Analysis of Radiation Toxicity in Mammalian Cells Stably Transduced with Mitochondrial Stat3
Abstract
1. Introduction
2. Results
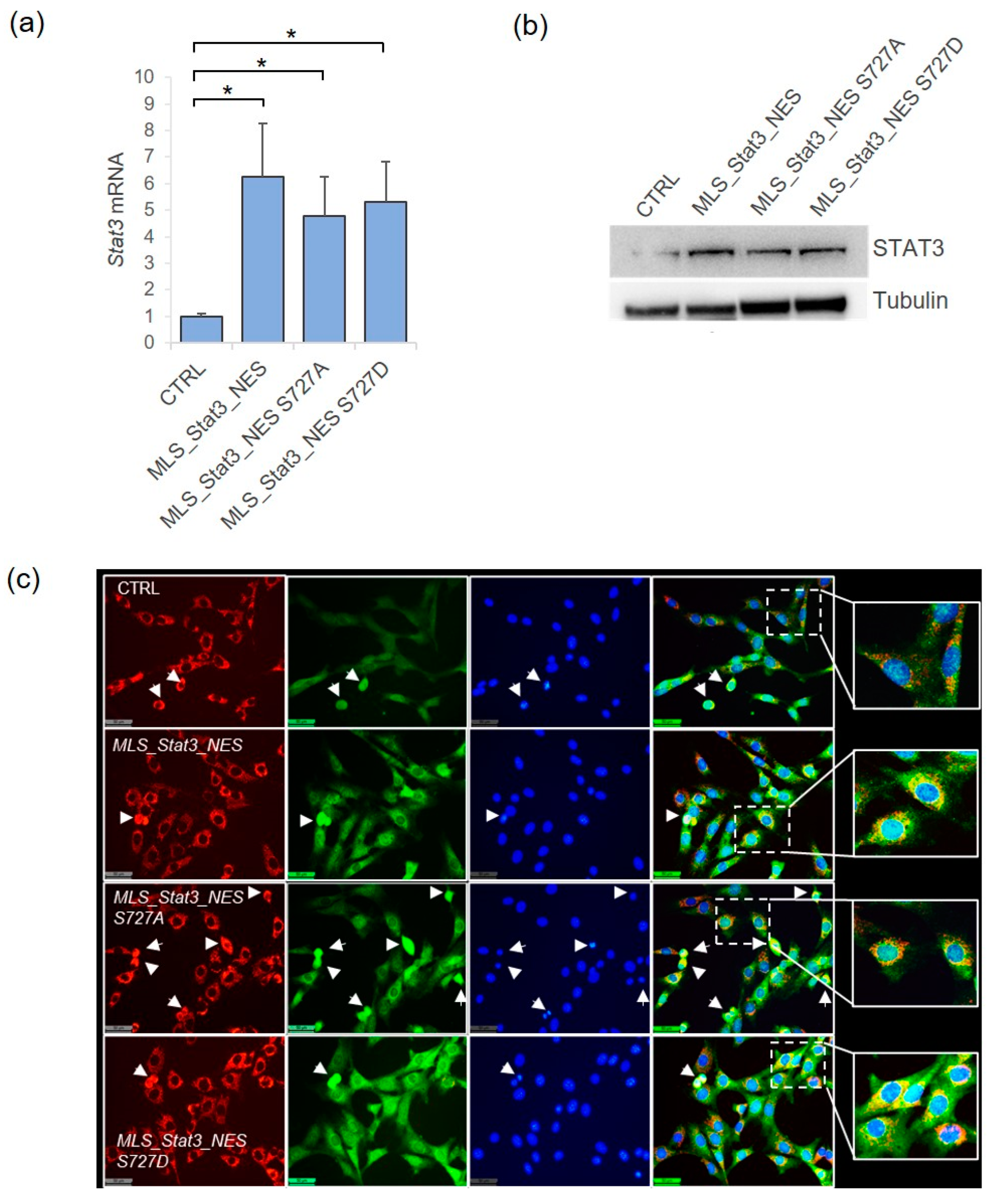
2.1. MitoSTAT3 Can Modulate the Proliferation of NIH-3T3 Mouse Cells
2.2. S727 Is Involved in the mitoSTAT3-Mediated Increase of Cell Viability upon UVC Stress
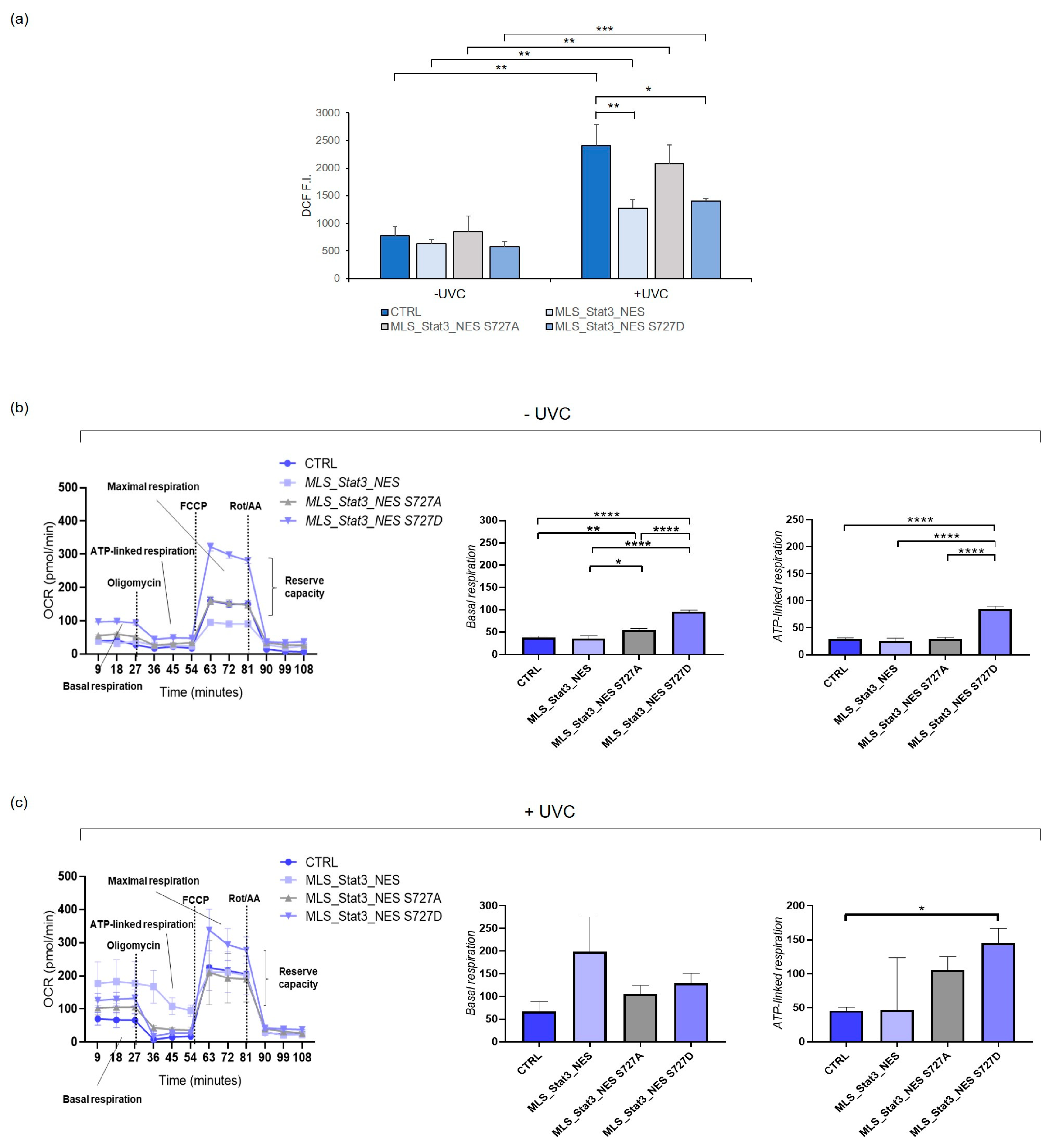
2.3. Oxidative Stress Induction and Mitochondrial Respiration in mitoStat3-Transduced Cells Irradiated with UVC
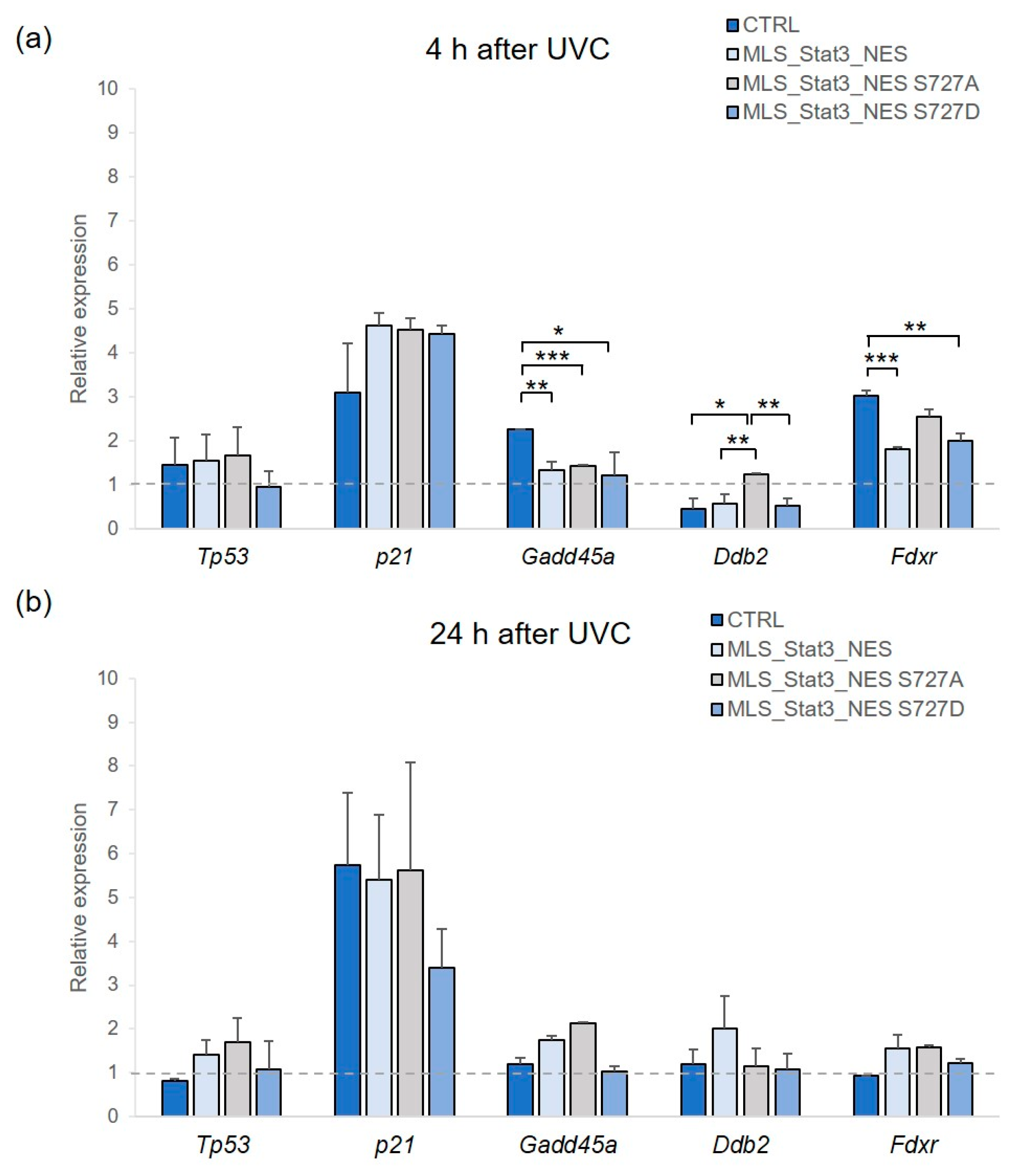
2.4. Gene Expression Analysis in mitoStat3-Transduced Cells Irradiated with UVC
2.5. UVC Radiation Toxicity in mitoStat3-Transduced Human HCT-116 Cells
3. Discussion
4. Materials and Methods
4.1. Cell Cultures and Reagents
4.2. Cell Irradiation
4.3. Generation of Lentiviral Constructs
4.4. Generation of Stable Cell Line Expressing mitoStat3
4.5. Cell Proliferation Assay
4.6. MTS Assay
4.7. Clonogenic Assay
4.8. Live-Dead Assay
4.9. Apoptosis Assay
4.10. Flow Cytometry Analyses
4.11. RNA Extraction and qRT-PCR
4.12. Cell Extracts and Western Blot
4.13. Immunofluorescence and Fluorescence Microscopy
4.14. Measurement of ROS Generation in Living Cells
4.15. Seahorse Assay
4.16. Establishing 3D Cultures
4.17. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hirano, T.; Ishihara, K.; Hibi, M. Roles of STAT3 in mediating the cell growth, differentiation and survival signals relayed through the IL-6 family of cytokine receptors. Oncogene 2000, 19, 2548–2556. [Google Scholar] [CrossRef]
- Yu, H.; Lee, H.; Herrmann, A.; Buettner, R.; Jove, R. Revisiting STAT3 signalling in cancer: New and unexpected biological functions. Nat. Rev. Cancer 2014, 14, 736–746. [Google Scholar] [CrossRef]
- Tošić, I.; Frank, D.A. STAT3 as a mediator of oncogenic cellular metabolism: Pathogenic and therapeutic implications. Neoplasia 2021, 23, 1167–1178. [Google Scholar] [CrossRef]
- Yu, H.; Pardoll, D.; Jove, R. STATs in cancer inflammation and immunity: A leading role for STAT3. Nat. Rev. Cancer 2009, 9, 798–809. [Google Scholar] [CrossRef]
- Yang, J.; Liao, X.; Agarwal, M.K.; Barnes, L.; Auron, P.E.; Stark, G.R. Unphosphorylated STAT3 accumulates in response to IL-6 and activates transcription by binding to NFkappaB. Genes Dev. 2007, 21, 1396–1408. [Google Scholar] [CrossRef]
- Wegrzyn, J.; Potla, R.; Chwae, Y.-J.; Sepuri, N.B.V.; Zhang, Q.; Koeck, T.; Derecka, M.; Szczepanek, K.; Szelag, M.; Gornicka, A.; et al. Function of Mitochondrial Stat3 in Cellular Respiration. Science 2009, 323, 793–797. [Google Scholar] [CrossRef]
- Garama, D.J.; White, C.L.; Balic, J.J.; Gough, D.J. Mitochondrial STAT3: Powering up a potent factor. Cytokine 2016, 87, 20–25. [Google Scholar] [CrossRef]
- Tesoriere, A.; Dinarello, A.; Argenton, F. The Roles of Post-Translational Modifications in STAT3 Biological Activities and Functions. Biomedicines 2021, 9, 956. [Google Scholar] [CrossRef]
- Gough, D.J.; Corlett, A.; Schlessinger, K.; Wegrzyn, J.; Larner, A.C.; Levy, D.E. Mitochondrial STAT3 Supports Ras-Dependent Oncogenic Transformation. Science 2009, 324, 1713–1716. [Google Scholar] [CrossRef]
- Szczepanek, K.; Chen, Q.; Derecka, M.; Salloum, F.N.; Zhang, Q.; Szelag, M.; Cichy, J.; Kukreja, R.C.; Dulak, J.; Lesnefsky, E.J.; et al. Mitochondrial-targeted Signal Transducer and Activator of Transcription 3 (STAT3) Protects against Ischemia-induced Changes in the Electron Transport Chain and the Generation of Reactive Oxygen Species. J. Biol. Chem. 2011, 286, 29610–29620. [Google Scholar] [CrossRef]
- Macias, E.; Rao, D.; Carbajal, S.; Kiguchi, K.; DiGiovanni, J. Stat3 Binds to mtDNA and Regulates Mitochondrial Gene Expression in Keratinocytes. J. Investig. Dermatol. 2014, 134, 1971–1980. [Google Scholar] [CrossRef]
- Carbognin, E.; Betto, R.M.; E Soriano, M.; Smith, A.G.; Martello, G. Stat3 promotes mitochondrial transcription and oxidative respiration during maintenance and induction of naive pluripotency. EMBO J. 2016, 35, 618–634. [Google Scholar] [CrossRef]
- Ploeger, C.; Huth, T.; Sugiyanto, R.N.; Pusch, S.; Goeppert, B.; Singer, S.; Tabti, R.; Hausser, I.; Schirmacher, P.; Désaubry, L.; et al. Prohibitin, STAT3 and SH2D4A physically and functionally interact in tumor cell mitochondria. Cell Death Dis. 2020, 11, 1023. [Google Scholar] [CrossRef]
- Peron, M.; Dinarello, A.; Meneghetti, G.; Martorano, L.; Betto, R.M.; Facchinello, N.; Tesoriere, A.; Tiso, N.; Martello, G.; Argenton, F. Y705 and S727 are required for the mitochondrial import and transcriptional activities of STAT3, and for regulation of stem cell proliferation. Development 2021, 148, dev199477. [Google Scholar] [CrossRef]
- Barry, S.P.; Townsend, P.A.; Knight, R.A.; Scarabelli, T.M.; Latchman, D.S.; Stephanou, A. STAT3 modulates the DNA damage response pathway. Int. J. Exp. Pathol. 2010, 91, 506–514. [Google Scholar] [CrossRef]
- Carballo, M.; Conde, M.; El Bekay, R.; Martín-Nieto, J.; Camacho, M.J.; Monteseirín, J.; Conde, J.; Bedoya, F.J.; Sobrino, F. Oxidative Stress Triggers STAT3 Tyrosine Phosphorylation and Nuclear Translocation in Human Lymphocytes. J. Biol. Chem. 1999, 274, 17580–17586. [Google Scholar] [CrossRef]
- Ng, I.H.; Yeap, Y.Y.; Ong, L.S.; Jans, D.A.; Bogoyevitch, M.A. Oxidative stress impairs multiple regulatory events to drive persistent cytokine-stimulated STAT3 phosphorylation. Biochim. Biophys. Acta 2014, 1843, 483–494. [Google Scholar] [CrossRef]
- Bito, T.; Sumita, N.; Masaki, T.; Shirakawa, T.; Ueda, M.; Yoshiki, R.; Tokura, Y.; Nishigori, C. Ultraviolet light induces Stat3 activation in human keratinocytes and fibroblasts through reactive oxygen species and DNA damage. Exp. Dermatol. 2010, 19, 654–660. [Google Scholar] [CrossRef]
- Ahsan, H.; Aziz, M.H.; Ahmad, N. Ultraviolet B exposure activates Stat3 signaling via phosphorylation at tyrosine705 in skin of SKH1 hairless mouse: A target for the management of skin cancer? Biochem. Biophys. Res. Commun. 2005, 333, 241–246. [Google Scholar] [CrossRef]
- Wen, Z.; Zhong, Z.; Darnell, J.E., Jr. Maximal activation of transcription by stat1 and stat3 requires both tyrosine and serine phosphorylation. Cell 1995, 82, 241–250. [Google Scholar] [CrossRef]
- Meier, J.A.; Larner, A.C. Toward a new STATe: The role of STATs in mitochondrial function. Semin. Immunol. 2014, 26, 20–28. [Google Scholar] [CrossRef]
- Lim, C.P.; Cao, X. Serine Phosphorylation and Negative Regulation of Stat3 by JNK. J. Biol. Chem. 1999, 274, 31055–31061. [Google Scholar] [CrossRef]
- Friedberg, E.C.; Walker, G.C.; Siede, W. DNA Repair and Mutagenesis; W. H. Freeman and Company: San Francisco, CA, USA, 1995. [Google Scholar]
- Dunkern, T.R.; Fritz, G.; Kaina, B. Ultraviolet light-induced DNA damage triggers apoptosis in nucleotide excision repair-deficient cells via Bcl-2 decline and caspase-3/-8 activation. Oncogene 2001, 20, 6026–6038. [Google Scholar] [CrossRef]
- Canman, C.E.; Kastan, M.B. Three paths to stress relief. Nature 1996, 384, 213–214. [Google Scholar] [CrossRef]
- Tai, M.-H.; Weng, C.-H.; Mon, D.-P.; Hu, C.-Y.; Wu, M.-H. Ultraviolet C Irradiation Induces Different Expression of Cyclooxygenase 2 in NIH 3T3 Cells and A431 Cells: The Roles of COX-2 Are Different in Various Cell Lines. Int. J. Mol. Sci. 2012, 13, 4351–4366. [Google Scholar] [CrossRef]
- Debacq-Chainiaux, F.; Leduc, C.; Verbeke, A.; Toussaint, O. UV, stress and aging. Dermato-Endocrinology 2012, 4, 236–240. [Google Scholar] [CrossRef]
- Childs, B.G.; Baker, D.J.; Kirkland, J.L.; Campisi, J.; Van Deursen, J.M. Senescence and apoptosis: Dueling or complementary cell fates? EMBO Rep. 2014, 15, 1139–1153. [Google Scholar] [CrossRef]
- Feng, R.; Han, J.; Ziegler, J.; Yang, M.; Castranova, V. Apaf-1 deficiency confers resistance to ultraviolet-induced apoptosis in mouse embryonic fibroblasts by disrupting reactive oxygen species amplification production and mitochondrial pathway. Free. Radic. Biol. Med. 2012, 52, 889–897. [Google Scholar] [CrossRef]
- Carpenter, R.L.; Lo, H.-W. STAT3 Target Genes Relevant to Human Cancers. Cancers 2014, 6, 897–925. [Google Scholar] [CrossRef]
- Fotedar, R.; Bendjennat, M.; Fotedar, A. Role of p21WAF1 in the cellular response to UV. Cell Cycle 2004, 3, 132–135. [Google Scholar] [CrossRef]
- Jung, H.J.; Kim, E.H.; Mun, J.-Y.; Park, S.; Smith, M.L.; Han, S.S.; Seo, Y.R. Base excision DNA repair defect in Gadd45a-deficient cells. Oncogene 2007, 26, 7517–7525. [Google Scholar] [CrossRef]
- Kiuchi, N.; Nakajima, K.; Ichiba, M.; Fukada, T.; Narimatsu, M.; Mizuno, K.; Hibi, M.; Hirano, T. STAT3 Is Required for the gp130-mediated Full Activation of the c-myc Gene. J. Exp. Med. 1999, 189, 63–73. [Google Scholar] [CrossRef]
- El-Deiry, W.S.; Tokino, T.; Velculescu, V.E.; Levy, D.B.; Parsons, R.; Trent, J.M.; Lin, D.; Mercer, W.E.; Kinzler, K.W.; Vogelstein, B. WAF1, a potential mediator of p53 tumor suppression. Cell 1993, 75, 817–825. [Google Scholar] [CrossRef]
- Smith, M.L.; Ford, J.M.; Hollander, M.C.; Bortnick, R.A.; Amundson, S.A.; Seo, Y.R.; Deng, C.-X.; Hanawalt, P.C.; Fornace, A.J., Jr. p53-Mediated DNA Repair Responses to UV Radiation: Studies of Mouse Cells Lacking p53, p21, and/or gadd45 Genes. Mol. Cell. Biol. 2000, 20, 3705–3714. [Google Scholar] [CrossRef]
- Li, H.; Zhang, X.-P.; Liu, F. Coordination between p21 and DDB2 in the Cellular Response to UV Radiation. PLOS ONE 2013, 8, e80111. [Google Scholar] [CrossRef]
- Niu, G.; Wright, K.L.; Ma, Y.; Wright, G.M.; Huang, M.; Irby, R.; Briggs, J.; Karras, J.; Cress, W.D.; Pardoll, D.; et al. Role of Stat3 in Regulating p53 Expression and Function. Mol. Cell. Biol. 2005, 25, 7432–7440. [Google Scholar] [CrossRef]
- Sammons, M.A.; Nguyen, T.-A.T.; McDade, S.S.; Fischer, M. Tumor suppressor p53: From engaging DNA to target gene regulation. Nucleic Acids Res. 2020, 48, 8848–8869. [Google Scholar] [CrossRef]
- Kastan, M.B.; I Radin, A.; Kuerbitz, S.J.; Onyekwere, O.; A Wolkow, C.; I Civin, C.; Stone, K.D.; Woo, T.; Ravindranath, Y.; Craig, R.W. Levels of p53 protein increase with maturation in human hematopoietic cells. Cancer Res 1991, 51, 4279–4286. [Google Scholar]
- Fu, L.; Benchimol, S. Participation of the human p53 3′UTR in translational repression and activation following gamma-irradiation. EMBO J. 1997, 16, 4117–4125. [Google Scholar] [CrossRef]
- Haronikova, L.; Olivares-Illana, V.; Wang, L.; Karakostis, K.; Chen, S.; Fåhraeus, R. The p53 mRNA: An integral part of the cellular stress response. Nucleic Acids Res. 2019, 47, 3257–3271. [Google Scholar] [CrossRef]
- Deng, C.; Zhang, P.; Harper, J.W.; Elledge, S.J.; Leder, P. Mice Lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell 1995, 82, 675–684. [Google Scholar] [CrossRef] [PubMed]
- Bunz, F.; Dutriaux, A.; Lengauer, C.; Waldman, T.; Zhou, S.; Brown, J.P.; Sedivy, J.M.; Kinzler, K.W.; Vogelstein, B. Requirement for p53 and p21 to Sustain G2 Arrest After DNA Damage. Science 1998, 282, 1497–1501. [Google Scholar] [CrossRef] [PubMed]
- Gorospe, M.; Wang, X.; Holbrook, N.J. Functional role of p21 during the cellular response to stress. Gene Expr. 1999, 7, 377–385. [Google Scholar]
- Stoyanova, T.; Yoon, T.; Kopanja, D.; Mokyr, M.B.; Raychaudhuri, P. The Xeroderma Pigmentosum Group E Gene Product DDB2 Activates Nucleotide Excision Repair by Regulating the Level of p21Waf1/Cip1. Mol. Cell. Biol. 2008, 28, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Phesse, T.; Myant, K.B.; Cole, A.M.; A Ridgway, R.; Pearson, H.; Muncan, V.; van den Brink, G.R.; Vousden, K.H.; Sears, R.; Vassilev, L.T.; et al. Endogenous c-Myc is essential for p53-induced apoptosis in response to DNA damage in vivo. Cell Death Differ. 2014, 21, 956–966. [Google Scholar] [CrossRef]
- Blattner, C.; Kannouche, P.; Litfin, M.; Bender, K.; Rahmsdorf, H.J.; Angulo, J.F.; Herrlich, P. UV-Induced Stabilization of c-fos and Other Short-Lived mRNAs. Mol. Cell. Biol. 2000, 20, 3616–3625. [Google Scholar] [CrossRef]
- Pedeux, R.; Lefort, K.; Cuenin, C.; Cortes, U.; Kellner, K.; Doré, J.-F.; Nakazawa, H. Specific induction ofgadd45in human melanocytes and melanoma cells after UVB irradiation. Int. J. Cancer 2002, 98, 811–816. [Google Scholar] [CrossRef]
- Cazzalini, O.; Perucca, P.; Mocchi, R.; Sommatis, S.; Prosperi, E.; Stivala, L.A. DDB2 association with PCNA is required for its degradation after UV-induced DNA damage. Cell Cycle 2014, 13, 240–248. [Google Scholar] [CrossRef]
- Zhang, Y.; Qian, Y.; Zhang, J.; Yan, W.; Jung, Y.-S.; Chen, M.; Huang, E.; Lloyd, K.; Duan, Y.; Wang, J.; et al. Ferredoxin reductase is critical for p53-dependent tumor suppression via iron regulatory protein 2. Genes Dev. 2017, 31, 1243–1256. [Google Scholar] [CrossRef]
- Liu, G.; Chen, X. The ferredoxin reductase gene is regulated by the p53 family and sensitizes cells to oxidative stress-induced apoptosis. Oncogene 2002, 21, 7195–7204. [Google Scholar] [CrossRef]
- Rieger, K.E.; Chu, G. Portrait of transcriptional responses to ultraviolet and ionizing radiation in human cells. Nucleic Acids Res. 2004, 32, 4786–4803. [Google Scholar] [CrossRef]
- Zoetemelk, M.; Rausch, M.; Colin, D.J.; Dormond, O.; Nowak-Sliwinska, P. Short-term 3D culture systems of various complexity for treatment optimization of colorectal carcinoma. Sci. Rep. 2019, 9, 7103. [Google Scholar] [CrossRef] [PubMed]
- Fede, C.; Selvestrel, F.; Compagnin, C.; Mognato, M.; Mancin, F.; Reddi, E.; Celotti, L. The toxicity outcome of silica nanoparticles (Ludox®) is influenced by testing techniques and treatment modalities. Anal. Bioanal. Chem. 2012, 404, 1789–1802. [Google Scholar] [CrossRef] [PubMed]
- Tajoli, F.; Dengo, N.; Mognato, M.; Dolcet, P.; Lucchini, G.; Faresin, A.; Grunwaldt, J.-D.; Huang, X.; Badocco, D.; Maggini, M.; et al. Microfluidic Crystallization of Surfactant-Free Doped Zinc Sulfide Nanoparticles for Optical Bioimaging Applications. ACS Appl. Mater. Interfaces 2020, 12, 44074–44087. [Google Scholar] [CrossRef] [PubMed]
- Canova, S.; Fiorasi, F.; Mognato, M.; Grifalconi, M.; Reddi, E.; Russo, A.; Celotti, L. “Modeled Microgravity” Affects Cell Response to Ionizing Radiation and Increases Genomic Damage. Radiat. Res. 2005, 163, 191–199. [Google Scholar] [CrossRef]
- Girardi, C.; De Pittà, C.; Casara, S.; Sales, G.; Lanfranchi, G.; Celotti, L.; Mognato, M. Analysis of miRNA and mRNA Expression Profiles Highlights Alterations in Ionizing Radiation Response of Human Lymphocytes under Modeled Microgravity. PLOS ONE 2012, 7, e31293. [Google Scholar] [CrossRef]
- Piotto, C.; Biscontin, A.; Millino, C.; Mognato, M. Functional validation of miRNAs targeting genes of DNA double-strand break repair to radiosensitize non-small lung cancer cells. Biochim. Biophys. Acta-Gene Regul. Mech. 2018, 1861, 1102–1118. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Bee, L.; Fabris, S.; Cherubini, R.; Mognato, M.; Celotti, L. The Efficiency of Homologous Recombination and Non-Homologous End Joining Systems in Repairing Double-Strand Breaks during Cell Cycle Progression. PLOS ONE 2013, 8, e69061. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zanin, A.; Meneghetti, G.; Menilli, L.; Tesoriere, A.; Argenton, F.; Mognato, M. Analysis of Radiation Toxicity in Mammalian Cells Stably Transduced with Mitochondrial Stat3. Int. J. Mol. Sci. 2023, 24, 8232. https://doi.org/10.3390/ijms24098232
Zanin A, Meneghetti G, Menilli L, Tesoriere A, Argenton F, Mognato M. Analysis of Radiation Toxicity in Mammalian Cells Stably Transduced with Mitochondrial Stat3. International Journal of Molecular Sciences. 2023; 24(9):8232. https://doi.org/10.3390/ijms24098232
Chicago/Turabian StyleZanin, Alisa, Giacomo Meneghetti, Luca Menilli, Annachiara Tesoriere, Francesco Argenton, and Maddalena Mognato. 2023. "Analysis of Radiation Toxicity in Mammalian Cells Stably Transduced with Mitochondrial Stat3" International Journal of Molecular Sciences 24, no. 9: 8232. https://doi.org/10.3390/ijms24098232
APA StyleZanin, A., Meneghetti, G., Menilli, L., Tesoriere, A., Argenton, F., & Mognato, M. (2023). Analysis of Radiation Toxicity in Mammalian Cells Stably Transduced with Mitochondrial Stat3. International Journal of Molecular Sciences, 24(9), 8232. https://doi.org/10.3390/ijms24098232





