Assembly of the Complete Mitochondrial Genome of Pereskia aculeata Revealed That Two Pairs of Repetitive Elements Mediated the Recombination of the Genome
Abstract
1. Introduction
2. Results
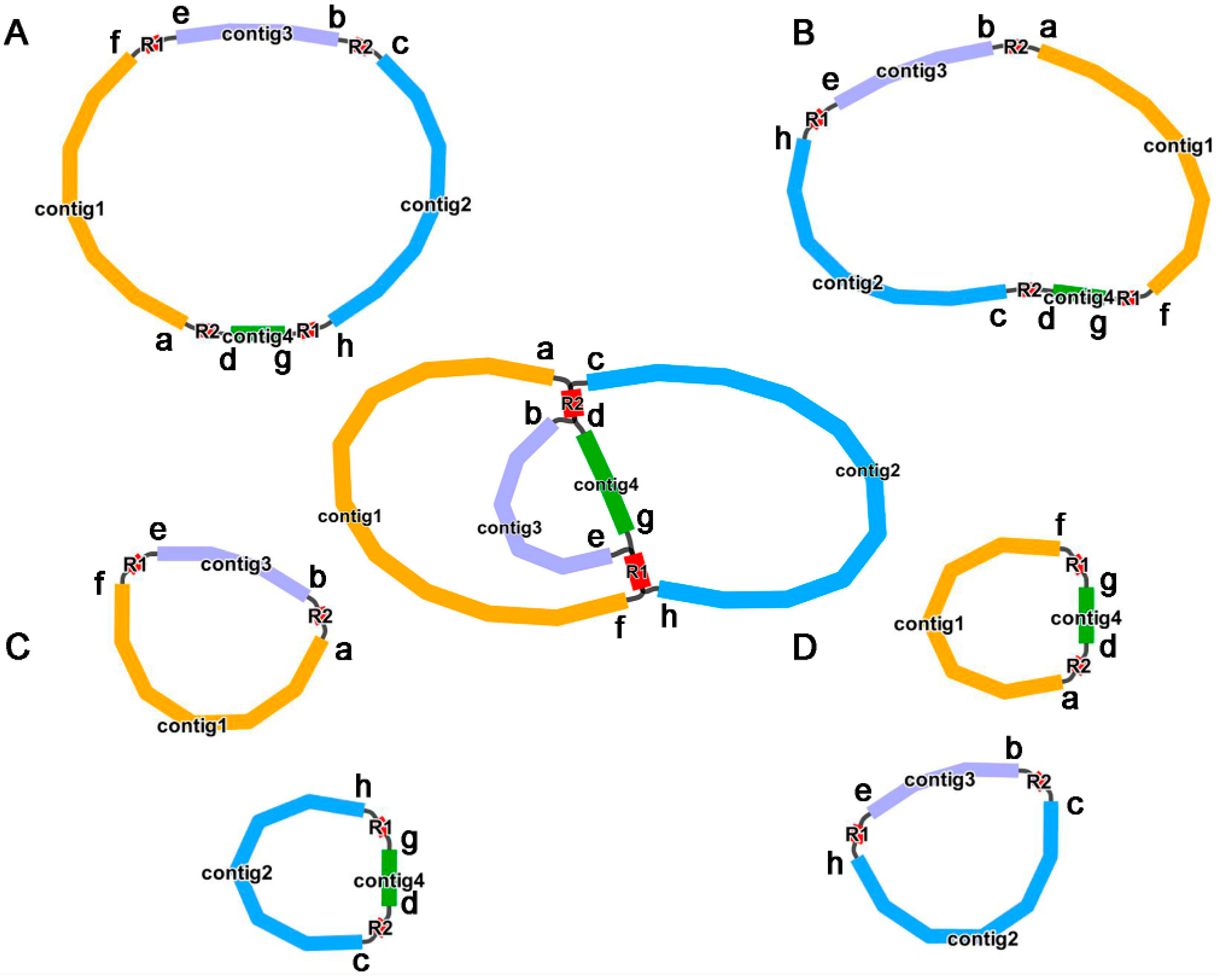
2.1. Mitogenome Structure of P. aculeate
2.2. Mitochondrial Gene Content
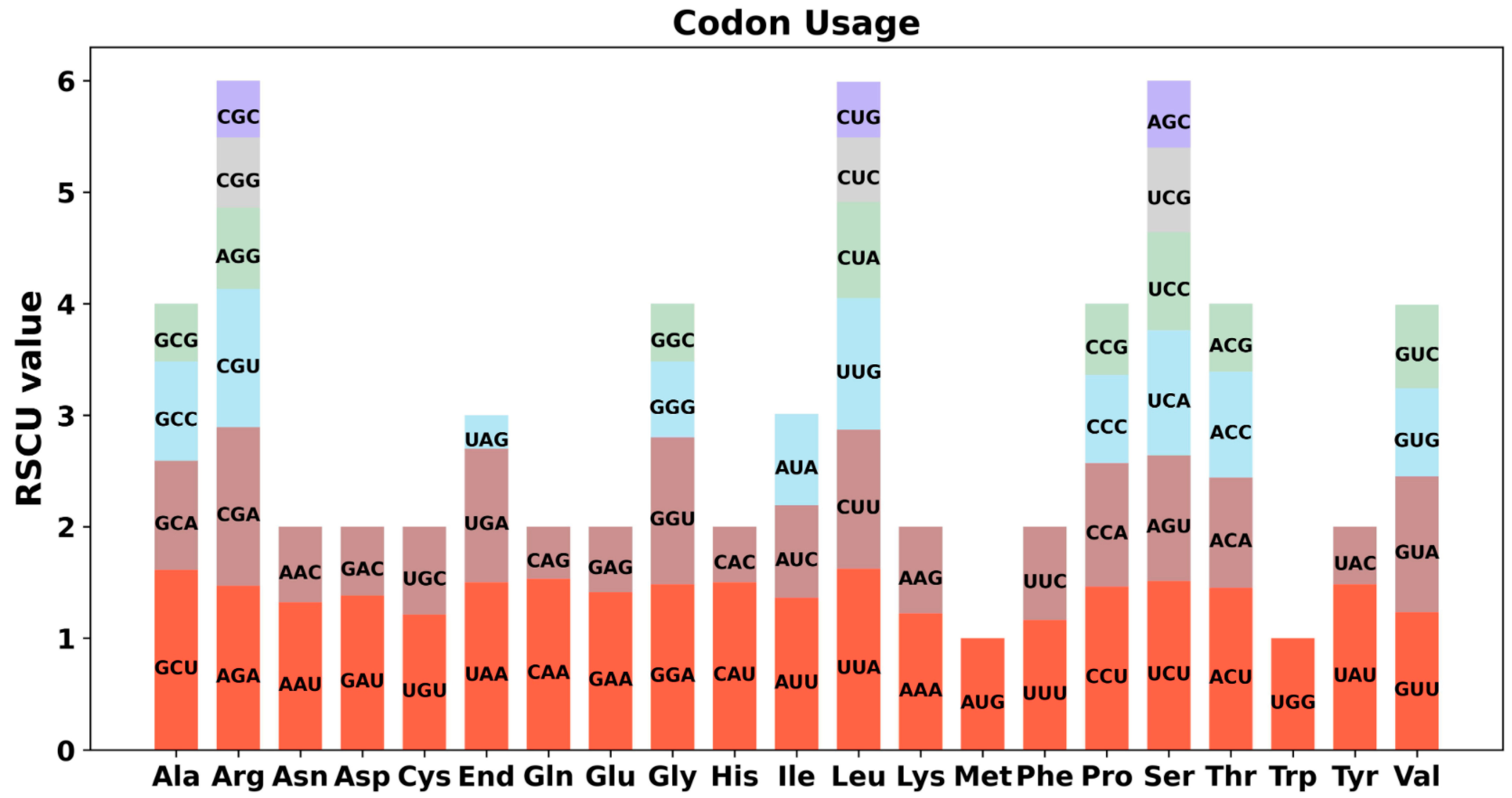
2.3. Codon Usage Analysis of PCGs
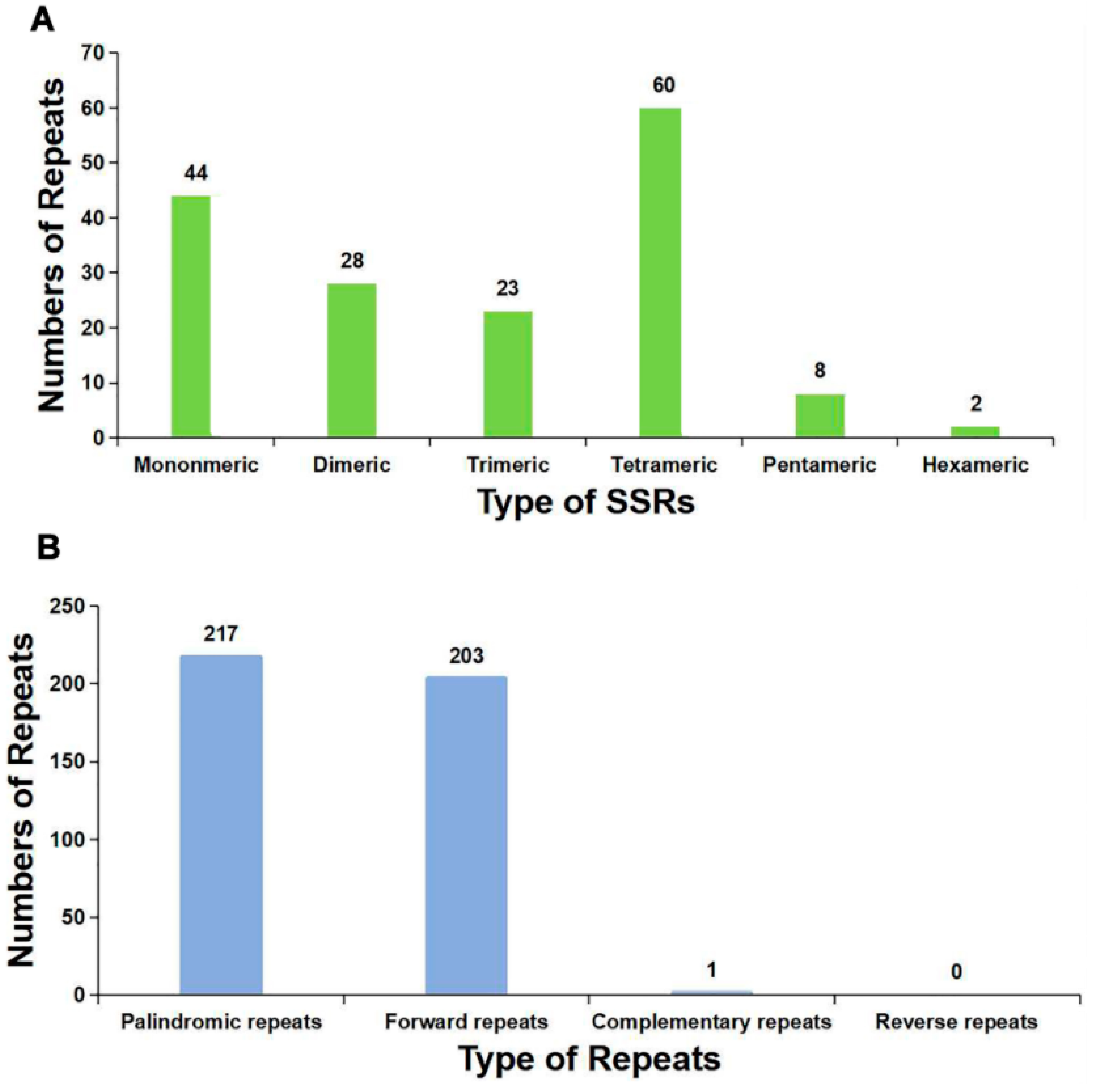
2.4. Repeat Elements
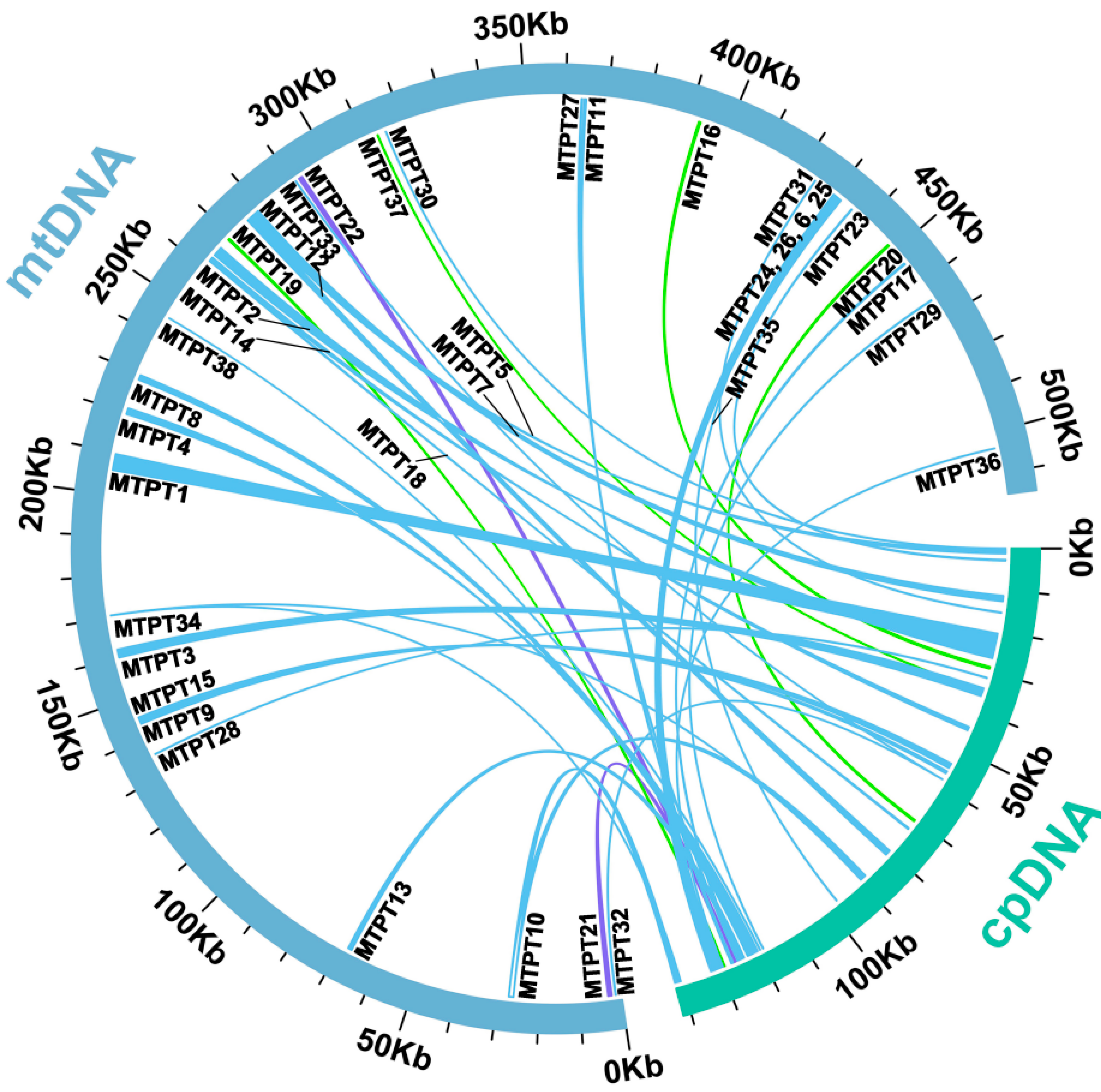
2.5. Plastid DNA Insertion in Mitogenome
2.6. RNA Editing
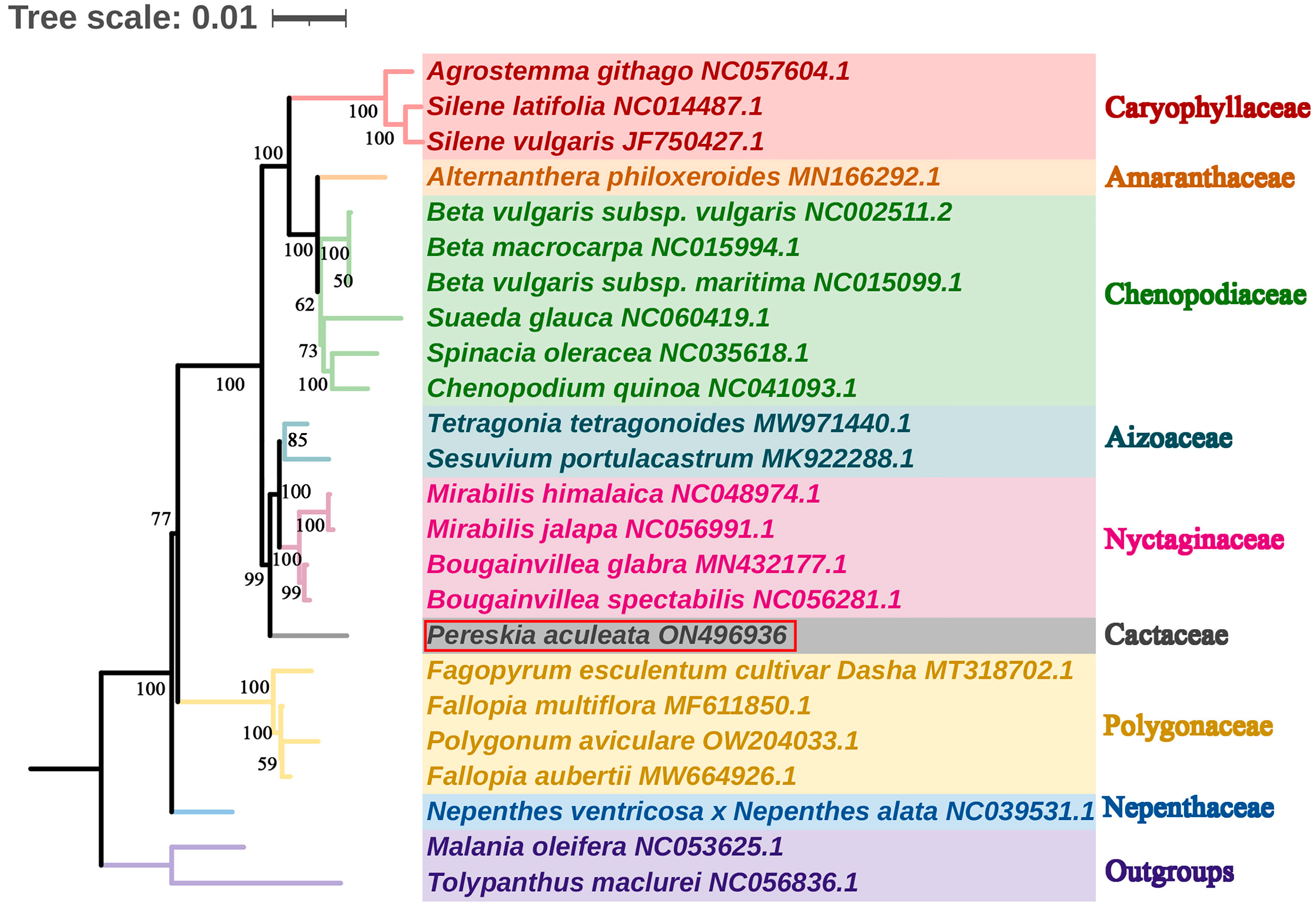
2.7. Phylogenetic Inference
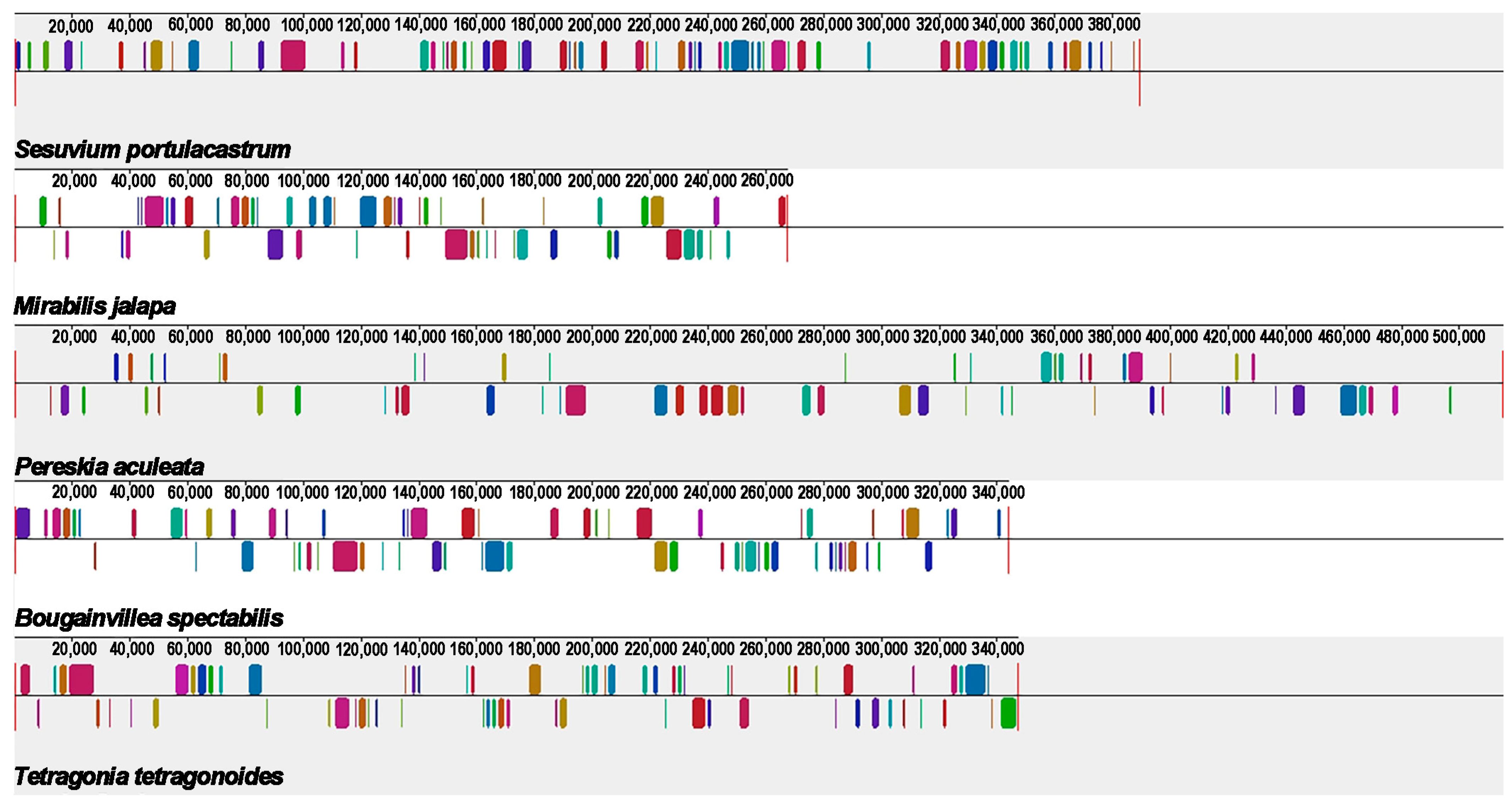
2.8. Synteny Analysis
3. Discussion
4. Materials and Methods
4.1. Plant Sampling, DNA Extraction, and Sequencing
4.2. Genome Assembly
4.3. Validating of Genome Structure
4.4. Genome Annotation
4.5. Codon Usage of Mitochondrial Genes
4.6. Analysis of Repeat Elements
4.7. Identification of Homologous Sequences among Organelle Genomes
4.8. The Prediction of RNA Editing Sites
4.9. Phylogenetic Inference
4.10. Synteny Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PCR | Polymerase chain reaction |
| DR | Direct repeat |
| SSR | Simple sequence repeat |
| ML | Maximum likelihood |
| NCBI | National Center for Biotechnology Information |
| BLAST | Basic Local Alignment Search Tool |
| PCGs | Protein-coding gene sequences |
| MTPT | Mitochondrial plastid DNA |
References
- Duarte, M.R.; Hayashi, S.S. Estudo anatômico de folha e caule de Pereskia aculeata Mill. (Cactaceae). Rev. Bras. Farmacogn. 2005, 15, 103–109. [Google Scholar] [CrossRef]
- Paterson, I.D.; Downie, D.A.; Hill, M.P. Using molecular methods to determine the origin of weed populations of Pereskia aculeata in South Africa and its relevance to biological control. Biol. Control. 2009, 48, 84–91. [Google Scholar] [CrossRef]
- Pinto, N.D.C.; Duque, A.P.D.; Pacheco, N.R.; Mendes, R.D.; Motta, E.V.D.; Bellozi, P.M.Q.; Ribeiro, A.; Salvador, M.J.; Scio, E. Pereskia aculeata: A plant food with antinociceptive activity. Pharm. Biol. 2015, 53, 1780–1785. [Google Scholar] [CrossRef]
- Merce, A.L.R.; Landaluze, J.S.; Mangrich, A.S.; Szpoganicz, B.; Sierakowski, M.R. Complexes of arabinogalactan of Pereskia aculeata and Co2+, Cu2+, Mn2+, and Ni2+. Bioresour. Technol. 2001, 76, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Takeiti, C.Y.; Antonio, G.C.; Motta, E.M.P.; Collares-Queiroz, F.P.; Park, K.J. Nutritive evaluation of a non-conventional leafy vegetable (Pereskia aculeata Miller). Int. J. Food Sci. Nutr. 2009, 60, 148–160. [Google Scholar] [CrossRef] [PubMed]
- Pinto, N.C.C.S.R.; Machado, D.C. Cytotoxic and antioxidant activity of Pereskia aculeata Miller. Pharmacologyonline 2012, 3, 63–69. [Google Scholar]
- Cruz-Rubio, J.M.; Mueller, M.; Viernstein, H.; Loeppert, R.; Praznik, W. Prebiotic potential and chemical characterization of the poly and oligosaccharides present in the mucilage of Opuntia ficus-indica and Opuntia joconostle. Food Chem. 2021, 362, 130167. [Google Scholar] [CrossRef]
- de Andrade Vieira, E.; Alves Alcantara, M.; Albuquerque Dos Santos, N.; Duarte Gondim, A.; Iacomini, M.; Mellinger, C.; Tribuzy de Magalhaes Cordeiro, A.M. Mucilages of cacti from Brazilian biodiversity: Extraction, physicochemical and technological properties. Food Chem. 2021, 346, 128892. [Google Scholar] [CrossRef]
- Silva, S.H.; Neves, I.C.O.; Oliveira, N.L.; de Oliveira, A.C.F.; Lago, A.M.T.; Giarola, T.M.D.; de Resende, J.V. Extraction processes and characterization of the mucilage obtained from green fruits of Pereskia aculeata Miller. Ind. Crops Prod. 2019, 140, 111716. [Google Scholar] [CrossRef]
- Torres-Acosta, A.A.; González-Calderón, P.Y. Opuntia ficus-Indica (OFI) Mucilage as Corrosion Inhibitor of Steel in CO2-Contaminated Mortar. Materials 2021, 14, 1316. [Google Scholar] [CrossRef]
- Bonner, J.; Millerd, A. Oxidative phosphorylation by plant mitochondria. Arch. Biochem. Biophys. 1953, 42, 135–148. [Google Scholar] [CrossRef] [PubMed]
- Daloso, D.M.; Müller, K.; Obata, T.; Florian, A.; Tohge, T.; Bottcher, A.; Riondet, C.; Bariat, L.; Carrari, F.; Nunes-Nesi, A.; et al. Thioredoxin, a master regulator of the tricarboxylic acid cycle in plant mitochondria. Proc. Natl. Acad. Sci. USA 2015, 112, E1392–E1400. [Google Scholar] [CrossRef] [PubMed]
- Valero, T. Mitochondrial Biogenesis: Pharmacological Approaches. Curr. Pharm. Des. 2014, 20, 5507–5509. [Google Scholar] [CrossRef] [PubMed]
- Levings, C.S.; Pring, D.R. Restriction Endonuclease Analysis of Mitochondrial-DNA from Normal and Texas Cytoplasmic Male-Sterile Maize. Science 1976, 193, 158–160. [Google Scholar] [CrossRef]
- Guo, J.; Wang, P.; Cheng, Q.; Sun, L.; Wang, H.; Wang, Y.; Kao, L.; Li, Y.; Qiu, T.; Yang, W.; et al. Proteomic analysis reveals strong mitochondrial involvement in cytoplasmic male sterility of pepper (Capsicum annuum L.). J. Proteom. 2017, 168, 15–27. [Google Scholar] [CrossRef]
- Bi, C.W.; Lu, N.; Xu, Y.Q.; He, C.P.; Lu, Z.H. Characterization and Analysis of the Mitochondrial Genome of Common Bean (Phaseolus vulgaris) by Comparative Genomic Approaches. Int. J. Mol. Sci. 2020, 21, 3778. [Google Scholar] [CrossRef]
- Cheng, Y.; He, X.; Priyadarshani, S.V.G.N.; Wang, Y.; Ye, L.; Shi, C.; Ye, K.; Zhou, Q.; Luo, Z.; Deng, F.; et al. Assembly and comparative analysis of the complete mitochondrial genome of Suaeda glauca. BMC Genom. 2021, 22, 167. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Guo, H.; Zhu, J.; Qu, K.; Chen, Y.; Guo, Y.; Ding, P.; Yang, H.; Xu, T.; Jing, Q.; et al. Complex Physical Structure of Complete Mitochondrial Genome of Quercus acutissima (Fagaceae): A Significant Energy Plant. Genes 2022, 13, 1321. [Google Scholar] [CrossRef]
- Gualberto, J.M.; Mileshina, D.; Wallet, C.; Niazi, A.K.; Weber-Lotfi, F.; Dietrich, A. The plant mitochondrial genome: Dynamics and maintenance. Biochimie 2014, 100, 107–120. [Google Scholar] [CrossRef]
- Heyer, W.D.; Ehmsen, K.T.; Liu, J. Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 2010, 44, 113–139. [Google Scholar] [CrossRef]
- Rocha, E.P.; Cornet, E.; Michel, B. Comparative and evolutionary analysis of the bacterial homologous recombination systems. PLoS Genet. 2005, 1, e15. [Google Scholar] [CrossRef] [PubMed]
- Kreuzer, K.N. Recombination-dependent DNA replication in phage T4. Trends Biochem. Sci. 2000, 25, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Yeeles, J.T.; Poli, J.; Marians, K.J.; Pasero, P. Rescuing stalled or damaged replication forks. Cold Spring Harb. Perspect. Biol. 2013, 5, a012815. [Google Scholar] [CrossRef] [PubMed]
- Cappadocia, L.; Marechal, A.; Parent, J.S.; Lepage, E.; Sygusch, J.; Brisson, N. Crystal structures of DNA-Whirly complexes and their role in Arabidopsis organelle genome repair. Plant Cell 2010, 22, 1849–1867. [Google Scholar] [CrossRef]
- Puchta, H. Double-strand break-induced recombination between ectopic homologous sequences in somatic plant cells. Genetics 1999, 152, 1173–1181. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.H.Y.; Pannunzio, N.R.; Adachi, N.; Lieber, M.R. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell. Biol. 2017, 18, 495–506. [Google Scholar] [CrossRef] [PubMed]
- Roger, A.J.; Munoz-Gomez, S.A.; Kamikawa, R. The Origin and Diversification of Mitochondria. Curr. Biol. 2017, 27, R1177–R1192. [Google Scholar] [CrossRef]
- Morimatsu, K.; Kowalczykowski, S.C. RecQ helicase and RecJ nuclease provide complementary functions to resect DNA for homologous recombination. Proc. Natl. Acad. Sci. USA 2014, 111, E5133–E5142. [Google Scholar] [CrossRef]
- Edmondson, A.C.; Song, D.; Alvarez, L.A.; Wall, M.K.; Almond, D.; McClellan, D.A.; Maxwell, A.; Nielsen, B.L. Characterization of a mitochondrially targeted single-stranded DNA-binding protein in Arabidopsis thaliana. Mol. Genet. Genom. 2005, 273, 115–122. [Google Scholar] [CrossRef]
- Garcia-Medel, P.L.; Baruch-Torres, N.; Peralta-Castro, A.; Trasvina-Arenas, C.H.; Torres-Larios, A.; Brieba, L.G. Plant organellar DNA polymerases repair double-stranded breaks by microhomology-mediated end-joining. Nucleic Acids Res. 2019, 47, 3028–3044. [Google Scholar] [CrossRef]
- Christensen, A.C. Plant Mitochondrial Genome Evolution Can Be Explained by DNA Repair Mechanisms. Genome Biol. Evol. 2013, 5, 1079–1086. [Google Scholar] [CrossRef] [PubMed]
- Richardson, A.O.; Rice, D.W.; Young, G.J.; Alverson, A.J.; Palmer, J.D. The “fossilized” mitochondrial genome of Liriodendron tulipifera: Ancestral gene content and order, ancestral editing sites, and extraordinarily low mutation rate. BMC Biol. 2013, 11, 29. [Google Scholar] [CrossRef] [PubMed]
- Kitazaki, K.; Kubo, T.; Kagami, H.; Matsumoto, T.; Fujita, A.; Matsuhira, H.; Matsunaga, M.; Mikami, T. A horizontally transferred tRNA(Cys) gene in the sugar beet mitochondrial genome: Evidence that the gene is present in diverse angiosperms and its transcript is aminoacylated. Plant J. 2011, 68, 262–272. [Google Scholar] [CrossRef] [PubMed]
- Rice, D.W.; Alverson, A.J.; Richardson, A.O.; Young, G.J.; Sanchez-Puerta, M.V.; Munzinger, J.; Barry, K.; Boore, J.L.; Zhang, Y.; DePamphilis, C.W.; et al. Horizontal Transfer of Entire Genomes via Mitochondrial Fusion in the Angiosperm amborella. Science 2013, 342, 1468–1473. [Google Scholar] [CrossRef]
- Mower, J.P. Variation in protein gene and intron content among land plant mitogenomes. Mitochondrion 2020, 53, 203–213. [Google Scholar] [CrossRef]
- Yang, H.X.; Li, W.H.; Yu, X.L.; Zhang, X.Y.; Zhang, Z.Y.; Liu, Y.X.; Wang, W.X.; Tian, X.X. Insights into molecular structure, genome evolution and phylogenetic implication through mitochondrial genome sequence of Gleditsia sinensis. Sci. Rep. 2021, 11, 14850. [Google Scholar] [CrossRef]
- Sau, K.; Gupta, S.K.; Sau, S.; Mandal, S.C.; Ghosh, T.C. Factors influencing synonymous codon and amino acid usage biases in Mimivirus. Biosystems 2006, 85, 107–113. [Google Scholar] [CrossRef]
- Lovin, D.D.; Washington, K.O.; deBruyn, B.; Hemme, R.R.; Mori, A.; Epstein, S.R.; Harker, B.W.; Streit, T.G.; Severson, D.W. Genome-based polymorphic microsatellite development and validation in the mosquito Aedes aegypti and application to population genetics in Haiti. BMC Genom. 2009, 10, 590. [Google Scholar] [CrossRef]
- Morgante, M.; Hanafey, M.; Powell, W. Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nat. Genet. 2002, 30, 194–200. [Google Scholar] [CrossRef]
- Naranpanawa, D.N.U.; Chandrasekara, C.H.W.M.R.B.; Bandaranayake, P.C.G.; Bandaranayake, A.U. Raw transcriptomics data to gene specific SSRs: A validated free bioinformatics workflow for biologists. Sci. Rep. 2020, 10, 18236. [Google Scholar] [CrossRef]
- Kawata, M.; Harada, T.; Shimamoto, Y.; Oono, K.; Takaiwa, F. Short inverted repeats function as hotspots of intermolecular recombination giving rise to oligomers of deleted plastid DNAs (ptDNAs). Curr. Genet. 1997, 31, 179–184. [Google Scholar] [CrossRef] [PubMed]
- Lopez, L.; Barreiro, R.; Fischer, M.; Koch, M.A. Mining microsatellite markers from public expressed sequence tags databases for the study of threatened plants. BMC Genom. 2015, 16, 781. [Google Scholar] [CrossRef] [PubMed]
- Simpson, L.; Emerson, R.B. RNA editing. Annu. Rev. Neurosci. 1996, 19, 27–52. [Google Scholar] [CrossRef] [PubMed]
- Edera, A.A.; Gandini, C.L.; Sanchez-Puerta, M.V. Towards a comprehensive picture of C-to-U RNA editing sites in angiosperm mitochondria. Plant Mol. Biol. 2018, 97, 215–231. [Google Scholar] [CrossRef]
- Chateigner-Boutin, A.L.; Small, I. Plant RNA editing. Rna Biol. 2010, 7, 213–219. [Google Scholar] [CrossRef]
- Darling, A.C.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef]
- Carson, A.R.; Scherer, S.W. Identifying concerted evolution and gene conversion in mammalian gene pairs lasting over 100 million years. BMC Evol. Biol. 2009, 9, 156. [Google Scholar] [CrossRef]
- Xia, C.; Li, J.; Zuo, Y.; He, P.; Zhang, H.; Zhang, X.; Wang, B.; Zhang, J.; Yu, J.; Deng, H. Complete mitochondrial genome of Thuja sutchuenensis and its implications on evolutionary analysis of complex mitogenome architecture in Cupressaceae. BMC Plant. Biol. 2023, 23, 84. [Google Scholar] [CrossRef]
- Jiang, M.; Ni, Y.; Li, J.; Liu, C. Characterisation of the complete mitochondrial genome of Taraxacum mongolicum revealed five repeat-mediated recombinations. Plant Cell Rep. 2023, 42, 775–789. [Google Scholar] [CrossRef]
- Yang, Z.; Ni, Y.; Lin, Z.; Yang, L.; Chen, G.; Nijiati, N.; Hu, Y.; Chen, X. De novo assembly of the complete mitochondrial genome of sweet potato (Ipomoea batatas [L.] Lam) revealed the existence of homologous conformations generated by the repeat-mediated recombination. BMC Plant Biol. 2022, 22, 285. [Google Scholar] [CrossRef]
- Dong, S.; Zhao, C.; Chen, F.; Liu, Y.; Zhang, S.; Wu, H.; Zhang, L.; Liu, Y. The complete mitochondrial genome of the early flowering plant Nymphaea colorata is highly repetitive with low recombination. BMC Genom. 2018, 19, 614. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Chen, H.; Ni, Y.; Li, J.; Cai, Y.; Ma, B.; Yu, J.; Wang, J.; Liu, C. De Novo Hybrid Assembly of the Salvia miltiorrhiza Mitochondrial Genome Provides the First Evidence of the Multi-Chromosomal Mitochondrial DNA Structure of Salvia Species. Int. J. Mol. Sci. 2022, 23, 14267. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Grewe, F.; Fan, W.; Young, G.J.; Knoop, V.; Palmer, J.D.; Mower, J.P. Ginkgo and Welwitschia Mitogenomes Reveal Extreme Contrasts in Gymnosperm Mitochondrial Evolution. Mol. Biol. Evol. 2016, 33, 1448–1460. [Google Scholar] [CrossRef] [PubMed]
- Sloan, D.B.; Alverson, A.J.; Chuckalovcak, J.P.; Wu, M.; McCauley, D.E.; Palmer, J.D.; Taylor, D.R. Rapid evolution of enormous, multichromosomal genomes in flowering plant mitochondria with exceptionally high mutation rates. PLoS Biol. 2012, 10, e1001241. [Google Scholar] [CrossRef]
- Mower, J.P.; Case, A.L.; Floro, E.R.; Willis, J.H. Evidence against equimolarity of large repeat arrangements and a predominant master circle structure of the mitochondrial genome from a monkeyflower (Mimulus guttatus) lineage with cryptic CMS. Genome Biol. Evol. 2012, 4, 670–686. [Google Scholar] [CrossRef]
- Alverson, A.J.; Wei, X.X.; Rice, D.W.; Stern, D.B.; Barry, K.; Palmer, J.D. Insights into the Evolution of Mitochondrial Genome Size from Complete Sequences of Citrullus lanatus and Cucurbita pepo (Cucurbitaceae). Mol. Biol. Evol. 2010, 27, 1436–1448. [Google Scholar] [CrossRef] [PubMed]
- Adams, K.L.; Ong, H.C.; Palmer, J.D. Mitochondrial gene transfer in pieces: Fission of the ribosomal protein gene rpl2 and partial or complete gene transfer to the nucleus. Mol. Biol. Evol. 2001, 18, 2289–2297. [Google Scholar] [CrossRef] [PubMed]
- Rabah, S.O.; Lee, C.; Hajrah, N.H.; Makki, R.M.; Alharby, H.F.; Alhebshi, A.M.; Sabir, J.S.M.; Jansen, R.K.; Ruhlman, T.A. Plastome Sequencing of Ten Nonmodel Crop Species Uncovers a Large Insertion of Mitochondrial DNA in Cashew. Plant Genome 2017, 10, 1–14. [Google Scholar] [CrossRef]
- Sprinzl, M.; Vassilenko, K.S. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 2005, 33, D139–D140. [Google Scholar] [CrossRef]
- Lukes, J.; Kaur, B.; Speijer, D. RNA Editing in Mitochondria and Plastids: Weird and Widespread. Trends Genet. 2021, 37, 99–102. [Google Scholar] [CrossRef]
- Brennicke, A.; Marchfelder, A.; Binder, S. RNA editing. FEMS Microbiol. Rev. 1999, 23, 297–316. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Wang, Y.; Li, S.; Wen, J.; Zhu, L.; Yan, K.; Du, Y.; Ren, J.; Li, S.; Chen, Z.; et al. Assembly and comparative analysis of the first complete mitochondrial genome of Acer truncatum Bunge: A woody oil-tree species producing nervonic acid. BMC Plant Biol. 2022, 22, 29. [Google Scholar] [CrossRef] [PubMed]
- Gerke, P.; Szovenyi, P.; Neubauer, A.; Lenz, H.; Gutmann, B.; McDowell, R.; Small, I.; Schallenberg-Rudinger, M.; Knoop, V. Towards a plant model for enigmatic U-to-C RNA editing: The organelle genomes, transcriptomes, editomes and candidate RNA editing factors in the hornwort Anthoceros agrestis. New Phytol. 2020, 225, 1974–1992. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.J.; Xiu, Z.H.; Meeley, R.; Tan, B.C. Empty Pericarp5 Encodes a Pentatricopeptide Repeat Protein That Is Required for Mitochondrial RNA Editing and Seed Development in Maize. Plant Cell 2013, 25, 868–883. [Google Scholar] [CrossRef]
- Grewe, F.; Edger, P.P.; Keren, I.; Sultan, L.; Pires, J.C.; Ostersetzer-Biran, O.; Mower, J.P. Comparative analysis of 11 Brassicales mitochondrial genomes and the mitochondrial transcriptome of Brassica oleracea. Mitochondrion 2014, 19, 135–143. [Google Scholar] [CrossRef]
- Kovar, L.; Nageswara-Rao, M.; Ortega-Rodriguez, S.; Dugas, D.V.; Straub, S.; Cronn, R.; Strickler, S.R.; Hughes, C.E.; Hanley, K.A.; Rodriguez, D.N.; et al. PacBio-Based Mitochondrial Genome Assembly of Leucaena trichandra (Leguminosae) and an Intrageneric Assessment of Mitochondrial RNA Editing. Genome Biol. Evol. 2018, 10, 2501–2517. [Google Scholar] [CrossRef]
- Arseneau, J.R.; Steeves, R.; Laflamme, M. Modified low-salt CTAB extraction of high-quality DNA from contaminant-rich tissues. Mol. Ecol. Resour. 2017, 17, 686–693. [Google Scholar] [CrossRef]
- Emerman, A.B.; Bowman, S.K.; Barry, A.; Henig, N.; Patel, K.M.; Gardner, A.F.; Hendrickson, C.L. NEBNext Direct: A Novel, Rapid, Hybridization-Based Approach for the Capture and Library Conversion of Genomic Regions of Interest. Curr. Protoc. Mol. Biol. 2017, 119, 7–30. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef]
- Chen, Y.; Ye, W.C.; Zhang, Y.D.; Xu, Y.S. High speed BLASTN: An accelerated MegaBLAST search tool. Nucleic Acids Res. 2015, 43, 7762–7768. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Schultz, M.B.; Zobel, J.; Holt, K.E. Bandage: Interactive visualization of de novo genome assemblies. Bioinformatics 2015, 31, 3350–3352. [Google Scholar] [CrossRef]
- Yu, J.; Li, J.; Zuo, Y.; Qin, Q.; Zeng, S.; Rennenberg, H.; Deng, H. Plastome variations reveal the distinct evolutionary scenarios of plastomes in the subfamily Cereoideae (Cactaceae). BMC Plant Biol. 2023, 23, 132. [Google Scholar] [CrossRef]
- Shi, L.C.; Chen, H.M.; Jiang, M.; Wang, L.Q.; Wu, X.; Huang, L.F.; Liu, C. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 2019, 47, W65–W73. [Google Scholar] [CrossRef]
- Tillich, M.; Lehwark, P.; Pellizzer, T.; Ulbricht-Jones, E.S.; Fischer, A.; Bock, R.; Greiner, S. GeSeq—Versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017, 45, W6–W11. [Google Scholar] [CrossRef]
- Lewis, S.E.; Searle, S.M.J.; Harris, N.; Gibson, M.; Iyer, V.; Richter, J.; Wiel, C.; Bayraktaroglu, L.; Birney, E.; Crosby, M.A.; et al. Apollo: A sequence annotation editor. Genome Biol. 2002, 3, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Chan, P.P.; Lowe, T.M. tRNAscan-SE: Searching for tRNA Genes in Genomic Sequences. Methods Mol. Biol. 2019, 1962, 1–14. [Google Scholar] [PubMed]
- Zhang, D.; Gao, F.L.; Jakovlic, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Beier, S.; Thiel, T.; Münch, T.; Scholz, U.; Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef]
- Kurtz, S.; Choudhuri, J.V.; Ohlebusch, E.; Schleiermacher, C.; Stoye, J.; Giegerich, R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001, 29, 4633–4642. [Google Scholar] [CrossRef]
- Zhang, H.E.; Meltzer, P.; Davis, S. RCircos: An R package for Circos 2D track plots. BMC Bioinform. 2013, 14, 244. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Edera, A.A.; Small, I.; Milone, D.H.; Sanchez-Puerta, M.V. Deepred-Mt: Deep representation learning for predicting C-to-U RNA editing in plant mitochondria. Comput. Biol. Med. 2021, 136, 104682. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [PubMed]



| Group of Genes | Name of Genes |
|---|---|
| ATP synthase | atp1, atp4, atp6, atp8, atp9 (2) |
| NADH dehydrogenase | nad1, nad2, nad3, nad4 (2), nad4L, nad5, nad6, nad7, nad9 |
| Ubichinol cytochrome c reductase | cob |
| Cytochrome c biogenesis | ccmB, ccmC, ccmFC, ccmFN |
| Cytochrome c oxidase | cox1, cox2, cox3 |
| Maturases | matR |
| Transport membrane protein | mttB |
| Large subunit of ribosome | rpl5, rpl16 |
| Small subunit of ribosome | rps1, rps3, rps4, rps12, rps13, rps7 |
| Succinate dehydrogenase | sdh4 |
| Ribosome RNAs | rrn18 (2), rrn26 (2), rrn5 (2) |
| Transfer RNAs | trnC-GCA (2), trnD-GUC, trnE-UUC, trnF-GAA, trnM-CAU (7), trnH-GUG, trnI-CAU(2), trnK-UUU, trnN-GUU, trnP-UGG (2), trnQ-UUG, trnS-GCU, trnS-UGA, trnV-GAC, trnW-CCA, trnY-GUA, trnG-GCC (2), trnT-UGU, trnfM-CAU |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Shan, Y.; Li, J.; Qin, Q.; Yu, J.; Deng, H. Assembly of the Complete Mitochondrial Genome of Pereskia aculeata Revealed That Two Pairs of Repetitive Elements Mediated the Recombination of the Genome. Int. J. Mol. Sci. 2023, 24, 8366. https://doi.org/10.3390/ijms24098366
Zhang X, Shan Y, Li J, Qin Q, Yu J, Deng H. Assembly of the Complete Mitochondrial Genome of Pereskia aculeata Revealed That Two Pairs of Repetitive Elements Mediated the Recombination of the Genome. International Journal of Molecular Sciences. 2023; 24(9):8366. https://doi.org/10.3390/ijms24098366
Chicago/Turabian StyleZhang, Xue, Yuanyu Shan, Jingling Li, Qiulin Qin, Jie Yu, and Hongping Deng. 2023. "Assembly of the Complete Mitochondrial Genome of Pereskia aculeata Revealed That Two Pairs of Repetitive Elements Mediated the Recombination of the Genome" International Journal of Molecular Sciences 24, no. 9: 8366. https://doi.org/10.3390/ijms24098366
APA StyleZhang, X., Shan, Y., Li, J., Qin, Q., Yu, J., & Deng, H. (2023). Assembly of the Complete Mitochondrial Genome of Pereskia aculeata Revealed That Two Pairs of Repetitive Elements Mediated the Recombination of the Genome. International Journal of Molecular Sciences, 24(9), 8366. https://doi.org/10.3390/ijms24098366






