NSD Overexpression in the Fat Body Increases Antimicrobial Peptide Production by the Immune Deficiency Pathway in Drosophila
Abstract
:1. Introduction
2. Results
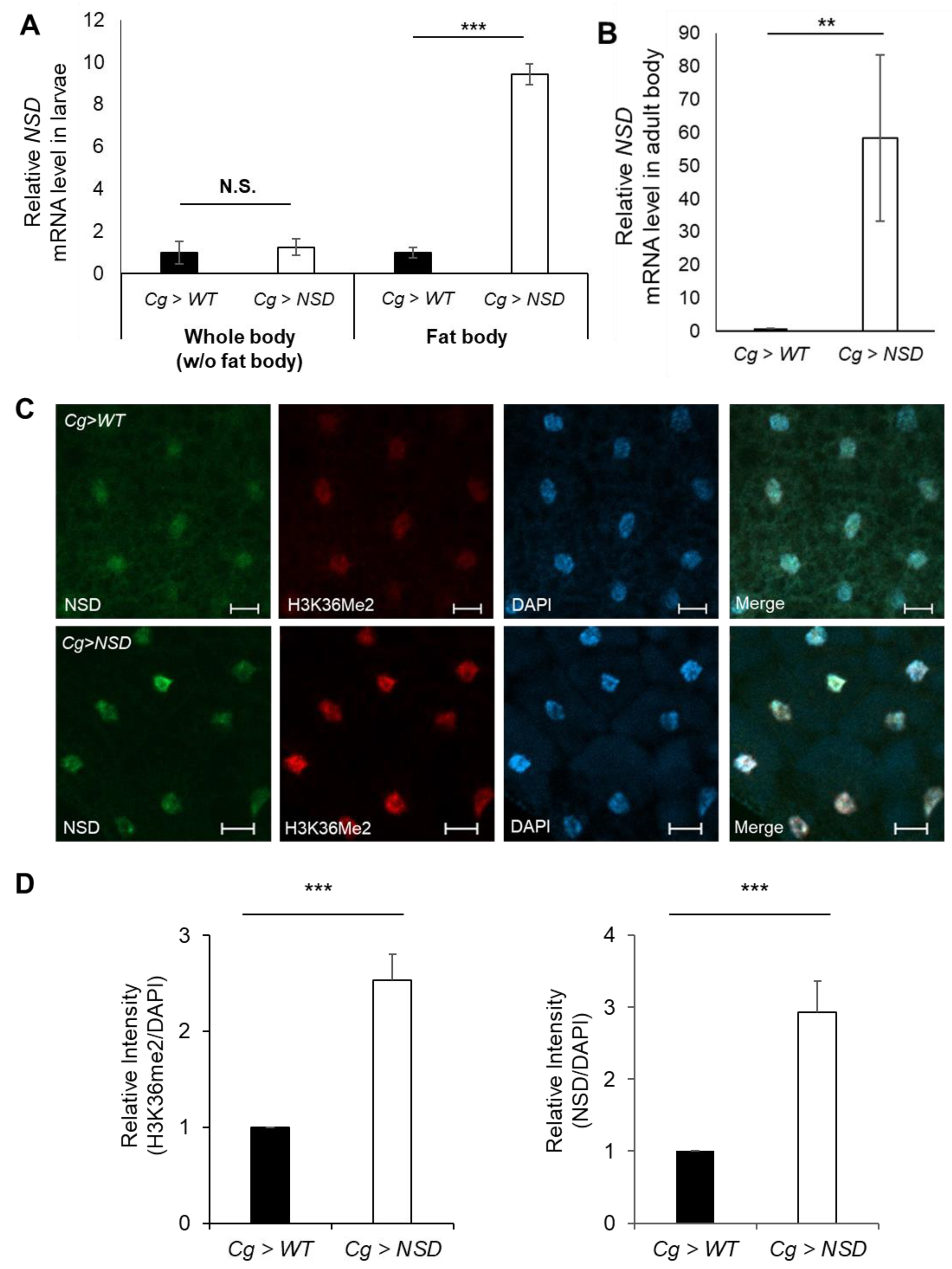
2.1. Generation of a Fat Body-Specific NSD-Overexpressing Fly Using the Gal4/UAS System
2.2. NSD Overexpression in the Drosophila Fat Body Affects the Innate Immune System via Relish
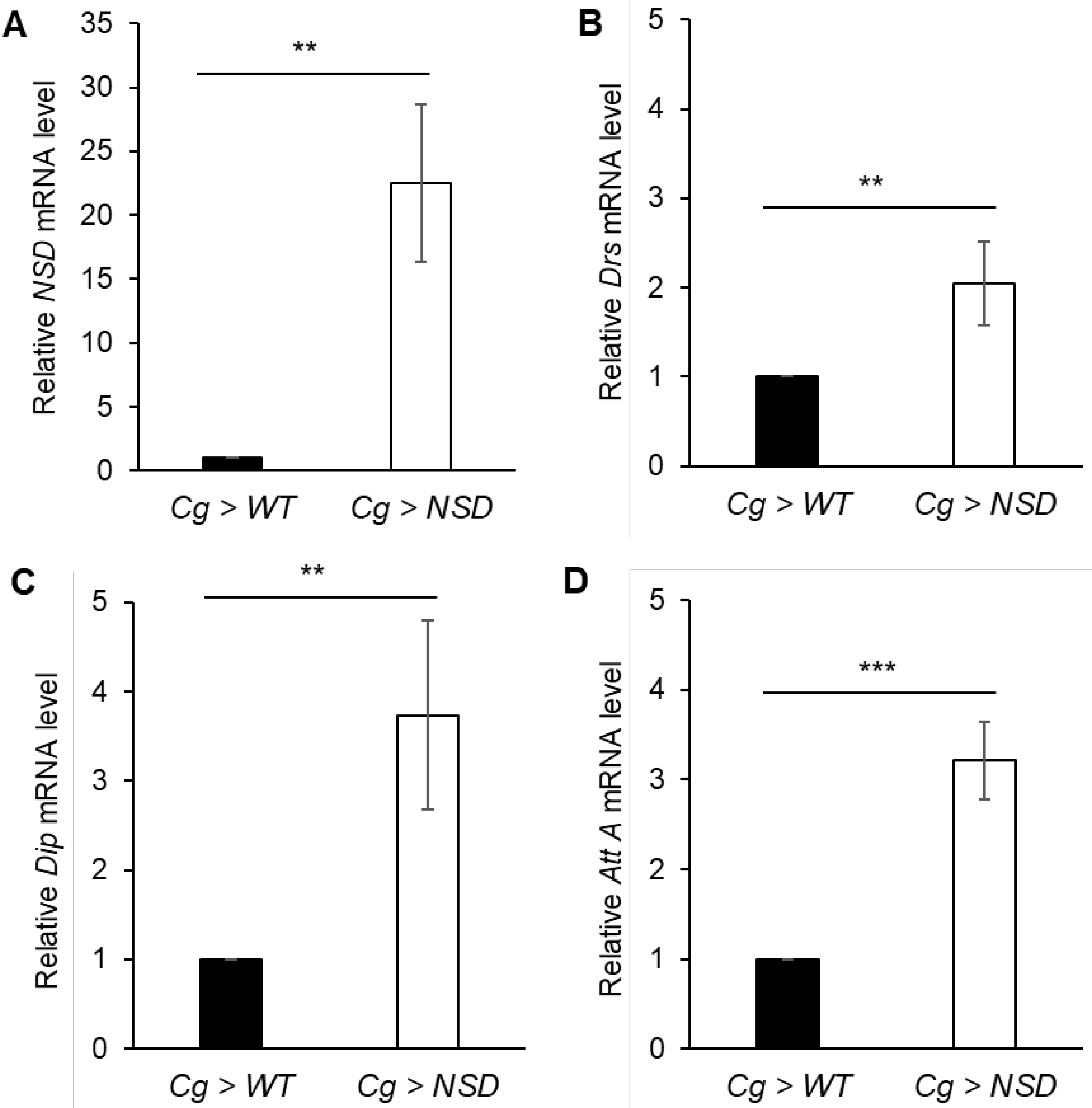
2.2.1. Effects of the Fat Body-Targeted NSD Overexpression on the Transcription of Various AMPs
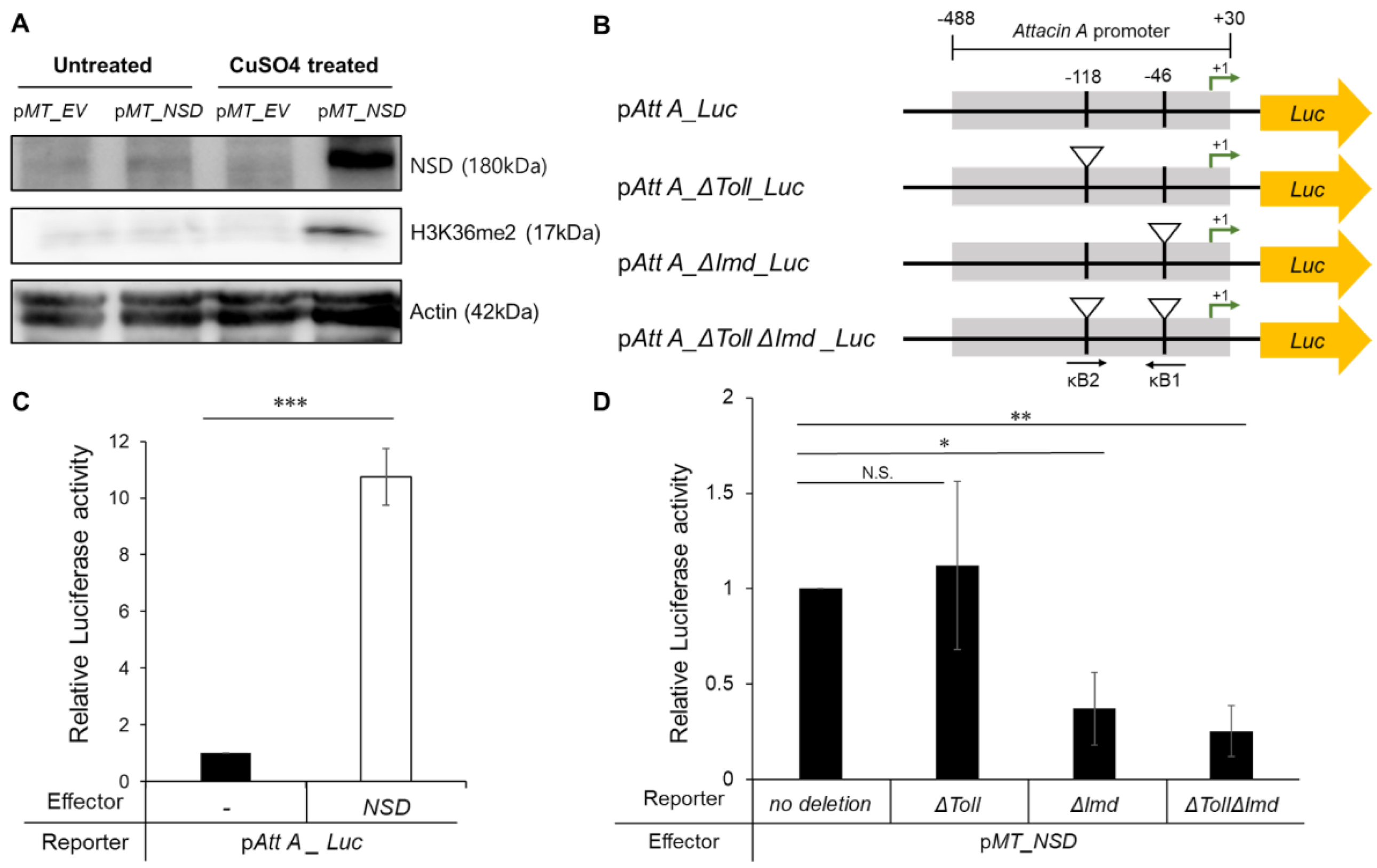
2.2.2. Activation of the NF-κB Activity by NSD Overexpression
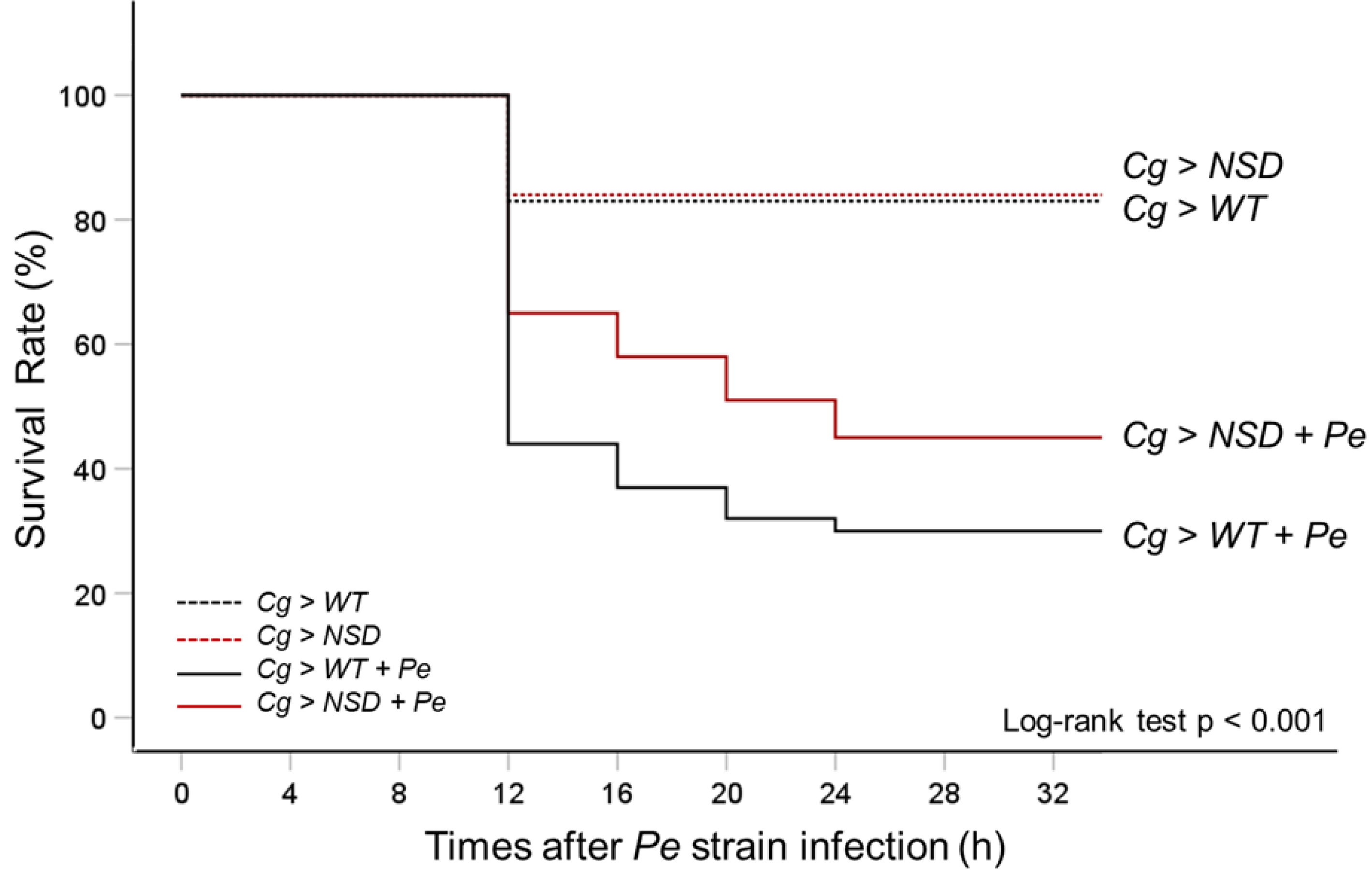
2.3. Enhanced Survival of NSD-Overexpressing Larvae upon Ingestion of Pathogenic Gram-Negative Pseudomonas Entomophila
3. Discussion
4. Materials and Methods
4.1. Fly Species and Bacterial Strains
4.2. RNA Preparation and qPCR
4.3. Immunohistochemistry
4.4. Plasmid Vector Cloning and Transient Transfection of S2 Cells
4.5. Luciferase Assay
4.6. Western Blot
4.7. Natural Pe Infection of Drosophila Larvae and its Effect on Larval Survival Rates
4.8. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wagner, E.J.; Carpenter, P.B. Understanding the Language of Lys36 Methylation at Histone H3. Nat. Rev. Mol. Cell Biol. 2012, 13, 115–126. [Google Scholar] [CrossRef] [PubMed]
- Tatton-Brown, K.; Douglas, J.; Coleman, K.; Ve Baujat, G.; Cole, T.R.P.; Das, S.; Horn, D.; Hughes, H.E.; Temple, I.K.; Faravelli, F.; et al. Genotype-Phenotype Associations in Sotos Syndrome: An Analysis of 266 Individuals with NSD1 Aberrations. Am. J. Hum. Genet. 2005, 77, 193–204. [Google Scholar] [CrossRef] [PubMed]
- Douglas, J.; Hanks, S.; Karen Temple, I.; Davies, S.; Murray, A.; Upadhyaya, M.; Tomkins, S.; Hughes, H.E.; Cole, T.R.P.; Rahman, N. NSD1 Mutations Are the Major Cause of Sotos Syndrome and Occur in Some Cases of Weaver Syndrome but Are Rare in Other Overgrowth Phenotypes. Am. J. Hum. Genet. 2003, 72, 132–143. [Google Scholar] [CrossRef] [PubMed]
- Nagai, T.; Matsumoto, N.; Kurotaki, N.; Harada, N.; Niikawa, N.; Ogata, T.; Imaizumi, K.; Kurosawa, K.; Kondoh, T.; Ohashi, H.; et al. Sotos Syndrome and Haploinsufficiency of NSD1: Clinical Features of Intragenic Mutations and Submicroscopic Deletions. J. Med. Genet. 2003, 40, 285–289. [Google Scholar] [CrossRef]
- Dikow, N.; Maas, B.; Gaspar, H.; Kreiss-Nachtsheim, M.; Engels, H.; Kuechler, A.; Garbes, L.; Netzer, C.; Neuhann, T.M.; Koehler, U.; et al. The Phenotypic Spectrum of Duplication 5q35.2–Q35.3 Encompassing NSD1: Is It Really a Reversed Sotos Syndrome? Am. J. Med. Genet. A 2013, 161, 2158–2166. [Google Scholar] [CrossRef]
- Rosenfeld, J.A.; Kim, K.H.; Angle, B.; Troxell, R.; Gorski, J.L.; Westemeyer, M.; Frydman, M.; Senturias, Y.; Earl, D.; Torchia, B.; et al. Further Evidence of Contrasting Phenotypes Caused by Reciprocal Deletions and Duplications: Duplication of NSD1 Causes Growth Retardation and Microcephaly. Mol. Syndromol. 2012, 3, 247–254. [Google Scholar] [CrossRef]
- Zhang, H.; Lu, X.; Beasley, J.; Mulvihill, J.J.; Liu, R.; Li, S.; Lee, J.-Y. Reversed Clinical Phenotype Due to a Microduplication of Sotos Syndrome Region Detected by Array CGH: Microcephaly, Developmental Delay and Delayed Bone Age. Am. J. Med. Genet. A 2011, 155, 1374–1378. [Google Scholar] [CrossRef]
- Franco, L.M.; de Ravel, T.; Graham, B.H.; Frenkel, S.M.; van Driessche, J.; Stankiewicz, P.; Lupski, J.R.; Vermeesch, J.R.; Cheung, S.W. A Syndrome of Short Stature, Microcephaly and Speech Delay Is Associated with Duplications Reciprocal to the Common Sotos Syndrome Deletion. Eur. J. Hum. Genet. 2010, 18, 258–261. [Google Scholar] [CrossRef]
- Baujat, G.; Cormier-Daire, V. Sotos Syndrome. Orphanet. J. Rare Dis. 2007, 2, 36. [Google Scholar] [CrossRef]
- Brennan, K.; Shin, J.H.; Tay, J.K.; Prunello, M.; Gentles, A.J.; Sunwoo, J.B.; Gevaert, O. NSD1 Inactivation Defines an Immune Cold, DNA Hypomethylated Subtype in Squamous Cell Carcinoma. Sci. Rep. 2017, 7, 17064. [Google Scholar] [CrossRef]
- Mukherjee, K.; Twyman, R.M.; Vilcinskas, A. Insects as Models to Study the Epigenetic Basis of Disease. Prog. Biophys. Mol. Biol. 2015, 118, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Ferrandon, D.; Imler, J.-L.; Hetru, C.; Hoffmann, J.A. The Drosophila Systemic Immune Response: Sensing and Signalling during Bacterial and Fungal Infections. Nat. Rev. Immunol. 2007, 7, 862–874. [Google Scholar] [CrossRef]
- Hoffmann, J.A.; Reichhart, J.-M. Drosophila Innate Immunity: An Evolutionary Perspective. Nat. Immunol. 2002, 3, 121–126. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Yang, X.; Xi, Y. Fat Body Remodeling and Homeostasis Control in Drosophila. Life Sci. 2016, 167, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Su, F.; Li, Q.; Zhang, J.; Li, Y.; Tang, T.; Hu, Q.; Yu, X.-Q. Pattern Recognition Receptors in Drosophila Immune Responses. Dev. Comp. Immunol. 2020, 102, 103468. [Google Scholar] [CrossRef]
- Hetru, C.; Hoffmann, J.A. NF-KappaB in the Immune Response of Drosophila. Cold Spring Harb. Perspect. Biol. 2009, 1, a000232. [Google Scholar] [CrossRef] [PubMed]
- Jeong, Y.; Kim, T.; Kim, S.; Hong, Y.-K.; Cho, K.S.; Lee, I.-S. Overexpression of Histone Methyltransferase NSD in Drosophila Induces Apoptotic Cell Death via the Jun-N-Terminal Kinase Pathway. Biochem. Biophys. Res. Commun. 2018, 496, 1134–1140. [Google Scholar] [CrossRef]
- Kim, T.; Shin, H.; Song, B.; Won, C.; Yoshida, H.; Yamaguchi, M.; Cho, K.S.; Lee, I.-S. Overexpression of H3K36 Methyltransferase NSD in Glial Cells Affects Brain Development in Drosophila. Glia 2020, 68, 2503–2516. [Google Scholar] [CrossRef]
- Gruntenko, N.E.; Rauschenbach, I.Y. The Role of Insulin Signalling in the Endocrine Stress Response in Drosophila Melanogaster: A Mini-Review. Gen. Comp. Endocrinol. 2018, 258, 134–139. [Google Scholar] [CrossRef]
- Asha, H.; Nagy, I.; Kovacs, G.; Stetson, D.; Ando, I.; Dearolf, C.R. Analysis of Ras-Induced Overproliferation in Drosophila Hemocytes. Genetics 2003, 163, 203–215. [Google Scholar] [CrossRef]
- Takata, K.; Yoshida, H.; Yamaguchi, M.; Sakaguchi, K. Drosophila Damaged DNA-Binding Protein 1 Is an Essential Factor for Development. Genetics 2004, 168, 855–865. [Google Scholar] [CrossRef] [PubMed]
- Khush, R.S.; Leulier, F.; Lemaitre, B. Drosophila Immunity: Two Paths to NF-ΚB. Trends Immunol. 2001, 22, 260–264. [Google Scholar] [CrossRef] [PubMed]
- Tapadia, M.G.; Verma, P. Immune Response and Anti-Microbial Peptides Expression in Malpighian Tubules of Drosophila Melanogaster Is under Developmental Regulation. PLoS ONE 2012, 7, e40714. [Google Scholar] [CrossRef] [PubMed]
- Vodovar, N.; Vinals, M.; Liehl, P.; Basset, A.; Degrouard, J.; Spellman, P.; Dé Ric Boccard, F.; Lemaitre, B. Drosophila Host Defense after Oral Infection by an Entomopathogenic Pseudomonas Species. Proc. Natl. Acad. Sci. USA 2005, 102, 11414–11419. [Google Scholar] [CrossRef] [PubMed]
- Badinloo, M.; Nguyen, E.; Suh, W.; Alzahrani, F.; Castellanos, J.; Klichko, V.I.; Orr, W.C.; Radyuk, S.N. Overexpression of Antimicrobial Peptides Contributes to Aging through Cytotoxic Effects in Drosophila Tissues. Arch. Insect. Biochem. Physiol. 2018, 98, e21464. [Google Scholar] [CrossRef]
- Busse, M.S.; Arnold, C.P.; Towb, P.; Katrivesis, J.; Wasserman, S.A. A ΚB Sequence Code for Pathway-Specific Innate Immune Responses. EMBO J. 2007, 26, 3826–3835. [Google Scholar] [CrossRef]
- Wu, P.-J.; Yan, S.-J. HP1a-mediated heterochromatin formation promotes antimicrobial responses against Pseudomonas aeruginosa infection. BMC Biol. 2022, 20, 234. [Google Scholar] [CrossRef]
- Alekseyenko, A.A.; Gorchakov, A.A.; Zee, B.M.; Fuchs, S.M.; Kharchenko, P.V.; Kuroda, M.I. Heterochromatin-associated interactions of Drosophila HP1a with dADD1, HIPP1, and repetitive RNAs. Genes Dev. 2014, 28, 1445–1460. [Google Scholar] [CrossRef]
- Ganesan, S.; Aggarwal, K.; Paquette, N.; Silverman, N. NF-ΚB/Rel Proteins and the Humoral Immune Responses of Drosophila Melanogaster. In NF-kB in Health and Disease; Springer: Heidelberg, Germany, 2010; pp. 25–60. [Google Scholar]
- Tanji, T.; Yun, E.-Y.; Ip, Y.T. Heterodimers of NF-KappaB Transcription Factors DIF and Relish Regulate Antimicrobial Peptide Genes in Drosophila. Proc. Natl. Acad. Sci. USA 2010, 107, 14715–14720. [Google Scholar] [CrossRef]
- Tanji, T.; Hu, X.; Weber, A.N.R.; Ip, Y.T. Toll and IMD Pathways Synergistically Activate an Innate Immune Response in Drosophila Melanogaster. Mol. Cell Biol. 2007, 27, 4578–4588. [Google Scholar] [CrossRef]
- Pettersson, M.; Schaffner, W. Synergistic Activation of Transcription by Multiple Binding Sites for NF-ΚB Even in Absence of Co-Operative Factor Binding to DNA. J. Mol. Biol. 1990, 214, 373–380. [Google Scholar] [CrossRef] [PubMed]
- Dushay, M.S.; Roethele, J.B.; Chaverri, J.M.; Dulek, D.E.; Syed, S.K.; Kitami, T.; Eldon, E.D. Two Attacin Antibacterial Genes of Drosophila Melanogaster. Gene 2000, 246, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Kurotaki, N.; Imaizumi, K.; Harada, N.; Masuno, M.; Kondoh, T.; Nagai, T.; Ohashi, H.; Naritomi, K.; Tsukahara, M.; Makita, Y.; et al. Haploinsufficiency of NSD1 Causes Sotos Syndrome. Nat. Genet. 2002, 30, 365–366. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Network. Comprehensive Genomic Characterization of Head and Neck Squamous Cell Carcinomas. Nature 2015, 517, 576–582. [Google Scholar] [CrossRef] [PubMed]
- Aerts, H.J.W.L.; Velazquez, E.R.; Leijenaar, R.T.H.; Parmar, C.; Grossmann, P.; Carvalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Decoding Tumour Phenotype by Noninvasive Imaging Using a Quantitative Radiomics Approach. Nat. Commun. 2014, 5, 4006. [Google Scholar] [CrossRef]
- Seiwert, T.Y.; Zuo, Z.; Keck, M.K.; Khattri, A.; Pedamallu, C.S.; Stricker, T.; Brown, C.; Pugh, T.J.; Stojanov, P.; Cho, J.; et al. Integrative and Comparative Genomic Analysis of HPV-Positive and HPV-Negative Head and Neck Squamous Cell Carcinomas. Clin. Cancer Res. 2015, 21, 632–641. [Google Scholar] [CrossRef]
- Chuang, S.-C.; Scelo, G.; Tonita, J.M.; Tamaro, S.; Jonasson, J.G.; Kliewer, E.V.; Hemminki, K.; Weiderpass, E.; Pukkala, E.; Tracey, E.; et al. Risk of Second Primary Cancer among Patients with Head and Neck Cancers: A Pooled Analysis of 13 Cancer Registries. Int. J. Cancer 2008, 123, 2390–2396. [Google Scholar] [CrossRef]
- Lu, T.; Jackson, M.W.; Wang, B.; Yang, M.; Chance, M.R.; Miyagi, M.; Gudkov, A.V.; Stark, G.R. Regulation of NF-ΚB by NSD1/FBXL11-Dependent Reversible Lysine Methylation of P65. Proc. Natl. Acad. Sci. USA 2010, 107, 46–51. [Google Scholar] [CrossRef]
- Chen, Y.; Tang, W.; Zhu, X.; Zhang, L.; Zhu, Y.; Xiao, H.; Xu, J.; Fang, Y.; Li, X.; Tang, C.; et al. Nuclear Receptor Binding SET Domain Protein 1 Promotes Epithelial-Mesenchymal Transition in Paclitaxel-Resistant Breast Cancer Cells via Regulating Nuclear Factor Kappa B and F-Box and Leucine-Rich Repeat Protein 11. Bioengineered 2021, 12, 11506–11519. [Google Scholar] [CrossRef]

| Name | Sequence (5′→3′) |
|---|---|
| NSD_F | TCCATCGTGTGGGCATATT |
| NSD_R | TGCATCATCCTTGAGTTTC |
| Drs_F | GTTCGCCCTCTTCGCTGTCC |
| Drs_R | CCACTGGAGCGTCCCTCCTC |
| Dip_F | GCTGCGCAATCGCTTCTACT |
| Dip_R | TGGTGGAGTGGGCTTCATG |
| Att_F | CACAATGTGGTGGGTCAGG |
| Att_R | GGCACCATGACCAGCATT |
| rp49_F | TACAGGCCCAAGATCGTGAA |
| rp49_R | GTTCGATCCGTAACCGATGT |
| ΔToll_F | GCTTTGATAAGGCATCCAGGCC |
| ΔToll_R | GCTCAGATGTGATGGTGGTTTACTTC |
| ΔImd_F | GCATCTTGAGGTATAAAACCGATGCATTG |
| ΔImd_R | CTGATGATTGACATGATTCATCTGATTGC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Won, C.; Nam, K.; Ko, D.; Kang, B.; Lee, I.-S. NSD Overexpression in the Fat Body Increases Antimicrobial Peptide Production by the Immune Deficiency Pathway in Drosophila. Int. J. Mol. Sci. 2023, 24, 8443. https://doi.org/10.3390/ijms24098443
Won C, Nam K, Ko D, Kang B, Lee I-S. NSD Overexpression in the Fat Body Increases Antimicrobial Peptide Production by the Immune Deficiency Pathway in Drosophila. International Journal of Molecular Sciences. 2023; 24(9):8443. https://doi.org/10.3390/ijms24098443
Chicago/Turabian StyleWon, Chihyun, Kyungju Nam, Donghee Ko, Byungjun Kang, and Im-Soon Lee. 2023. "NSD Overexpression in the Fat Body Increases Antimicrobial Peptide Production by the Immune Deficiency Pathway in Drosophila" International Journal of Molecular Sciences 24, no. 9: 8443. https://doi.org/10.3390/ijms24098443





