, M.P.; Reichler, I.M.; Balogh, O.; Goericke-Pesch, S. Do uterine PTGS2, PGFS, and PTGFR expression play a role in canine uterine inertia? Cell Tissue Res. 2021, 385, 251–264. [Google Scholar] [CrossRef] [PubMed]
| Cell Line 1 | Histological Classification | Time to Resistance | IC50 Parental Doxorubicin 3 | IC50 Resistant Doxorubicin | IC50 Parental Carboplatin 3 | IC50 Resistant Carboplatin |
|---|---|---|---|---|---|---|
| TihoDProAdcarc1258 | PAC of the prostate | N/A 2 | 0.35 µM | N/A 2 | 97.7 µM | N/A 2 |
| TihoDProAdcarc0846 | PAC of the prostate | 30 weeks | 0.18 µM | 18.40 µM | 106.0 µM | 115.3 µM |
| TihoDProAdcarc1508 | PAC of the prostate | 28 weeks | 0.35 µM | X 4 | 67.7 µM | 87.9 µM |
| TihoDProAdcarc1511.1 | PAC of the prostate | 48 weeks | 1.31 µM | X 4 | 86.1 µM | 94.6 µM |
| TihoDProMetadcarc1511.2 | PAC metastasis | N/A 2 | >2 µM | N/A 2 | 38.3 µM | N/A 2 |
| TihoDProMetadcarc1511.3 | PAC metastasis | N/A 2 | >2 µM | N/A 2 | 46.1 µM | N/A 2 |
| TihoDProCarc/TCC0840 | TCC of the prostate | 64 weeks | 0.49 µM | 1.03 µM | 129.3 µM | 70.1 µM |
| TihoDProTCC1509 | TCC of the prostate | 32 weeks | 0.06 µM | 2.10 µM | 33.6 µM | 30.7 µM |
| TihoDUrtTCC1506 | TCC of the urinary bladder | 40 weeks | 0.03 µM | 7.26 µM | 39.8 µM | 78.6 µM |
| Cell Line | IC50 Doxorubicin with Tariquidar |
|---|---|
| Adcarc0846 | 0.13 µM |
| Adcarc0846doxo | 0.29 µM |
| Adcarc1508 | 0.14 µM |
| Adcarc1508doxo | 0.54 µM |
| Adcarc1511.1 | 0.23 µM |
| Adcarc1511.1doxo | 0.06 µM |
| TCC0840 | 0.24 µM |
| TCC0840doxo | 0.28 µM |
| TCC1509 | 0.05 µM |
| TCC1509doxo | 0.05 µM |
| TCC1506 | 0.07 µM |
| TCC1506doxo | 0.46 µM |
| Gene | Sequence (5′-3′) | Amplicon Length | Efficiency | Temperature | Accession Number |
|---|---|---|---|---|---|
| ABCB1 | for GACTCGGGAGCAGAAGTTTGA rev ACCCCGAAGATGTGTGCTTT | 90 bp | 1.97 | 57 °C | NM_001003215.2 |
| RALBP1 | for TGGCATGAAGTGTGAAGGCA rev TCCTCTCTGTCATAGGCTGCT | 84 bp | 1.94 | 57 °C | XM_038672657.1 |
| TOP2A | for TCAGCCCTTTGGCTCGGTTA rev TTGCAGGACCACCCAGTACC | 160 bp | 1.93 | 60 °C | XM_038676554.1 XM_038676553.1 |
| GAPDH [57] | for GGCCAAGAGGGTCATCATCTC rev GGGGCCGTCCACGGTCTTCT | 228 bp | 1.99 | 60 °C | NM_001003142 |
| ACTB [57] | for GCTGTGCTGTCCCTGTATG rev GCGTACCCCTCATAGATGG | 98 bp | 1.98 | 60 °C | NM_001195845.3 |
| HPRT | for TGACACTGGGAAAACAATGCA rev GGTCCTTTTCACCAGCAAGCT | 94 bp | 2.05 | 60 °C | NM_001003357.2 NM_001313818.1 |
| Target | Host | Clone | Manufacturer | Dilution |
|---|---|---|---|---|
| ABCB1 | rabbit | monoclonal EPR10364-57 (ab170904) | Abcam plc, Cambridge, UK | 1/1000 |
| RALBP1 | mouse | monoclonal H-10 (sc-48337) | Santa Cruz Biotechnology, Santa Cruz, TX, USA | 1/200 |
| TOP2A | mouse | monoclonal F-12 (sc-365916) | Santa Cruz Biotechnology, Santa Cruz, TX, USA | 1/200 |
| ACTB | rabbit | monoclonal 13E5 (#4970) | Cell Signaling Technology, Leiden, Netherlands | 1/1000 |
| rabbit IgG | goat | polyclonal BA-1000 | Vector Laboratories, Burlingame, CA, USA | 1/1000 |
| mouse IgG | horse | polyclonal BA-9200 | Vector Laboratories, Burlingame, CA, USA | 1/1000 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Packeiser, E.-M.; Engels, L.; Nolte, I.; Goericke-Pesch, S.; Murua Escobar, H. MDR1 Inhibition Reverses Doxorubicin-Resistance in Six Doxorubicin-Resistant Canine Prostate and Bladder Cancer Cell Lines. Int. J. Mol. Sci. 2023, 24, 8136. https://doi.org/10.3390/ijms24098136
Packeiser E-M, Engels L, Nolte I, Goericke-Pesch S, Murua Escobar H. MDR1 Inhibition Reverses Doxorubicin-Resistance in Six Doxorubicin-Resistant Canine Prostate and Bladder Cancer Cell Lines. International Journal of Molecular Sciences. 2023; 24(9):8136. https://doi.org/10.3390/ijms24098136
Chicago/Turabian StylePackeiser, Eva-Maria, Leoni Engels, Ingo Nolte, Sandra Goericke-Pesch, and Hugo Murua Escobar. 2023. "MDR1 Inhibition Reverses Doxorubicin-Resistance in Six Doxorubicin-Resistant Canine Prostate and Bladder Cancer Cell Lines" International Journal of Molecular Sciences 24, no. 9: 8136. https://doi.org/10.3390/ijms24098136
APA StylePackeiser, E. -M., Engels, L., Nolte, I., Goericke-Pesch, S., & Murua Escobar, H. (2023). MDR1 Inhibition Reverses Doxorubicin-Resistance in Six Doxorubicin-Resistant Canine Prostate and Bladder Cancer Cell Lines. International Journal of Molecular Sciences, 24(9), 8136. https://doi.org/10.3390/ijms24098136





