Computational and Enzymatic Studies of Sartans in SARS-CoV-2 Spike RBD-ACE2 Binding: The Role of Tetrazole and Perspectives as Antihypertensive and COVID-19 Therapeutics
Abstract
1. Introduction
2. Results
2.1. Computational Studies on the Interactions of Ligands with Spike Protein/ACE2
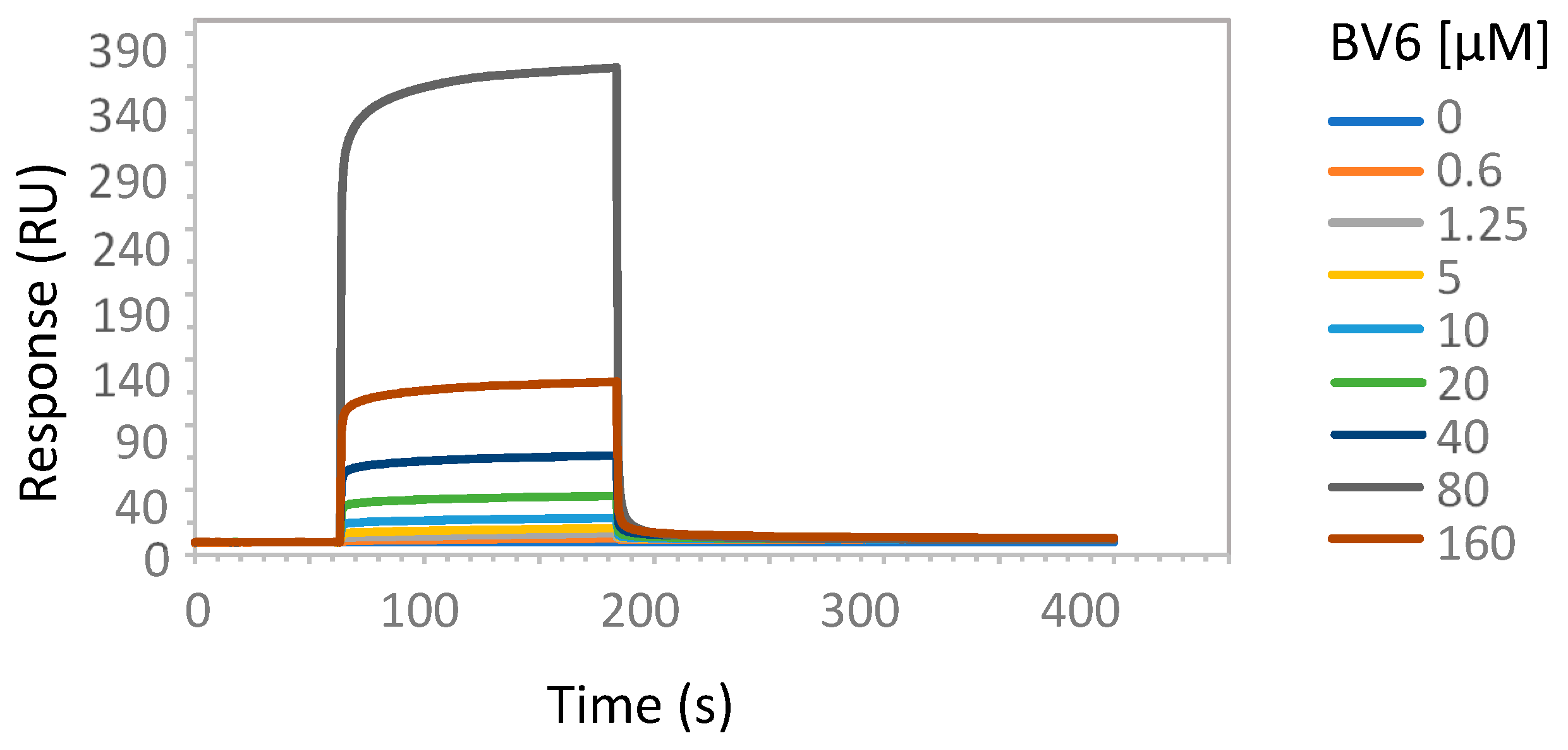
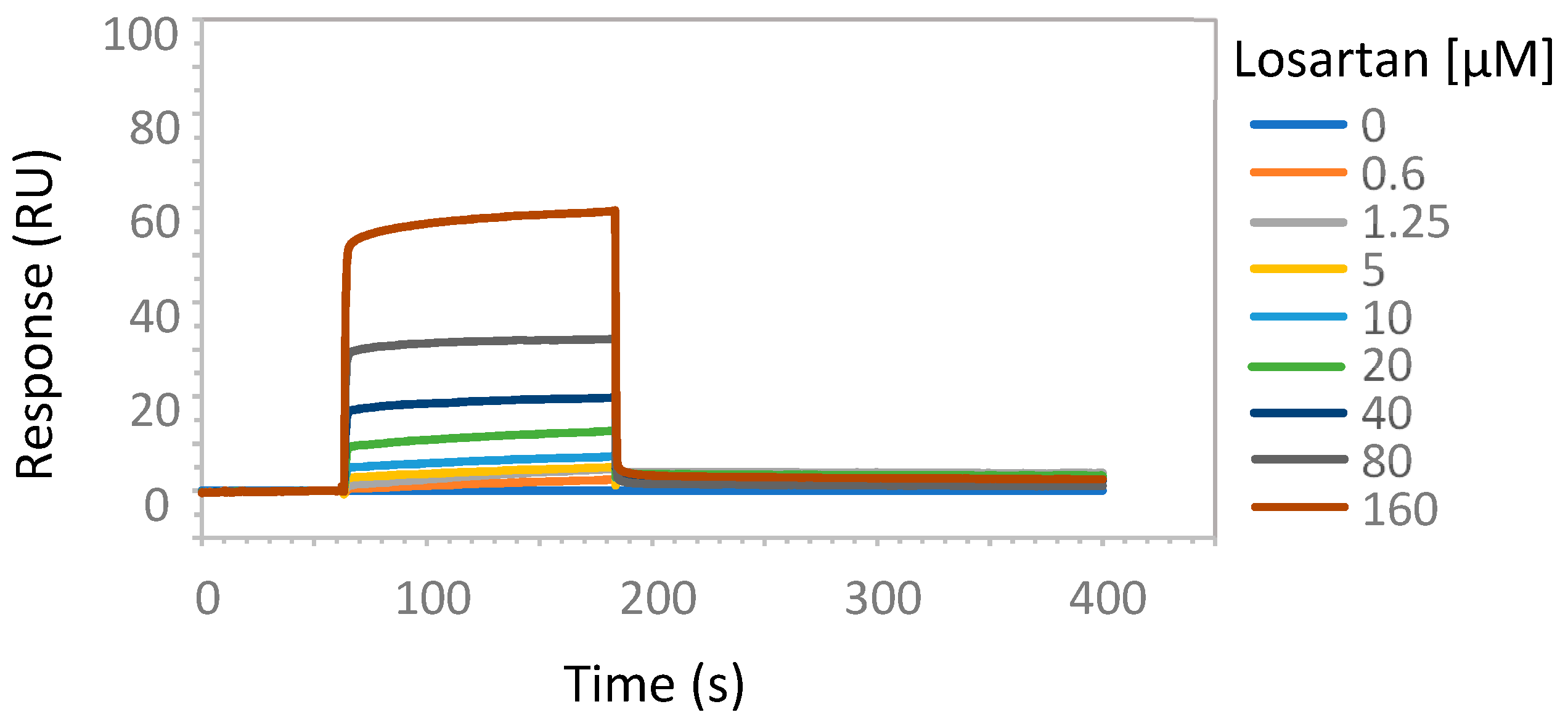
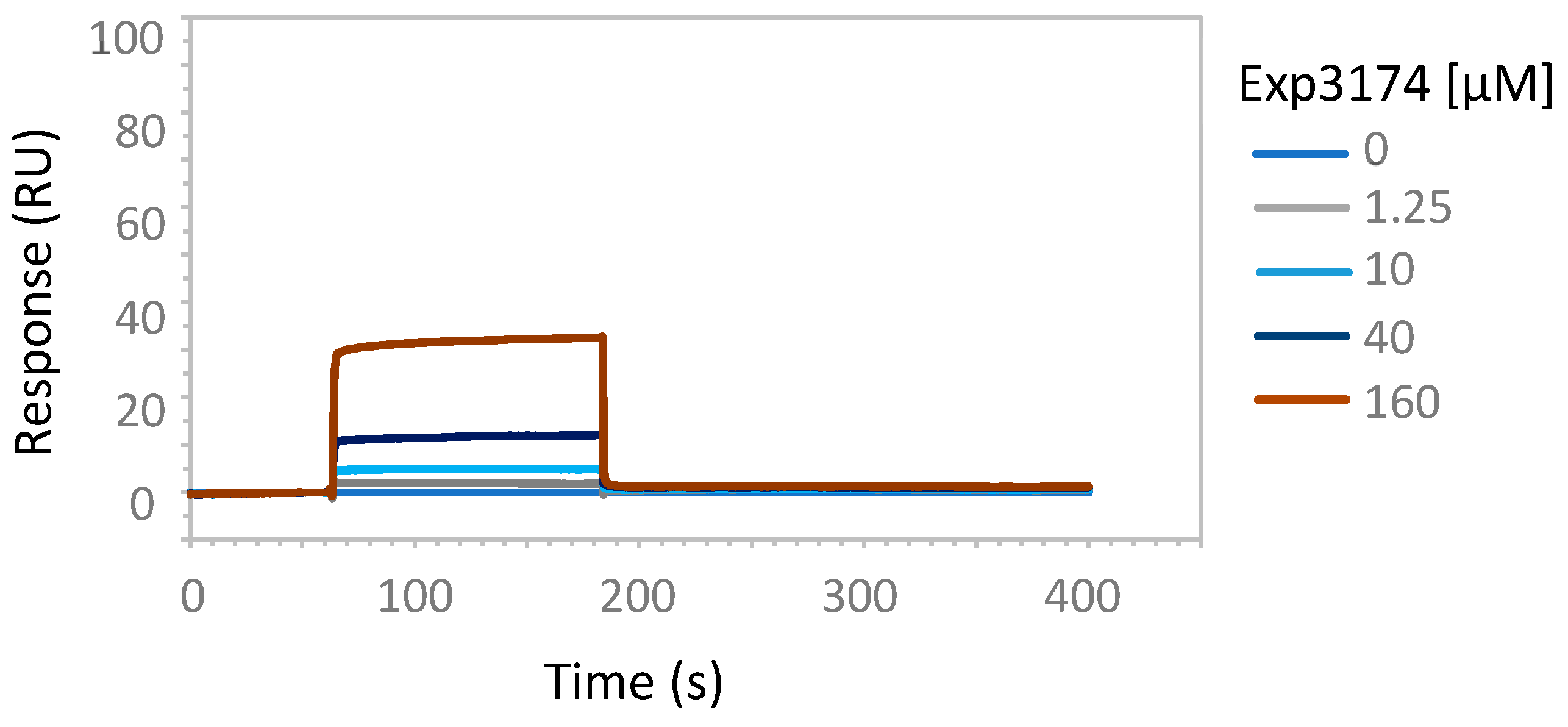
2.2. Enzymatic Studies Supporting the Interaction of Sartans with ACE2
3. Discussion
3.1. Computational Approaches
3.2. Hypertension and COVID-19 Mechanisms Are Similar
3.3. Anionic Groups of ANGII and ARBs Interact with Positive Sites of AT1R and ACE2
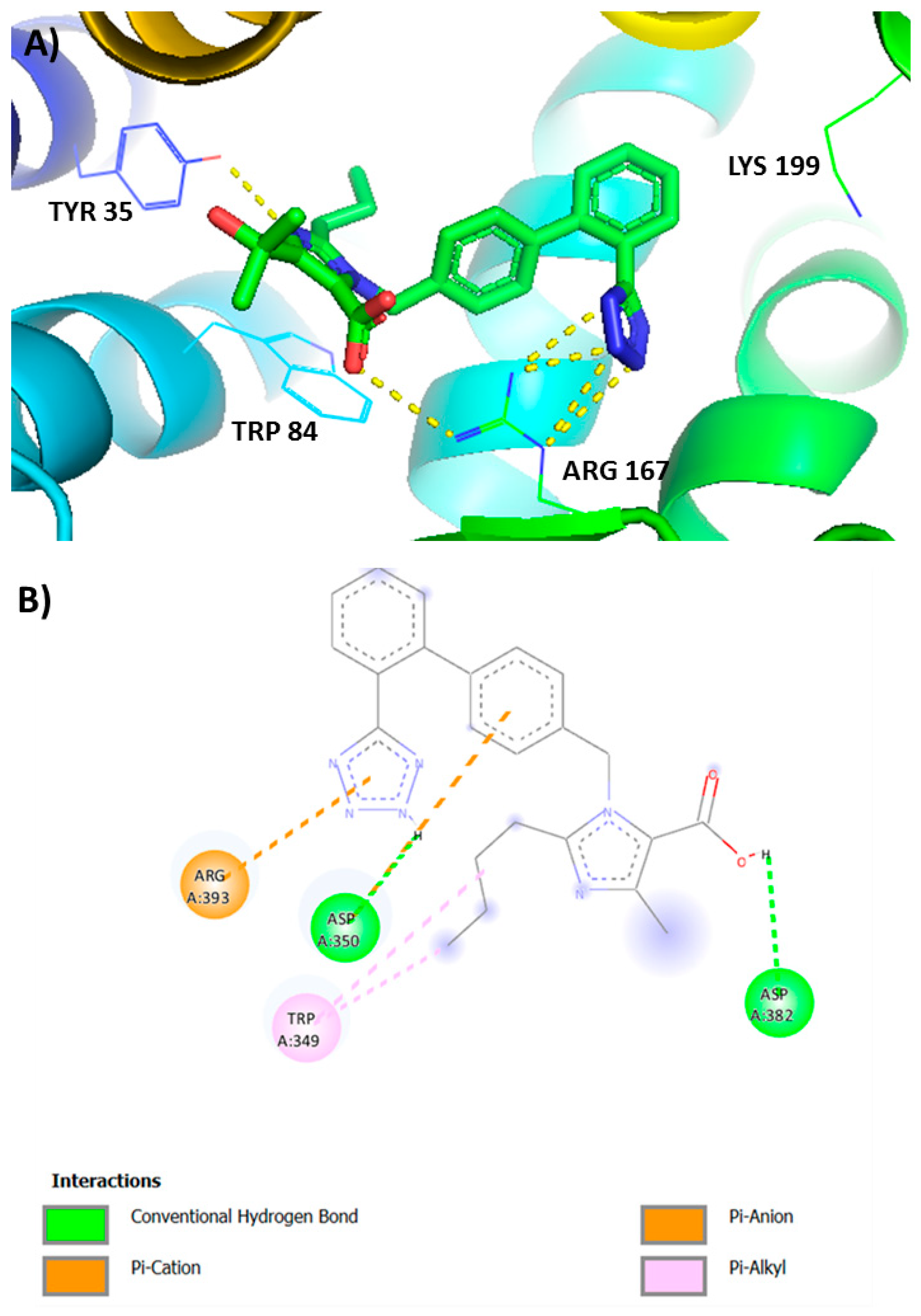
3.4. Tetrazole of Sartans Interact with AT1R (Arg 167) and Spike 681–686 Arginines
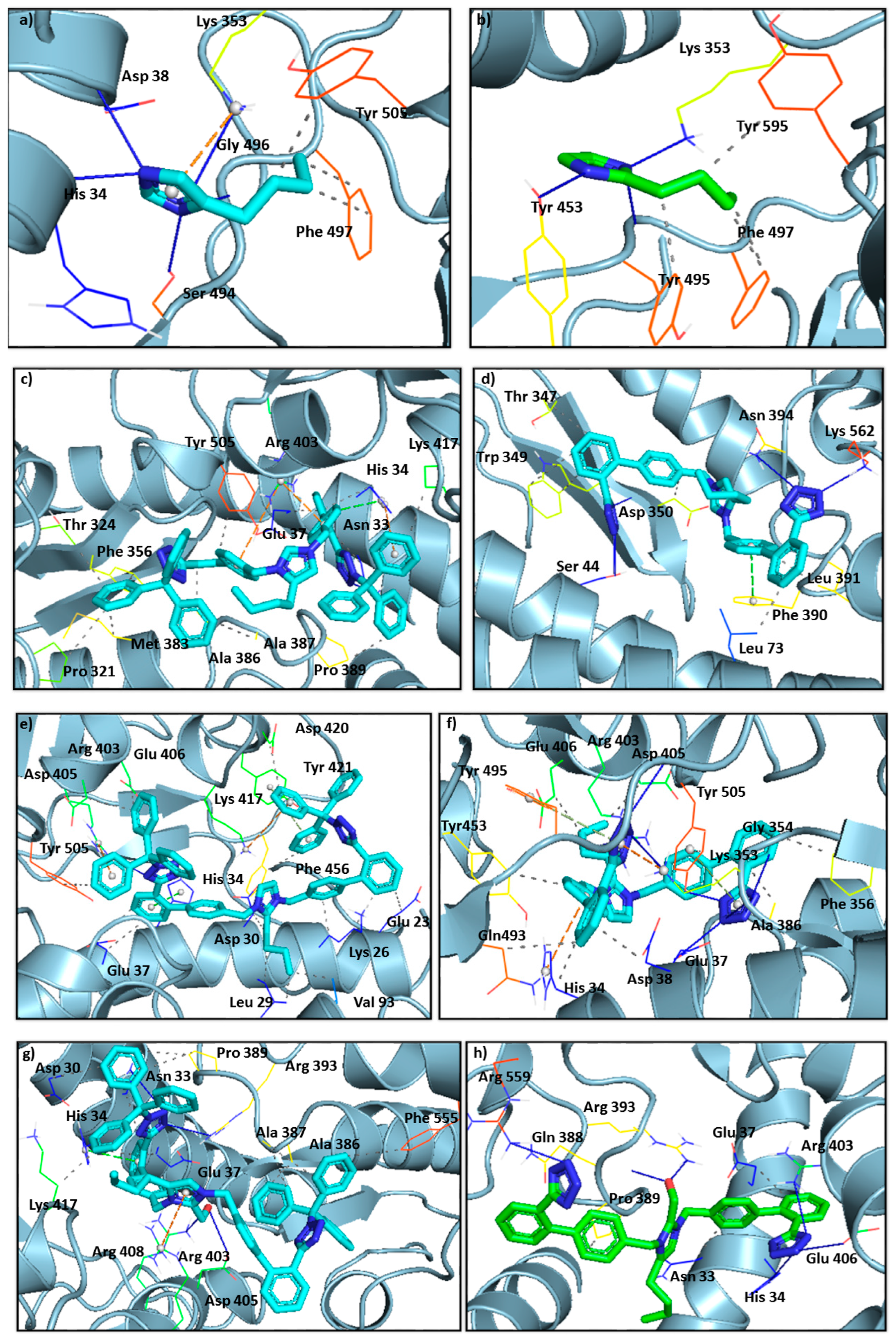
3.5. Ligands Interacting with RBD/ACE2 Complex
3.6. Imidazole and Benzimidazole Based Sartans
3.7. Possible Target of Sartans as Revealed by Enzymatic Assays
3.8. Clinical Perspectives of Bisartans as COVID-19 Antivirals
4. Materials and Methods
4.1. In Silico Methodology and Ligand Preparation
4.2. Molecular Docking
4.3. Docking Parameters
4.4. Organic Synthesis of Bisartans
4.5. ACE2 Protein Purification
4.6. Surface Plasmon Resonance Analysis
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Gadanec, L.K.; McSweeney, K.R.; Qaradakhi, T.; Ali, B.; Zulli, A.; Apostolopoulos, V. Can SARS-CoV-2 Virus Use Multiple Receptors to Enter Host Cells? Int. J. Mol. Sci. 2021, 22, 992. [Google Scholar] [CrossRef] [PubMed]
- Kate Gadanec, L.; Qarad, T.; Renee McSweeney, K.; Ashiana Ali, B.; Zulli, A.; Apostolopoulos, V. Dual targeting of Toll-like receptor 4 and angiotensin-converting enzyme 2: A proposed approach to SARS-CoV-2 treatment. Future Microbiol. 2021, 16, 205–209. [Google Scholar] [CrossRef] [PubMed]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Lemmin, T.; Kalbermatter, D.; Harder, D.; Plattet, P.; Fotiadis, D. Structures and dynamics of the novel S1/S2 protease cleavage site loop of the SARS-CoV-2 spike glycoprotein. J. Struct. Biol. X 2020, 4, 100038. [Google Scholar] [CrossRef]
- Peacock, T.P.; Goldhill, D.H.; Zhou, J.; Baillon, L.; Frise, R.; Swann, O.C.; Kugathasan, R.; Penn, R.; Brown, J.C.; Sanchez-David, R.Y.; et al. The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat. Microbiol. 2021, 6, 899–909. [Google Scholar] [CrossRef]
- Li, W.; Moore, M.J.; Vasilieva, N.; Sui, J.; Wong, S.K.; Berne, M.A.; Somasundaran, M.; Sullivan, J.L.; Luzuriaga, K.; Greenough, T.C.; et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 2003, 426, 450–454. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.K.; Hassanzadeganroudsari, M.; Feehan, J.; Apostolopoulos, V. The race for a COVID-19 vaccine: Where are we up to? Expert Rev. Vaccines 2021, 21, 355–376. [Google Scholar] [CrossRef] [PubMed]
- Chavda, V.P.; Kapadia, C.; Soni, S.; Prajapati, R.; Chauhan, S.C.; Yallapu, M.M.; Apostolopoulos, V. A global picture: Therapeutic perspectives for COVID-19. Immunotherapy 2022, 14, 351–371. [Google Scholar] [CrossRef]
- Feehan, J.; Apostolopoulos, V. Is COVID-19 the worst pandemic? Maturitas 2021, 149, 56–58. [Google Scholar] [CrossRef]
- Burki, T. The future of Paxlovid for COVID-19. Lancet Respir. Med. 2022, 10, e68. [Google Scholar] [CrossRef]
- Hammond, J.; Leister-Tebbe, H.; Gardner, A.; Abreu, P.; Bao, W.; Wisemandle, W.; Baniecki, M.; Hendrick, V.M.; Damle, B.; Simón-Campos, A.; et al. Oral Nirmatrelvir for High-Risk, Nonhospitalized Adults with COVID-19. N. Engl. J. Med. 2022, 386, 1397–1408. [Google Scholar] [CrossRef]
- Fischer, W.; Eron, J.J.; Holman, W.; Cohen, M.S.; Fang, L.; Szewczyk, L.J.; Sheahan, T.P.; Baric, R.; Mollan, K.R.; Wolfe, C.R.; et al. Molnupiravir, an Oral Antiviral Treatment for COVID-19. medRxiv 2021. [Google Scholar] [CrossRef]
- Jayk Bernal, A.; Gomes da Silva, M.M.; Musungaie, D.B.; Kovalchuk, E.; Gonzalez, A.; Delos Reyes, V.; Martín-Quirós, A.; Caraco, Y.; Williams-Diaz, A.; Brown, M.L.; et al. Molnupiravir for Oral Treatment of COVID-19 in Nonhospitalized Patients. N. Engl. J. Med. 2022, 386, 509–520. [Google Scholar] [CrossRef] [PubMed]
- Tian, L.; Pang, Z.; Li, M.; Lou, F.; An, X.; Zhu, S.; Song, L.; Tong, Y.; Fan, H.; Fan, J. Molnupiravir and Its Antiviral Activity Against COVID-19. Front. Immunol. 2022, 13, 855496. [Google Scholar] [CrossRef] [PubMed]
- Owen, D.R.; Allerton, C.M.N.; Anderson, A.S.; Aschenbrenner, L.; Avery, M.; Berritt, S.; Boras, B.; Cardin, R.D.; Carlo, A.; Coffman, K.J.; et al. An oral SARS-CoV-2 Mpro inhibitor clinical candidate for the treatment of COVID-19. Science 2021, 374, 1586–1593. [Google Scholar] [CrossRef]
- Jeong, J.H.; Chokkakula, S.; Min, S.C.; Kim, B.K.; Choi, W.-S.; Oh, S.; Yun, Y.S.; Kang, D.H.; Lee, O.-J.; Kim, E.-G.; et al. Combination therapy with nirmatrelvir and molnupiravir improves the survival of SARS-CoV-2 infected mice. BioRxiv 2022. [Google Scholar] [CrossRef]
- Manns, M.P.; Maasoumy, B. Breakthroughs in hepatitis C research: From discovery to cure. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 533–550. [Google Scholar] [CrossRef]
- Tsai, E. Review of Current and Potential Treatments for Chronic Hepatitis B Virus Infection. Gastroenterol. Hepatol. 2021, 17, 367–376. [Google Scholar]
- Moore, G.J.; Pires, J.M.; Kelaidonis, K.; Gadanec, L.K.; Zulli, A.; Apostolopoulos, V.; Matsoukas, J.M. Receptor Interactions of Angiotensin II and Angiotensin Receptor Blockers—Relevance to COVID-19. Biomolecules 2021, 11, 979. [Google Scholar] [CrossRef]
- Moore, G.J.; Ridgway, H.; Kelaidonis, K.; Chasapis, C.T.; Ligielli, I.; Mavromoustakos, T.; Bojarska, J.; Matsoukas, J.M. Actions of Novel Angiotensin Receptor Blocking Drugs, Bisartans, Relevant for COVID-19 Therapy: Biased Agonism at Angiotensin Receptors and the Beneficial Effects of Neprilysin in the Renin Angiotensin System. Molecules 2022, 27, 4854. [Google Scholar] [CrossRef]
- Ridgway, H.; Chasapis, C.T.; Kelaidonis, K.; Ligielli, I.; Moore, G.J.; Gadanec, L.K.; Zulli, A.; Apostolopoulos, V.; Mavromoustakos, T.; Matsoukas, J.M. Understanding the Driving Forces That Trigger Mutations in SARS-CoV-2: Mutational Energetics and the Role of Arginine Blockers in COVID-19 Therapy. Viruses 2022, 14, 1029. [Google Scholar] [CrossRef] [PubMed]
- Ridgway, H.; Moore, G.J.; Mavromoustakos, T.; Tsiodras, S.; Ligielli, I.; Kelaidonis, K.; Chasapis, C.T.; Gadanec, L.K.; Zulli, A.; Apostolopoulos, V.; et al. Discovery of a new generation of angiotensin receptor blocking drugs: Receptor mechanisms and in silico binding to enzymes relevant to SARS-CoV-2. Comput. Struct. Biotechnol. J. 2022, 20, 2091–2111. [Google Scholar] [CrossRef] [PubMed]
- Matsoukas, J.; Apostolopoulos, V.; Zulli, A.; Moore, G.; Kelaidonis, K.; Moschovou, K.; Mavromoustakos, T. From Angiotensin II to Cyclic Peptides and Angiotensin Receptor Blockers (ARBs): Perspectives of ARBs in COVID-19 Therapy. Molecules 2021, 26, 618. [Google Scholar] [CrossRef] [PubMed]
- Ridgway, H.; Ntallis, C.; Chasapis, C.T.; Kelaidonis, K.; Matsoukas, M.-T.; Plotas, P.; Apostolopoulos, V.; Moore, G.; Tsiodras, S.; Paraskevis, D.; et al. Molecular Epidemiology of SARS-CoV-2: The Dominant Role of Arginine in Mutations and Infectivity. Viruses 2023, 15, 309. [Google Scholar] [CrossRef]
- Dalkas, G.A.; Chasapis, C.T.; Gkazonis, P.V.; Bentrop, D.; Spyroulias, G.A. Conformational Dynamics of the Anthrax Lethal Factor Catalytic Center. Biochemistry 2010, 49, 10767–10769. [Google Scholar] [CrossRef]
- Guo, J.; Huang, Z.; Lin, L.; Lv, J. Coronavirus Disease 2019 (COVID-19) and Cardiovascular Disease: A Viewpoint on the Potential Influence of Angiotensin-Converting Enzyme Inhibitors/Angiotensin Receptor Blockers on Onset and Severity of Severe Acute Respiratory Syndrome Coronavirus 2 Infection. J. Am. Heart Assoc. 2020, 9, e016219. [Google Scholar] [CrossRef]
- Nejat, R.; Sadr, A.S. Are losartan and imatinib effective against SARS-CoV2 pathogenesis? A pathophysiologic-based in silico study. Silico Pharmacol. 2020, 9, 1. [Google Scholar] [CrossRef]
- Zhang, H.; Unal, H.; Desnoyer, R.; Han, G.W.; Patel, N.; Katritch, V.; Karnik, S.S.; Cherezov, V.; Stevens, R.C. Structural Basis for Ligand Recognition and Functional Selectivity at Angiotensin Receptor. J. Biol. Chem. 2015, 290, 29127–29139. [Google Scholar] [CrossRef]
- Zhang, H.; Unal, H.; Gati, C.; Han, G.W.; Liu, W.; Zatsepin, N.A.; James, D.; Wang, D.; Nelson, G.; Weierstall, U.; et al. Structure of the Angiotensin Receptor Revealed by Serial Femtosecond Crystallography. Cell 2015, 161, 833–844. [Google Scholar] [CrossRef]
- Cui, X.; Chen, W.; Zhou, H.; Gong, Y.; Zhu, B.; Lv, X.; Guo, H.; Duan, J.; Zhou, J.; Marcon, E.; et al. Pulmonary Edema in COVID-19 Patients: Mechanisms and Treatment Potential. Front. Pharmacol. 2021, 12, 664349. [Google Scholar] [CrossRef]
- Meng, J.; Xiao, G.; Zhang, J.; He, X.; Ou, M.; Bi, J.; Yang, R.; Di, W.; Wang, Z.; Li, Z.; et al. Renin-angiotensin system inhibitors improve the clinical outcomes of COVID-19 patients with hypertension. Emerg. Microbes Infect. 2020, 9, 757–760. [Google Scholar] [CrossRef] [PubMed]
- Saravi, B.; Li, Z.; Lang, C.N.; Schmid, B.; Lang, F.K.; Grad, S.; Alini, M.; Richards, R.G.; Schmal, H.; Südkamp, N.; et al. The Tissue Renin-Angiotensin System and Its Role in the Pathogenesis of Major Human Diseases: Quo Vadis? Cells 2021, 10, 650. [Google Scholar] [CrossRef] [PubMed]
- Steckelings, U.M.; Widdop, R.E.; Sturrock, E.D.; Lubbe, L.; Hussain, T.; Kaschina, E.; Unger, T.; Hallberg, A.; Carey, R.M.; Sumners, C.; et al. The Angiotensin AT2 Receptor: From a Binding Site to a Novel Therapeutic Target. Pharmacol. Rev. 2022, 74, 1051–1135. [Google Scholar] [CrossRef] [PubMed]
- Stone, R.E.; Liu, S.; Levy, A.M.; Kashani, N.; Louie, S.G.; Rodgers, K.E.; Kelland, E.E.; Lund, B.T. Activation of the Protective Arm of the Renin Angiotensin System in Demyelinating Disease. J. Neuroimmune Pharmacol. 2019, 15, 249–263. [Google Scholar] [CrossRef] [PubMed]
- Stegbauer, J.; Lee, D.-H.; Seubert, S.; Ellrichmann, G.; Manzel, A.; Kvakan, H.; Muller, D.N.; Gaupp, S.; Rump, L.C.; Gold, R.; et al. Role of the renin-angiotensin system in autoimmune inflammation of the central nervous system. Proc. Natl. Acad. Sci. USA 2009, 106, 14942–14947. [Google Scholar] [CrossRef] [PubMed]
- Dargahi, N.; Matsoukas, J.; Apostolopoulos, V. Streptococcus thermophilus ST285 Alters Pro-Inflammatory to Anti-Inflammatory Cytokine Secretion against Multiple Sclerosis Peptide in Mice. Brain Sci. 2020, 10, 126. [Google Scholar] [CrossRef] [PubMed]
- Schiffrin, E.L.; Flack, J.M.; Ito, S.; Muntner, P.; Webb, R.C. Hypertension and COVID-19. Am. J. Hypertens. 2020, 33, 373–374. [Google Scholar] [CrossRef]
- Wang, D.; Hu, B.; Hu, C.; Zhu, F.; Liu, X.; Zhang, J.; Wang, B.; Xiang, H.; Cheng, Z.; Xiong, Y.; et al. Clinical Characteristics of 138 Hospitalized Patients with 2019 Novel Coronavirus–Infected Pneumonia in Wuhan, China. JAMA 2020, 323, 1061. [Google Scholar] [CrossRef]
- Zhang, W.; Zhao, Y.; Zhang, F.; Wang, Q.; Li, T.; Liu, Z.; Wang, J.; Qin, Y.; Zhang, X.; Yan, X.; et al. The use of anti-inflammatory drugs in the treatment of people with severe coronavirus disease 2019 (COVID-19): The Perspectives of clinical immunologists from China. Clin. Immunol. 2020, 214, 108393. [Google Scholar] [CrossRef]
- Zhou, F.; Yu, T.; Du, R.; Fan, G.; Liu, Y.; Liu, Z.; Xiang, J.; Wang, Y.; Song, B.; Gu, X.; et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet 2020, 395, 1054–1062. [Google Scholar] [CrossRef]
- Lanza, K.; Perez, L.G.; Costa, L.B.; Cordeiro, T.M.; Palmeira, V.A.; Ribeiro, V.T.; Simões e Silva, A.C. COVID-19: The renin–angiotensin system imbalance hypothesis. Clin. Sci. 2020, 134, 1259–1264. [Google Scholar] [CrossRef] [PubMed]
- Vieira, C.; Nery, L.; Martins, L.; Jabour, L.; Dias, R.; Simões e Silva, A.C. Downregulation of Membrane-bound Angiotensin Converting Enzyme 2 (ACE2) Receptor has a Pivotal Role in COVID-19 Immunopathology. Curr. Drug Targets 2021, 22, 254–281. [Google Scholar] [CrossRef] [PubMed]
- Costa, L.B.; Perez, L.G.; Palmeira, V.A.; Macedo e Cordeiro, T.; Ribeiro, V.T.; Lanza, K.; Simões e Silva, A.C. Insights on SARS-CoV-2 Molecular Interactions with the Renin-Angiotensin System. Front. Cell Dev. Biol. 2020, 8, 559841. [Google Scholar] [CrossRef] [PubMed]
- Agelis, G.; Resvani, A.; Koukoulitsa, C.; Tůmová, T.; Slaninová, J.; Kalavrizioti, D.; Spyridaki, K.; Afantitis, A.; Melagraki, G.; Siafaka, A.; et al. Rational design, efficient syntheses and biological evaluation of N, N ′-symmetrically bis-substituted butylimidazole analogs as a new class of potent Angiotensin II receptor blockers. Eur. J. Med. Chem. 2013, 62, 352–370. [Google Scholar] [CrossRef] [PubMed]
- Wingler, L.M.; McMahon, C.; Staus, D.P.; Lefkowitz, R.J.; Kruse, A.C. Distinctive Activation Mechanism for Angiotensin Receptor Revealed by a Synthetic Nanobody. Cell 2019, 176, 479–490.e412. [Google Scholar] [CrossRef]
- Courvoisier, E.; Williams, P.A.; Lim, G.K.; Hughes, C.E.; Harris, K.D.M. The crystal structure of l-arginine. Chem. Commun. 2012, 48, 2761. [Google Scholar] [CrossRef] [PubMed]
- Moore, G.J.; Matsoukas, J.M. Angiotensin as a model for hormone—Receptor interactions. Bioscience Reports 1985, 5, 407–416. [Google Scholar] [CrossRef]
- Turner, R.J.; Matsoukas, J.M.; Moore, G.J. Fluorescence properties of angiotensin II analogues in receptor-simulating environments: Relationship between tyrosinate fluorescence lifetime and biological activity. Biochim. Biophys. Acta (BBA)-Biomembr. 1991, 1065, 21–28. [Google Scholar] [CrossRef]
- Hondrelis, J.; Lonergan, G.; Voliotis, S.; Matsoukas, J. One pot synthesis and conformation of N-t-butyloxycarbonyl, O-Phenacyl derivatives of proline and other secondary amino acids. Tetrahedron 1990, 46, 565–576. [Google Scholar] [CrossRef]
- Matsoukas, J.; Cordopatis, P.; Belte, U.; Goghari, M.H.; Ganter, R.C.; Franklin, K.J.; Moore, G.J. Importance of the N-terminal domain of the type II angiotensin antagonist sarmesin for receptor blockade. J. Med. Chem. 1988, 31, 1418–1421. [Google Scholar] [CrossRef]
- Matsoukas, J.M.; Agelis, G.; Hondrelis, J.; Yamdagni, R.; Wu, Q.; Ganter, R.; Moore, D.; Moore, G.J.; Smith, J.R. Synthesis and biological activities of angiotensin II, sarilesin, and sarmesin analogs containing Aze or Pip at position 7. J. Med. Chem. 1993, 36, 904–911. [Google Scholar] [CrossRef] [PubMed]
- Matsoukas, M.-T.; Potamitis, C.; Plotas, P.; Androutsou, M.-E.; Agelis, G.; Matsoukas, J.; Zoumpoulakis, P. Insights into AT1 Receptor Activation through AngII Binding Studies. J. Chem. Inf. Model. 2013, 53, 2798–2811. [Google Scholar] [CrossRef] [PubMed]
- Matsoukas, J.M.; Hondrelis, J.; Keramida, M.; Mavromoustakos, T.; Makriyannis, A.; Yamdagni, R.; Wu, Q.; Moore, G.J. Role of the NH2-terminal domain of angiotensin II (ANG II) and [Sar1]angiotensin II on conformation and activity. NMR evidence for aromatic ring clustering and peptide backbone folding compared with [des-1,2,3]angiotensin II. J. Biol. Chem. 1994, 269, 5303–5312. [Google Scholar] [CrossRef]
- Polevaya, L.; Mavromoustakos, T.; Zoumboulakis, P.; Grdadolnik, S.G.; Roumelioti, P.; Giatas, N.; Mutule, I.; Keivish, T.; Vlahakos, D.V.; Iliodromitis, E.K.; et al. Synthesis and study of a cyclic angiotensin II antagonist analogue reveals the role of π*–π* interactions in the C-terminal aromatic residue for agonist activity and its structure resemblance with AT1 non-peptide antagonists. Bioorg. Med. Chem. 2001, 9, 1639–1647. [Google Scholar] [CrossRef] [PubMed]
- Friligou, I.; Papadimitriou, E.; Gatos, D.; Matsoukas, J.; Tselios, T. Microwave-assisted solid-phase peptide synthesis of the 60–110 domain of human pleiotrophin on 2-chlorotrityl resin. Amino Acids 2010, 40, 1431–1440. [Google Scholar] [CrossRef] [PubMed]
- Blow, D.M.; Birktoft, J.J.; Hartley, B.S. Role of a Buried Acid Group in the Mechanism of Action of Chymotrypsin. Nature 1969, 221, 337–340. [Google Scholar] [CrossRef] [PubMed]
- Duarte, M.; Pelorosso, F.; Nicolosi, L.N.; Victoria Salgado, M.; Vetulli, H.; Aquieri, A.; Azzato, F.; Castro, M.; Coyle, J.; Davolos, I.; et al. Telmisartan for treatment of COVID-19 patients: An open multicenter randomized clinical trial. EClinicalMedicine 2021, 37, 100962. [Google Scholar] [CrossRef]
- Walther, T.; Kuebler, W.M. Don’t judge too RAShly: The multifaceted role of the renin-angiotensin system and its therapeutic potential in COVID-19. Am. J. Physiol.-Lung Cell. Mol. Physiol. 2020, 318, L1023–L1024. [Google Scholar] [CrossRef]
- Fatouros, P.R.; Roy, U.; Sur, S. Modeling Substrate Coordination to Zn-Bound Angiotensin Converting Enzyme 2. bioRxiv 2021. preprint. [Google Scholar] [CrossRef]
- Abassi, Z.A.; Skorecki, K.; Heyman, S.N.; Kinaneh, S.; Armaly, Z. COVID-19 infection and mortality: A physiologist’s perspective enlightening clinical features and plausible interventional strategies. Am. J. Physiol.-Lung Cell. Mol. Physiol. 2020, 318, L1020–L1022. [Google Scholar] [CrossRef]
- Abassi, Z.A.; Skorecki, K.; Heyman, S.N.; Kinaneh, S.; Armaly, Z. Reply to Letter to the Editor: “Don’t judge too RAShly: The multifaceted role of the renin-angiotensin system and its therapeutic potential in COVID-19”. Am. J. Physiol.-Lung Cell. Mol. Physiol. 2020, 318, L1029–L1030. [Google Scholar] [CrossRef] [PubMed]
- Rico-Mesa, J.S.; White, A.; Anderson, A.S. Outcomes in Patients with COVID-19 Infection Taking ACEI/ARB. Curr. Cardiol. Rep. 2020, 22, 31. [Google Scholar] [CrossRef] [PubMed]
- Matsoukas, J.M.; Gadanec, L.K.; Zulli, A.; Apostolopoulos, V.; Kelaidonis, K.; Ligielli, I.; Moschovou, K.; Georgiou, N.; Plotas, P.; Chasapis, C.T.; et al. Diminazene Aceturate Reduces Angiotensin II Constriction and Interacts with the Spike Protein of Severe Acute Respiratory Syndrome Coronavirus 2. Biomedicines 2022, 10, 1731. [Google Scholar] [CrossRef] [PubMed]
- Muhamad, S.-A.; Ugusman, A.; Kumar, J.; Skiba, D.; Hamid, A.A.; Aminuddin, A. COVID-19 and Hypertension: The What, the Why, and the How. Front. Physiol. 2021, 12, 665064. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, A.; Narwal, M.; Majowicz, S.A.; Varricchio, C.; Toner, S.A.; Ballatore, C.; Brancale, A.; Murakami, K.S.; Jose, J. Identification of SARS-CoV-2 inhibitors targeting Mpro and PLpro using in-cell-protease assay. Commun. Biol. 2022, 5, 169. [Google Scholar] [CrossRef]
- Harder, E.; Damm, W.; Maple, J.; Wu, C.; Reboul, M.; Xiang, J.Y.; Wang, L.; Lupyan, D.; Dahlgren, M.K.; Knight, J.L.; et al. OPLS3: A Force Field Providing Broad Coverage of Drug-like Small Molecules and Proteins. J. Chem. Theory Comput. 2015, 12, 281–296. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2009, 31, 455–461. [Google Scholar] [CrossRef]
- Oulas, A.; Richter, J.; Zanti, M.; Tomazou, M.; Michailidou, K.; Christodoulou, K.; Christodoulou, C.; Spyrou, G.M. In depth analysis of Cyprus-specific mutations of SARS-CoV-2 strains using computational approaches. BMC Genom. Data 2021, 22, 48. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Islam, R.; Parves, M.R.; Paul, A.S.; Uddin, N.; Rahman, M.S.; Mamun, A.A.; Hossain, M.N.; Ali, M.A.; Halim, M.A. A molecular modeling approach to identify effective antiviral phytochemicals against the main protease of SARS-CoV-2. J. Biomol. Struct. Dyn. 2020, 39, 3213–3224. [Google Scholar] [CrossRef]
- Sarma, P.; Shekhar, N.; Prajapat, M.; Avti, P.; Kaur, H.; Kumar, S.; Singh, S.; Kumar, H.; Prakash, A.; Dhibar, D.P.; et al. In-silico homology assisted identification of inhibitor of RNA binding against 2019-nCoV N-protein (N terminal domain). J. Biomol. Struct. Dyn. 2020, 39, 2724–2732. [Google Scholar] [CrossRef] [PubMed]
- Lupala, C.S.; Li, X.; Lei, J.; Chen, H.; Qi, J.; Liu, H.; Su, X.-D. Computational simulations reveal the binding dynamics between human ACE2 and the receptor binding domain of SARS-CoV-2 spike protein. bioRxiv 2020. [Google Scholar] [CrossRef]
- Wang, Z.; Zheng, Q.; Liu, S.; Elofsson, A. Potential covalent drugs targeting the main protease of the SARS-CoV-2 coronavirus. Bioinformatics 2020, 36, 3295–3298. [Google Scholar] [CrossRef]
- Shang, J.; Ye, G.; Shi, K.; Wan, Y.; Luo, C.; Aihara, H.; Geng, Q.; Auerbach, A.; Li, F. Structural basis of receptor recognition by SARS-CoV-2. Nature 2020, 581, 221–224. [Google Scholar] [CrossRef] [PubMed]
- Pommié, C.; Levadoux, S.; Sabatier, R.; Lefranc, G.; Lefranc, M.-P. IMGT standardized criteria for statistical analysis of immunoglobulin V-REGION amino acid properties. J. Mol. Recognit. 2004, 17, 17–32. [Google Scholar] [CrossRef]
- Salentin, S.; Schreiber, S.; Haupt, V.J.; Adasme, M.F.; Schroeder, M. PLIP: Fully automated protein–ligand interaction profiler. Nucleic Acids Res. 2015, 43, W443–W447. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, Y.; Wu, L.; Niu, S.; Song, C.; Zhang, Z.; Lu, G.; Qiao, C.; Hu, Y.; Yuen, K.-Y.; et al. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell 2020, 181, 894–904.e899. [Google Scholar] [CrossRef]
- Allouche, A.-R. Gabedit-A graphical user interface for computational chemistry softwares. J. Comput. Chem. 2011, 32, 174–182. [Google Scholar] [CrossRef]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An open chemical toolbox. J. Chem. Inform. 2011, 3, 33. [Google Scholar] [CrossRef]
- Zhu, Z.-L.; Qiu, X.-D.; Wu, S.; Liu, Y.-T.; Zhao, T.; Sun, Z.-H.; Li, Z.-R.; Shan, G.-Z. Blocking Effect of Demethylzeylasteral on the Interaction between Human ACE2 Protein and SARS-CoV-2 RBD Protein Discovered Using SPR Technology. Molecules 2020, 26, 57. [Google Scholar] [CrossRef]

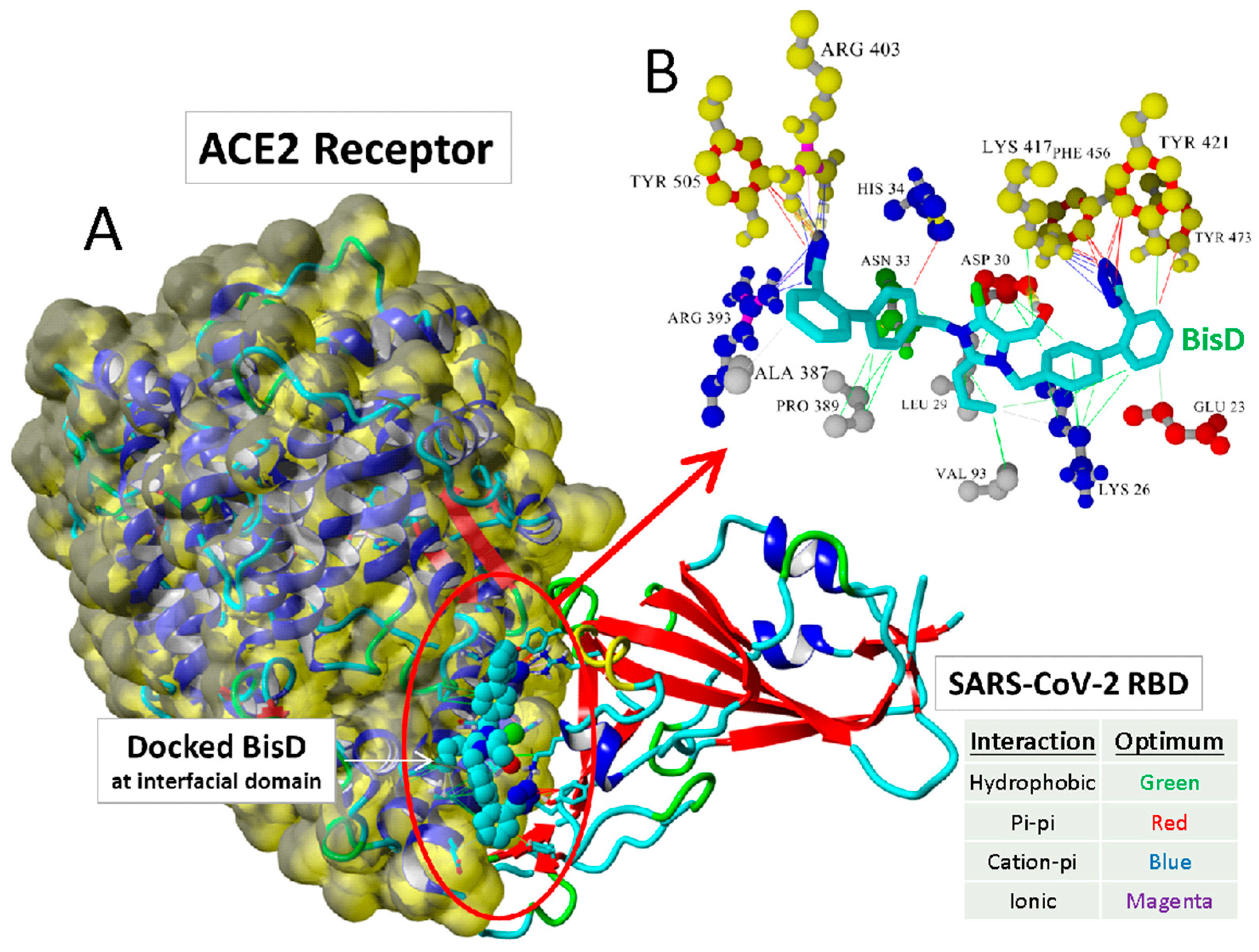

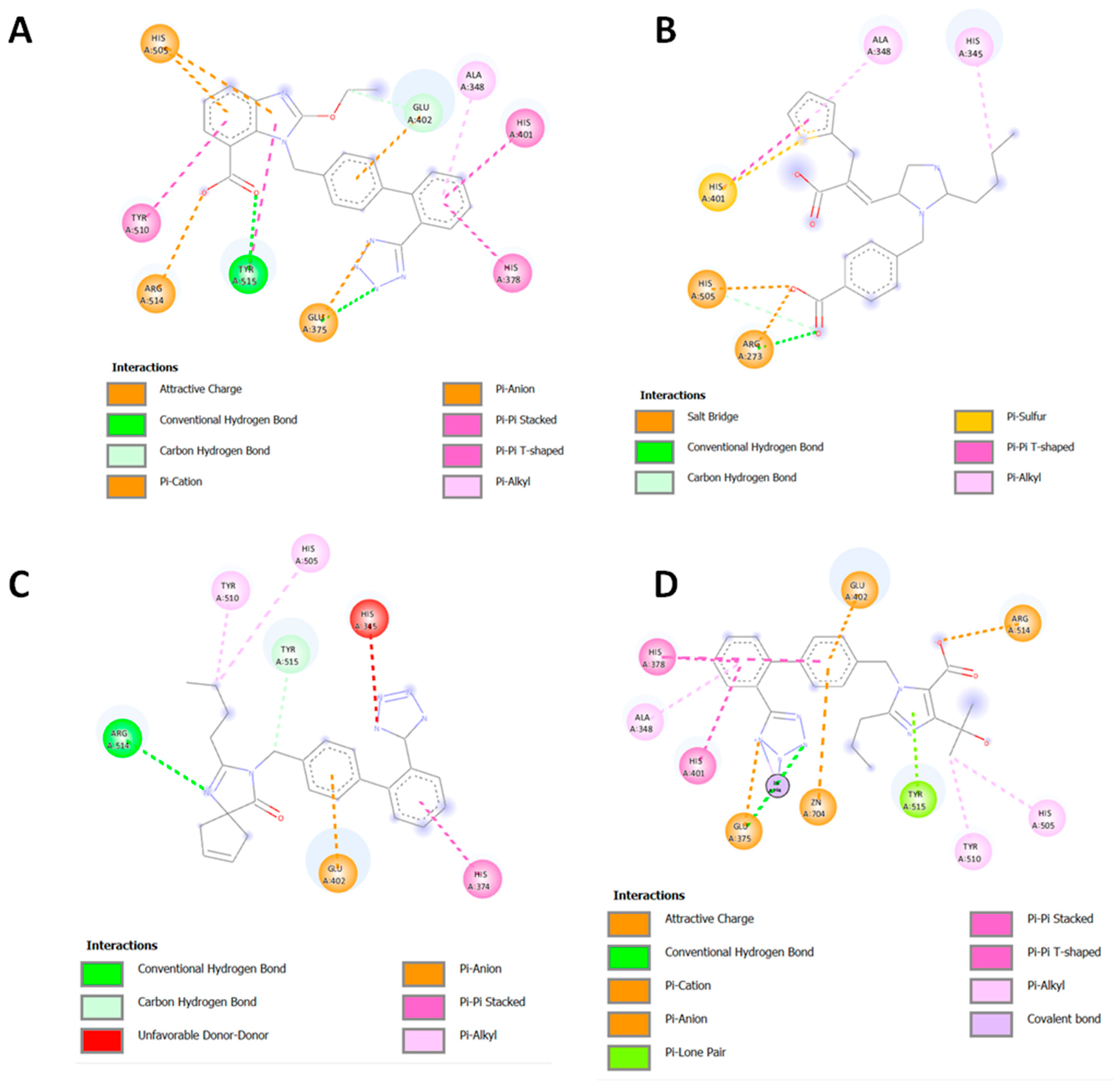
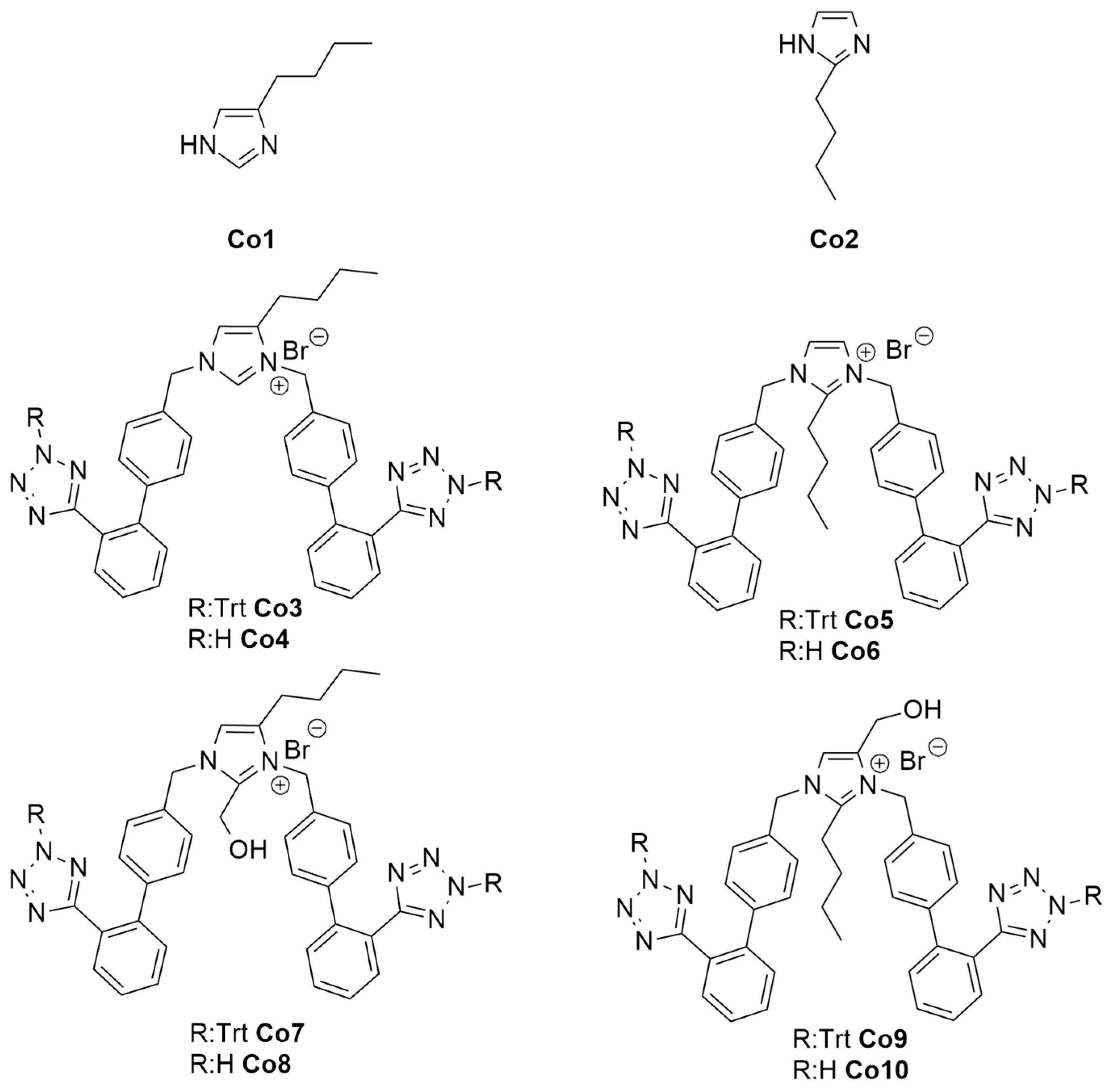
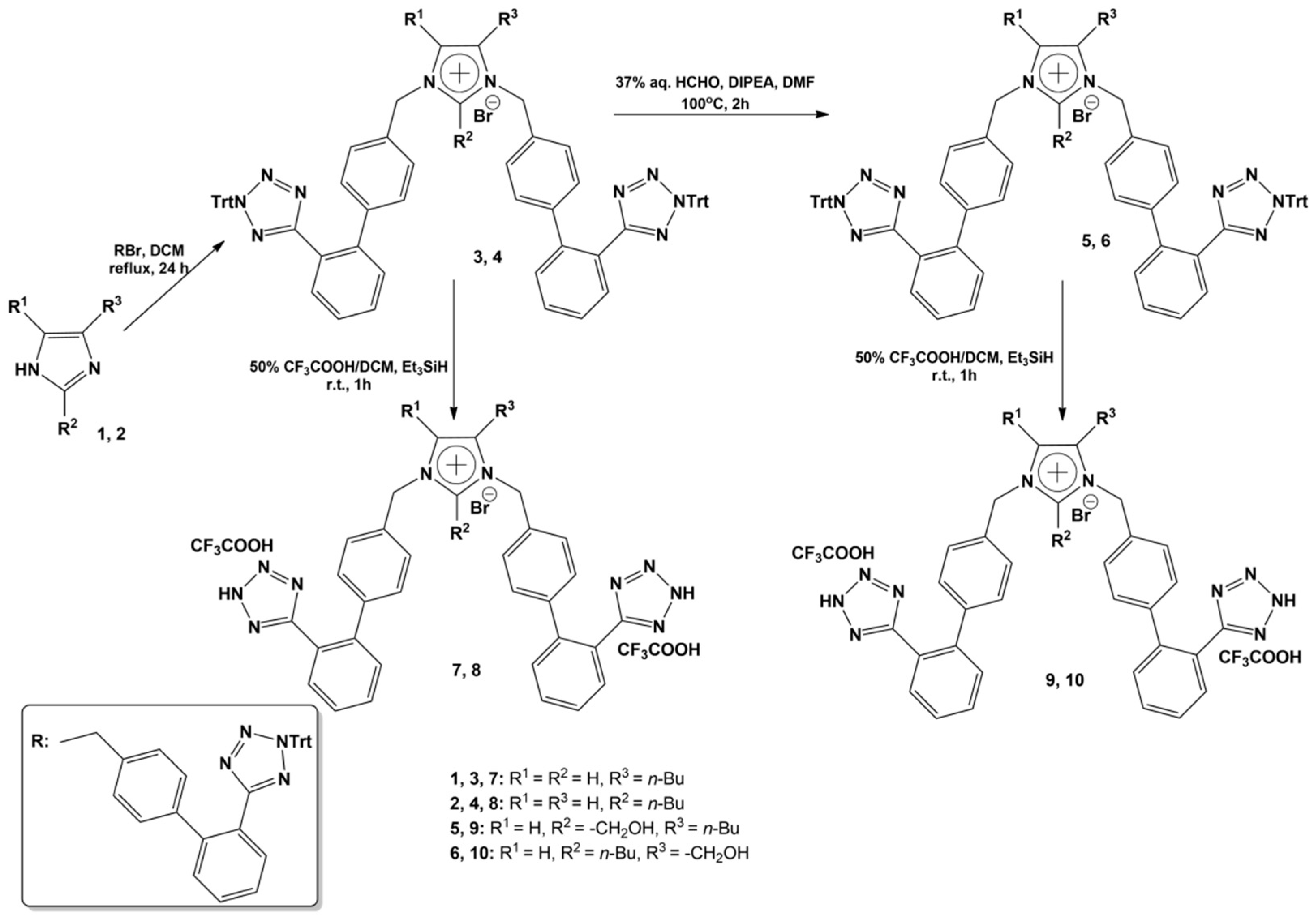
| Structures | Ligands | Best Scoring Pose Bound to ACE2-Spike RBD Complex (kcal/mol) | |
|---|---|---|---|
| 1 | Co1 | −4.73 | |
| 2 | Co2 | −4.87 | |
| 3 | Co3 | trityl protected | −9.4 |
| 4 | Co4 | free | −10.68 |
| 5 | Co5 | trityl protected | −9.71 |
| 6 | Co6 | free | −12.1 |
| 7 | Co7 | trityl protected | −6.81 |
| 8 | Co8 | free | −10.12 |
| 9 | Co9 | trityl protected | −6.8 |
| 10 | Co10 | free | −11.34 |
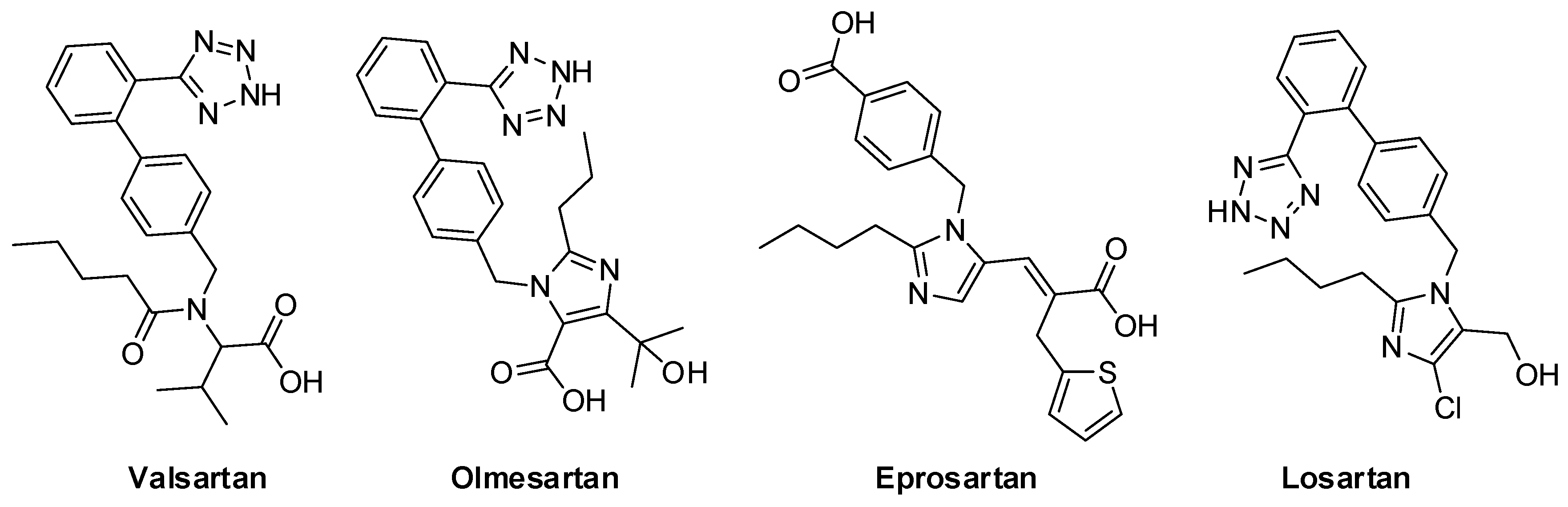
| 11 | valsartan | −8.53 | |
| 12 | olmesartan | −7.88 | |
| 13 | eprosartan | −6.98 | |
| 14 | losartan | −7.74 |
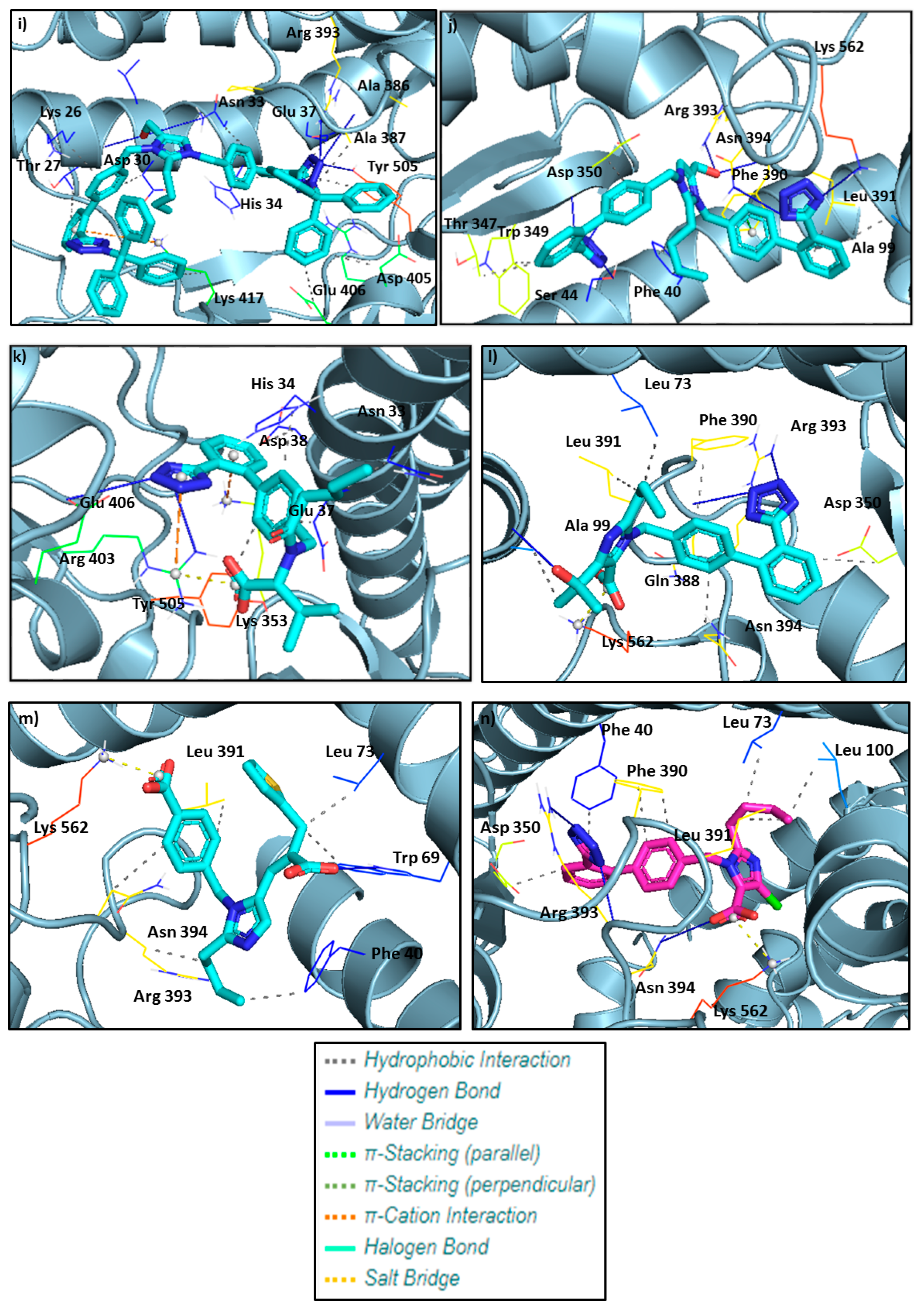
| Type of Interactions | Amino Acids of S-RBD Involved and Distance of Interactions (Å) | |||
|---|---|---|---|---|
| Valsartan | Olmesartan | Eprosartan | Losartan | |
| Hydrogen Bonds | 403B Arg 3.03 406B Glu 3.06 | 99A Ala 2.11 390A Phe 2.52 393A Arg 2.17 | 69A Trp 2.14 | 393A Arg 2.12 393A Arg 2.43 394A Asn 2.86 |
| Hydrophobic Interactions | 33A Asn 3.66 34A His 3.56 37A Glu 3.66 38A Asp 3.67 505B Tyr 3.20 | 73A Leu 3.11 99A Ala 3.79 350A Asp 3.67 390A Phe 3.59 391A Leu 3.34 394A Asn 3.61 | 0A Phe 3.23 69A Trp 3.28 73A Leu 3.25 391A Leu 3.34 393A Arg 3.18 394A Asn 3.85 | 40A Phe 3.90 73A Leu 3.48 100A Leu 3.55 350A Asp 3.33 390A Phe 3.33 390A Phe 3.83 391A Leu 3.30 |
| π–Cation Interactions | 353A Lys 3.89 403B Arg 4.26 | - | - | - |
| Salt Bridges | 403B Arg 3.35 | 562A Lys 2.68 | 562A Lys 2.74 | 562A Lys 3.19 |
| Type of Interactions | Amino Acids of S-RBD Involved and Distance of Interactions (Å) | |||||
|---|---|---|---|---|---|---|
| Co1 | Co2 | Co3 | Co5 | Co7 | Co9 | |
| Hydrogen Bonds | 34A His 2.13 38A Asp 3.57 353A Lys 3.04 494B Ser 2.05 496B Gly 2.72 | 353A Lys 2.20 453B Tyr 2.11 496B Gly 2.28 | 33A Asn 3.06 | 403B Arg 3.11 | 33A Asn 3.76 393A Arg 3.51 403B Arg 2.67 405B Asp 3.18 | 26A Lys 1.87 33A Asn 3.64 386A Ala 3.02 393A Arg 2.97 505B Tyr 3.26 |
| Hydrophobic Interactions | 497B Phe 3.41 497B Phe 3.68 505B Tyr 3.52 | 495B Tyr 3.10 497B Phe 3.42 497B Phe 3.57 505B Tyr 3.57 | 33A Asn 3.75 34A His 3.87 37A Glu 3.09 321A Pro 3.84 324A Thr 3.59 356A Phe 3.24 356A Phe 4.00 383A Met 3.75 386A Ala 3.04 387A Ala 3.27 389A Pro 3.26 417B Lys 3.39 505B Tyr 3.34 | 23A Glu 3.77 26A Lys 2.95 26A Lys 3.16 29A Leu 3.72 29A Leu 3.23 30A Asp 3.78 37A Glu 3.93 37A Glu 3.94 93A Val 3.20 405B Asp 3.63 406B Glu 3.61 420B Asp 3.76 456B Phe 3.98 505B Tyr 3.91 | 30A Asp 3.98 33A Asn 3.65 34A His 3.84 37A Glu 3.18 386A Ala 3.23 387A Ala 3.78 387A Ala 3.63 389A Pro 3.87 389A Pro 3.62 417B Lys 3.42 555A Phe 3.75 | 26A Lys 3.61 27A Thr 3.87 30A Asp 3.70 33A Asn 3.80 34A His 3.96 37A Glu 3.63 387A Ala 3.86 405B Asp 3.52 406B Glu 3.95 417B Lys 3.90 417B Lys 3.50 505B Tyr 3.81 |
| π–Stacking | 34A His 4.06 | 34A His 3.98 421B Tyr 5.30 | 34A His 3.90 | |||
| π–Cation Interactions | 353A Lys 3.86 | 34A His 5.61 403B Arg 3.89 403B Arg 5.91 | 403B Arg 5.21 417B Lys 5.49 | 408B Arg 5.55 | 417B Lys 5.39 | |
| Type of Interactions | Amino Acids of S-RBD Involved and Distance of Interactions (Å) | |||
|---|---|---|---|---|
| Co4 | Co6 | Co8 | Co10 | |
| Hydrogen Bonds | 44A Ser 1.92 44A Ser 3.19 350A Asp 2.45 394A Asn 2.74 562A Lys 2.13 | 37A Glu 3.16 353A Lys 3.07 353A Lys 3.26 354A Gly 2.85 403B Arg 3.01 496B Gly 2.23 505B Tyr 2.73 505B Tyr 1.84 | 34A His 3.05 388A Gln 1.99 393A Arg 2.14 403B Arg 2.64 406B Glu 2.49 559A Arg 2.32 | 44A Ser 3.58 350A Asp 2.49 350A Asp 2.14 390A Phe 2.22 393A ARg 2.82 394A Asn 2.79 562A Lys 2.28 |
| Hydrophobic Interactions | 73A Leu 3.28 347A Thr 3.95 349A Trp 3.20 349A Trp 3.89 350A Asp 3.25 391A Leu 3.17 | 34A His 3.52 38A Asp 3.52 356A Phe 3.53 386A Ala 3.41 405B Asp 3.71 406B Glu 3.74 453B Tyr 3.47 493B Gln 3.99 505B Tyr 3.40 | 33A Asn 3.38 37A Glu 3.35 389A Pro 3.45 | 40A Phe 3.22 40A Phe 3.56 99A Ala 3.44 347A Thr 3.65 349A Trp 3.70 350A Asp 3.18 391A Leu 3.13 |
| π–Stacking | 390A Phe 4.00 | 495B Tyr 5.21 505B Tyr 4.50 | 390A Phe 4.33 | |
| π–Cation Interactions | 34A His 4.65 353A Lys 3.23 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kelaidonis, K.; Ligielli, I.; Letsios, S.; Vidali, V.P.; Mavromoustakos, T.; Vassilaki, N.; Moore, G.J.; Hoffmann, W.; Węgrzyn, K.; Ridgway, H.; et al. Computational and Enzymatic Studies of Sartans in SARS-CoV-2 Spike RBD-ACE2 Binding: The Role of Tetrazole and Perspectives as Antihypertensive and COVID-19 Therapeutics. Int. J. Mol. Sci. 2023, 24, 8454. https://doi.org/10.3390/ijms24098454
Kelaidonis K, Ligielli I, Letsios S, Vidali VP, Mavromoustakos T, Vassilaki N, Moore GJ, Hoffmann W, Węgrzyn K, Ridgway H, et al. Computational and Enzymatic Studies of Sartans in SARS-CoV-2 Spike RBD-ACE2 Binding: The Role of Tetrazole and Perspectives as Antihypertensive and COVID-19 Therapeutics. International Journal of Molecular Sciences. 2023; 24(9):8454. https://doi.org/10.3390/ijms24098454
Chicago/Turabian StyleKelaidonis, Konstantinos, Irene Ligielli, Spiros Letsios, Veroniki P. Vidali, Thomas Mavromoustakos, Niki Vassilaki, Graham J. Moore, Weronika Hoffmann, Katarzyna Węgrzyn, Harry Ridgway, and et al. 2023. "Computational and Enzymatic Studies of Sartans in SARS-CoV-2 Spike RBD-ACE2 Binding: The Role of Tetrazole and Perspectives as Antihypertensive and COVID-19 Therapeutics" International Journal of Molecular Sciences 24, no. 9: 8454. https://doi.org/10.3390/ijms24098454
APA StyleKelaidonis, K., Ligielli, I., Letsios, S., Vidali, V. P., Mavromoustakos, T., Vassilaki, N., Moore, G. J., Hoffmann, W., Węgrzyn, K., Ridgway, H., Chasapis, C. T., & Matsoukas, J. M. (2023). Computational and Enzymatic Studies of Sartans in SARS-CoV-2 Spike RBD-ACE2 Binding: The Role of Tetrazole and Perspectives as Antihypertensive and COVID-19 Therapeutics. International Journal of Molecular Sciences, 24(9), 8454. https://doi.org/10.3390/ijms24098454







