Advances in Nucleic Acid Research: Exploring the Potential of Oligonucleotides for Therapeutic Applications and Biological Studies
Abstract
:1. Introduction
2. Oligonucleotides as Therapeutic Agents
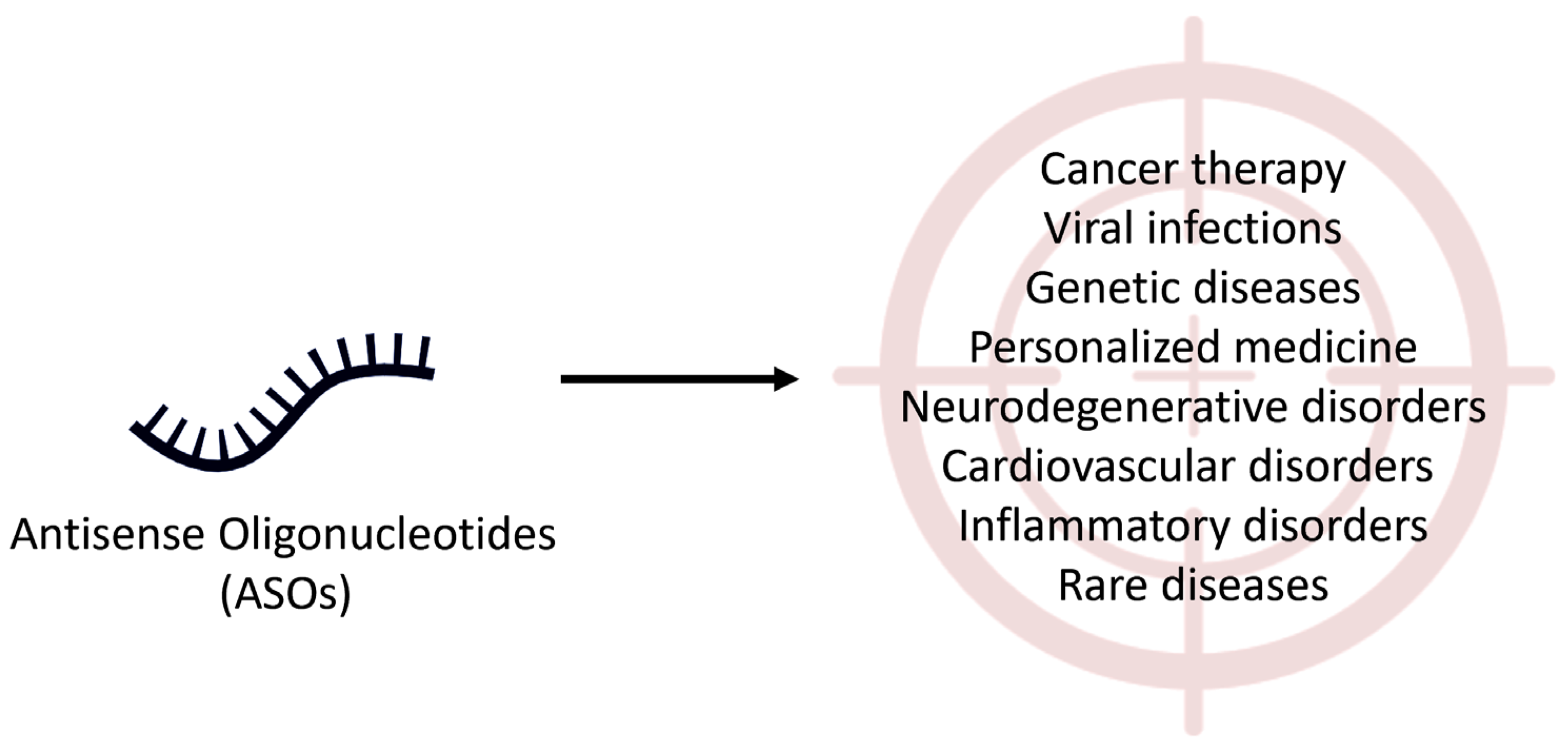
2.1. Therapeutic Applications of Antisense Oligonucleotides
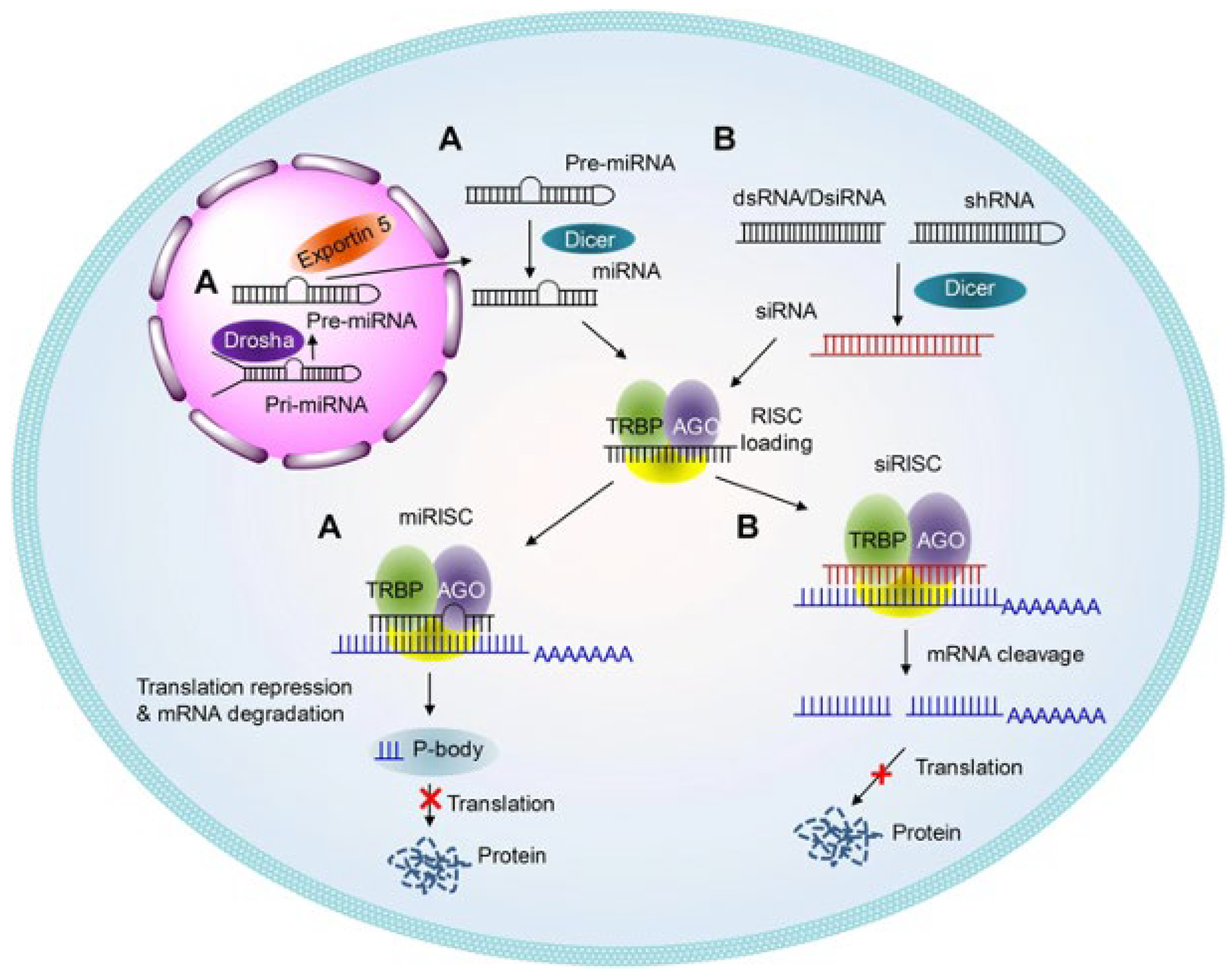
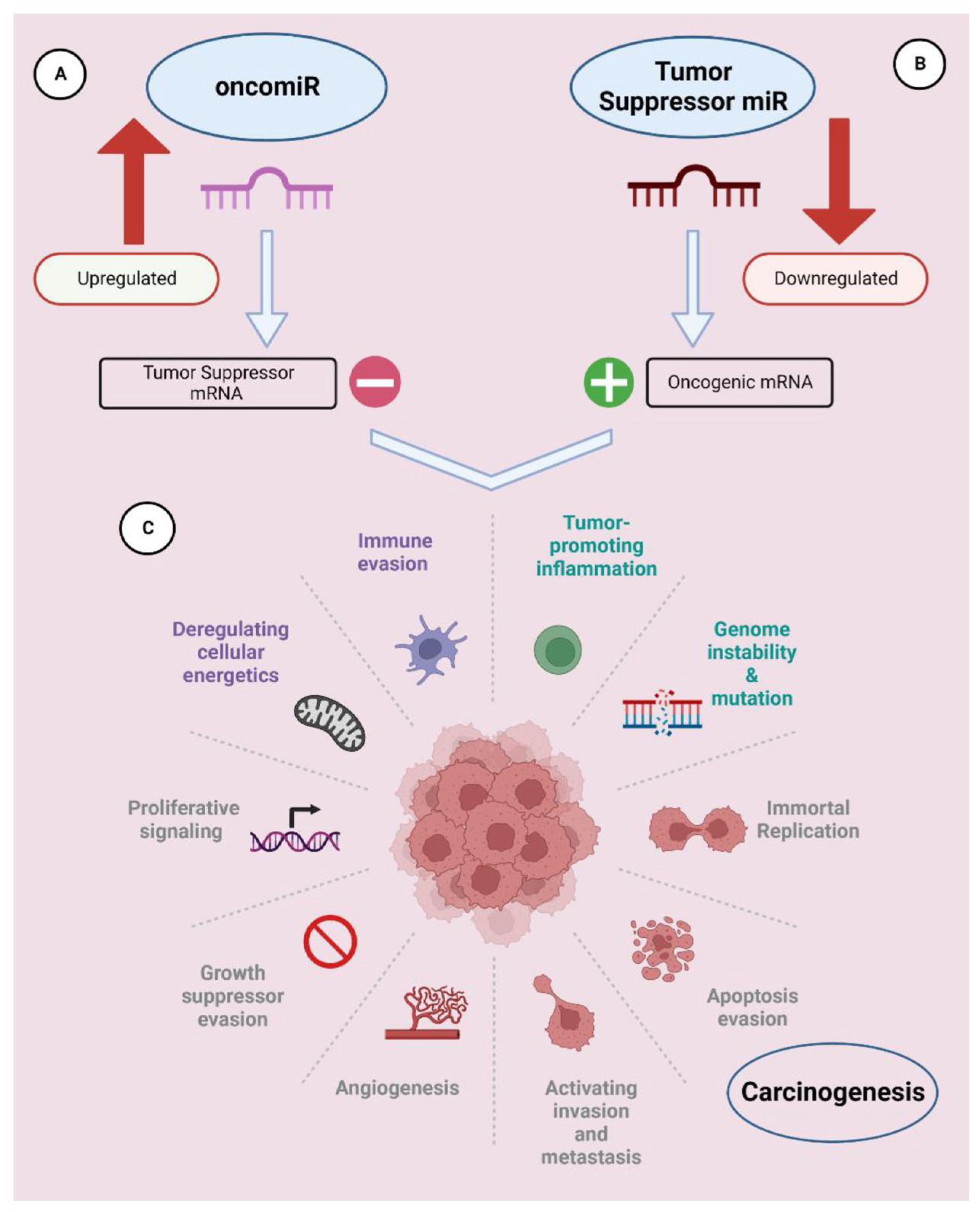
2.2. Therapeutic Applications of RNA Interference (RNAi)
3. Oligonucleotides as Biomimetic Models for Biological Studies
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Gubu, A.; Zhang, X.; Lu, A.; Zhang, B.; Ma, Y.; Zhang, G. Nucleic acid amphiphiles: Synthesis, properties and applications. Mol. Ther. Nucleic Acids 2023, 33, 144. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Li, P.; Chu, H.C.; Lo, P.K. Nucleic acids and their analogues for biomedical applications. Biosensors 2022, 12, 93. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.E.; Zain, R. Therapeutic oligonucleotides: State of the art. Annu. Rev. Pharmacol. Toxicol. 2019, 59, 605–630. [Google Scholar] [CrossRef] [PubMed]
- Roberts, T.C.; Langer, R.; Wood, M.J. Advances in oligonucleotide drug delivery. Nat. Rev. Drug Discov. 2020, 19, 673–694. [Google Scholar] [CrossRef] [PubMed]
- Thakur, S.; Sinhari, A.; Jain, P.; Jadhav, H.R. A perspective on oligonucleotide therapy: Approaches to patient customization. Front. Pharmacol. 2022, 13, 1006304. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Veroniaina, H.; Su, N.; Sha, K.; Jiang, F.; Wu, Z.; Qi, X. Applications and developments of gene therapy drug delivery systems for genetic diseases. Asian J. Pharm. Sci. 2021, 16, 687–703. [Google Scholar] [CrossRef]
- Kulkarni, J.A.; Witzigmann, D.; Thomson, S.B.; Chen, S.; Leavitt, B.R.; Cullis, P.R.; van der Meel, R. The current landscape of nucleic acid therapeutics. Nat. Nanotechnol. 2021, 16, 630–643. [Google Scholar] [CrossRef]
- Lares, M.R.; Rossi, J.J.; Ouellet, D.L. RNAi and small interfering RNAs in human disease therapeutic applications. Trends Biotechnol. 2010, 28, 570–579. [Google Scholar] [CrossRef]
- Hu, B.; Zhong, L.; Weng, Y.; Peng, L.; Huang, Y.; Zhao, Y.; Liang, X.J. Therapeutic siRNA: State of the art. Signal Transduct. Target. Ther. 2020, 5, 101. [Google Scholar] [CrossRef]
- Offord, C.; Cohen, J. Award honors pair for mRNA work key to COVID-19 vaccines. Science 2023, 382, 22. [Google Scholar] [CrossRef]
- Baghban, R.; Ghasemian, A.; Mahmoodi, S. Nucleic acid-based vaccine platforms against the coronavirus disease 19 (COVID-19). Arch. Microbiol. 2023, 205, 150. [Google Scholar] [CrossRef] [PubMed]
- Ning, Y.; Hu, J.; Lu, F. Aptamers used for biosensors and targeted therapy. Biomed. Pharmacother. 2020, 132, 110902. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Juhas, M.; Kwok, C.K. Aptamers targeting SARS-CoV-2: A promising tool to fight against COVID-19. Trends Biotechnol. 2023, 41, 528–544. [Google Scholar] [CrossRef]
- Weaver, S.; Mohammadi, M.H.; Nakatsuka, N. Aptamer-functionalized capacitive biosensors. Biosens. Bioelectron. 2023, 224, 115014. [Google Scholar] [CrossRef] [PubMed]
- Guan, C.; Zhu, X.; Feng, C. DNA nanodevice-based drug delivery systems. Biomolecules 2021, 11, 1855. [Google Scholar] [CrossRef] [PubMed]
- Nsairat, H.; AlShaer, W.; Odeh, F.; Essawi, E.; Khater, D.; Al Bawab, A.; El-Tanani, M.; Awidi, A.; Mubarak, M.S. Recent advances in using liposomes for delivery of nucleic acid-based therapeutics. OpenNano 2023, 11, 100132. [Google Scholar] [CrossRef]
- Benizri, S.; Gissot, A.; Martin, A.; Vialet, B.; Grinstaff, M.W.; Barthélémy, P. Bioconjugated oligonucleotides: Recent developments and therapeutic applications. Bioconjug. Chem. 2019, 30, 366–383. [Google Scholar] [CrossRef]
- Rinaldi, C.; Wood, M.J. Antisense oligonucleotides: The next frontier for treatment of neurological disorders. Nat. Rev. Neurol. 2018, 14, 9–21. [Google Scholar] [CrossRef]
- Egli, M.; Manoharan, M. Chemistry, structure and function of approved oligonucleotide therapeutics. Nucleic Acids Res. 2023, 51, 2529–2573. [Google Scholar] [CrossRef]
- Pabon-Martinez, Y.V.; Xu, Y.; Villa, A.; Lundin, K.E.; Geny, S.; Nguyen, C.H.; Pedersen, E.B.; Jørgensen, P.T.; Wengel, J.; Nilsson, L.; et al. LNA effects on DNA binding and conformation: From single strand to duplex and triplex structures. Sci. Rep. 2017, 7, 11043. [Google Scholar] [CrossRef]
- Winkle, M.; El-Daly, S.M.; Fabbri, M.; Calin, G.A. Noncoding RNA therapeutics—Challenges and potential solutions. Nat. Rev. Drug Discov. 2021, 20, 629–651. [Google Scholar] [CrossRef] [PubMed]
- Korkmaz, İ.; Taşkan, G.; Arslan, S. RNA-Based therapeutic oligonucleotide strategies. Adv. Eng. Inform. (AED) 2022, 5, 133–136. [Google Scholar]
- Kara, G.; Calin, G.A.; Ozpolat, B. RNAi-based therapeutics and tumor targeted delivery in cancer. Adv. Drug Deliv. Rev. 2022, 182, 114113. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, P.E.; Egholm, M.; Berg, R.H.; Buchardt, O. Sequence selective recognition of DNA by strand displacement with a thymine substituted polyamide. Science 1991, 254, 1497–1500. [Google Scholar] [CrossRef] [PubMed]
- Perera, J.D.R.; Carufe, K.E.; Glazer, P.M. Peptide nucleic acids and their role in gene regulation and editing. Biopolymers 2021, 112, e23460. [Google Scholar] [CrossRef] [PubMed]
- Montazersaheb, S.; Hejazi, M.S.; Charoudeh, H.N. Potential of peptide nucleic acids in future therapeutic applications. Adv. Pharm. Bull. 2018, 8, 551. [Google Scholar] [CrossRef] [PubMed]
- Demidov, V.V.; Potaman, V.N.; Frank-Kamenetskil, M.D.; Egholm, M.; Buchard, O.; Sönnichsen, S.H.; Nlelsen, P.E. Stability of peptide nucleic acids in human serum and cellular extracts. Biochem. Pharmacol. 1994, 48, 1310–1313. [Google Scholar] [CrossRef]
- Gupta, A.; Mishra, A.; Puri, N. Peptide nucleic acids: Advanced tools for biomedical applications. J. Biotechnol. 2017, 259, 148–159. [Google Scholar] [CrossRef]
- Moccia, M.; Antonacci, A.; Saviano, M.; Caratelli, V.; Arduini, F.; Scognamiglio, V. Emerging technologies in the design of peptide nucleic acids (PNAs) based biosensors. TrAC Trends Anal. Chem. 2020, 132, 116062. [Google Scholar] [CrossRef]
- Lima, J.F.; Cerqueira, L.; Figueiredo, C.; Oliveira, C.; Azevedo, N.F. Anti-miRNA oligonucleotides: A comprehensive guide for design. RNA Biol. 2018, 15, 338–352. [Google Scholar] [CrossRef]
- Zarrilli, F.; Amato, F.; Morgillo, C.M.; Pinto, B.; Santarpia, G.; Borbone, N.; D’Errico, S.; Catalanotti, B.; Piccialli, G.; Castaldo, G.; et al. Peptide nucleic acids as miRNA target protectors for the treatment of cystic fibrosis. Molecules 2017, 22, 1144. [Google Scholar] [CrossRef] [PubMed]
- Herdewijn, P. Oligonucleotide Synthesis: Methods and Applications; Springer Science & Business Media: Cham, Switzerland, 2008; Volume 288, 431p. [Google Scholar]
- Karwowski, B.T. FapydG in the Shadow of OXOdG—A Theoretical Study of Clustered DNA Lesions. Int. J. Mol. Sci. 2023, 24, 5361. [Google Scholar] [CrossRef] [PubMed]
- Brooks, P.J. The cyclopurine deoxynucleosides: DNA repair, biological effects, mechanistic insights, and unanswered questions. Free Radic. Biol. Med. 2017, 107, 90–100. [Google Scholar] [CrossRef] [PubMed]
- Chatgilialoglu, C.; Ferreri, C.; Geacintov, N.E.; Krokidis, M.G.; Liu, Y.; Masi, A.; Shafirovich, V.; Terzidis, M.A.; Tsegay, P.S. 5′,8-Cyclopurine Lesions in DNA Damage: Chemical, Analytical, Biological, and Diagnostic Significance. Cells 2019, 8, 513. [Google Scholar] [CrossRef] [PubMed]
- Chatgilialoglu, C.; Eriksson, L.A.; Krokidis, M.G.; Masi, A.; Wang, S.; Zhang, R. Oxygen dependent purine lesions in double-stranded oligodeoxynucleotides: Kinetic and computational studies highlight the mechanism for 5′,8-cyclopurine formation. J. Am. Chem. Soc. 2020, 142, 5825–5833. [Google Scholar] [CrossRef] [PubMed]
- Kropachev, K.; Ding, S.; Terzidis, M.A.; Masi, A.; Liu, Z.; Cai, Y.; Kolbanovskiy, M.; Chatgilialoglu, C.; Broyde, S.; Shafirovich, V. Structural basis for the recognition of diastereomeric 5′,8-cyclo-2′-deoxypurine lesions by the human nucleotide excision repair system. Nucleic Acids Res. 2014, 42, 5020–5032. [Google Scholar] [CrossRef] [PubMed]
- Platella, C.; Criscuolo, A.; Riccardi, C.; Gaglione, R.; Arciello, A.; Musumeci, D.; DellaGreca, M.; Montesarchio, D. Exploring the Binding of Natural Compounds to Cancer-Related G-Quadruplex Structures: From 9,10-Dihydrophenanthrenes to Their Dimeric and Glucoside Derivatives. Int. J. Mol. Sci. 2023, 24, 7765. [Google Scholar] [CrossRef] [PubMed]
- Fortini, P.; Pascucci, B.; Parlanti, E.; Sobol, R.W.; Wilson, S.H.; Dogliotti, E. Different DNA polymerases are involved in the short- and long-patch base excision repair in mammalian cells. Biochemistry 1998, 11, 3575–3580. [Google Scholar] [CrossRef]
- Stucki, M.; Pascucci, B.; Parlanti, E.; Fortini, P.; Wilson, S.H.; Hubsher, U.; Dogliotti, E. Mammalian base excision repair by DNA polymerases delta and epsilon. Oncogene 1998, 17, 835–843. [Google Scholar] [CrossRef]
- Gopi, C.; Dhanaraju, M.D.; Dhanaraju, K. Antisense oligonucleotides: Recent progress in the treatment of various diseases. Beni-Suef Univ. J. Basic Appl. Sci. 2022, 11, 19. [Google Scholar] [CrossRef]
- Dhuri, K.; Bechtold, C.; Quijano, E.; Pham, H.; Gupta, A.; Vikram, A.; Bahal, R. Antisense oligonucleotides: An emerging area in drug discovery and development. J. Clin. Med. 2020, 9, 2004. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Lee, J.Y.; Woo, J.Z.; Xu, L.; Nguyenla, X.; Yamashiro, L.H.; Ji, F.; Biering, S.B.; Van Dis, E.; Gonzalez, F.; et al. An intranasal ASO therapeutic targeting SARS-CoV-2. Nat. Commun. 2022, 13, 4503. [Google Scholar] [CrossRef] [PubMed]
- Langner, H.K.; Jastrzebska, K.; Caruthers, M.H. Synthesis and characterization of thiophosphoramidate morpholino oligonucleotides and chimeras. J. Am. Chem. Soc. 2020, 142, 16240–16253. [Google Scholar] [CrossRef] [PubMed]
- Hanvey, J.C.; Peffer, N.J.; Bisi, J.E.; Thomson, S.A.; Cadilla, R.; Josey, J.A.; Ricca, D.J.; Hassman, C.F.; Bonham, M.A.; Au, K.G.; et al. Antisense and antigene properties of peptide nucleic acids. Science 1992, 258, 1481–1485. [Google Scholar] [CrossRef] [PubMed]
- Swayze, E.E.; Siwkowski, A.M.; Wancewicz, E.V.; Migawa, M.T.; Wyrzykiewicz, T.K.; Hung, G.; Monia, B.P.; Bennett, A.C.F. Antisense oligonucleotides containing locked nucleic acid improve potency but cause significant hepatotoxicity in animals. Nucleic Acids Res. 2007, 35, 687–700. [Google Scholar] [CrossRef] [PubMed]
- Crooke, S.T. Molecular mechanisms of antisense oligonucleotides. Nucleic Acid Ther. 2017, 27, 70–77. [Google Scholar] [CrossRef] [PubMed]
- Finkel, R.S.; Chiriboga, C.A.; Vajsar, J.; Day, J.W.; Montes, J.; De Vivo, D.C.; Bishop, K.M.; Foster, R.; Liu, Y.; Ramirez-Schrempp, D.; et al. Treatment of infantile-onset spinal muscular atrophy with nusinersen: A phase 2, open-label, dose-escalation study. Lancet 2016, 388, 3017–3026. [Google Scholar] [CrossRef] [PubMed]
- Panda, S.; Chatterjee, O.; Chatterjee, S. Nucleic Acid-Based Strategies to Treat Neurodegenerative Diseases. In Nucleic Acid Biology and its Application in Human Diseases; Chatterjee, S., Chattopadhyay, S., Eds.; Springer Nature: Singapore, 2023; pp. 105–133. [Google Scholar]
- Amulya, E.; Sikder, A.; Vambhurkar, G.; Shah, S.; Khatri, D.K.; Raghuvanshi, R.S.; Singh, S.B.; Srivastava, S. Nanomedicine based strategies for oligonucleotide traversion across the blood–brain barrier. J. Control. Release 2023, 354, 554–571. [Google Scholar] [CrossRef]
- Gupta, R.; Salave, S.; Rana, D.; Karunakaran, B.; Butreddy, A.; Benival, D.; Kommineni, N. Versatility of Liposomes for Antisense Oligonucleotide Delivery: A Special Focus on Various Therapeutic Areas. Pharmaceutics 2023, 15, 1435. [Google Scholar] [CrossRef]
- Pollak, A.J.; Zhao, L.; Crooke, S.T. Systematic Analysis of Chemical Modifications of Phosphorothioate Antisense Oligonucleotides that Modulate Their Innate Immune Response. Nucleic Acid Ther. 2023, 33, 95–107. [Google Scholar] [CrossRef]
- Boros, B.D.; Schoch, K.M.; Kreple, C.J.; Miller, T.M. Antisense Oligonucleotides for the Study and Treatment of ALS. Neurotherapeutics 2022, 19, 1145–1158. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Hu, Y.; Ju, D. Gene therapy for neurodegenerative disorders: Advances, insights and prospects. Acta Pharm. Sin. B 2020, 10, 1347–1359. [Google Scholar] [CrossRef] [PubMed]
- Bennett, C.F.; Krainer, A.R.; Cleveland, D.W. Antisense oligonucleotide therapies for neurodegenerative diseases. Annu. Rev. Neurosci. 2019, 42, 385–406. [Google Scholar] [CrossRef] [PubMed]
- Evers, M.M.; Pepers, B.A.; van Deutekom, J.C.; Mulders, S.A.; den Dunnen, J.T.; Aartsma-Rus, A.; van Ommen, G.J.B.; van Roon-Mom, W.M. Targeting several CAG expansion diseases by a single antisense oligonucleotide. PLoS ONE 2011, 6, e24308. [Google Scholar] [CrossRef] [PubMed]
- Rook, M.E.; Southwell, A.L. Antisense oligonucleotide therapy: From design to the Huntington disease clinic. BioDrugs 2022, 36, 105–119. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, F.B.; Wild, E.J. Huntington’s disease clinical trials corner: February 2018. J. Huntington’s Dis. 2018, 7, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Murphy, M.P. Amyloid-beta solubility in the treatment of Alzheimer’s disease. N. Engl. J. Med. 2018, 378, 391–392. [Google Scholar] [CrossRef] [PubMed]
- DeVos, S.L.; Goncharoff, D.K.; Chen, G.; Kebodeaux, C.S.; Yamada, K.; Stewart, F.R.; Schuler, D.R.; Maloney, S.E.; Wozniak, D.F.; Rigo, F.; et al. Antisense reduction of tau in adult mice protects against seizures. J. Neurosci. 2013, 33, 12887–12897. [Google Scholar] [CrossRef]
- O’Neill, C.P.; Dwyer, R.M. Nanoparticle-based delivery of tumor suppressor microRNA for cancer therapy. Cells 2020, 9, 521. [Google Scholar] [CrossRef]
- Ciccone, G.; Ibba, M.L.; Coppola, G.; Catuogno, S.; Esposito, C.L. The Small RNA Landscape in NSCLC: Current Therapeutic Applications and Progresses. Int. J. Mol. Sci. 2023, 24, 6121. [Google Scholar] [CrossRef]
- Di Fusco, D.; Dinallo, V.; Marafini, I.; Figliuzzi, M.M.; Romano, B.; Monteleone, G. Antisense oligonucleotide: Basic concepts and therapeutic application in inflammatory bowel disease. Front. Pharmacol. 2019, 10, 305. [Google Scholar] [CrossRef] [PubMed]
- Karras, J.G.; Crosby, J.R.; Guha, M.; Tung, D.; Miller, D.A.; Gaarde, W.A.; Geary, R.S.; Monia, B.P.; Gregory, S.A. Anti-inflammatory activity of inhaled IL-4 receptor-α antisense oligonucleotide in mice. Am. J. Respir. Cell Mol. Biol. 2007, 36, 276–285. [Google Scholar] [CrossRef]
- Zorzi, F.; Angelucci, E.; Sedda, S.; Pallone, F.; Monteleone, G. Smad7 antisense oligonucleotide-based therapy for inflammatory bowel diseases. Dig. Liver Dis. 2013, 45, 552–555. [Google Scholar] [CrossRef] [PubMed]
- Pelisch, N.; Almanza, J.R.; Stehlik, K.E.; Aperi, B.V.; Kroner, A. Use of a self-delivering Anti-CCL3 FANA Oligonucleotide as an innovative approach to target inflammation after Spinal Cord Injury. Eneuro 2021, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Lam, J.K.; Chow, M.Y.; Zhang, Y.; Leung, S.W. siRNA versus miRNA as therapeutics for gene silencing. Mol. Ther. Nucleic Acids 2015, 4, e252. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Jing, Y.; Zhang, H.; Niu, Y.; Liu, C.; Wang, J.; Zen, K.; Zhang, C.Y.; Li, D. Comprehensive evolutionary analysis of the major RNA-induced silencing complex members. Sci. Rep. 2018, 8, 14189. [Google Scholar] [CrossRef]
- Dana, H.; Chalbatani, G.M.; Mahmoodzadeh, H.; Karimloo, R.; Rezaiean, O.; Moradzadeh, A.; Mehmandoost, N.; Moazzen, F.; Mazraeh, A.; Marmari, V.; et al. Molecular mechanisms and biological functions of siRNA. Int. J. Biomed. Sci. 2017, 13, 48. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- Chaudhary, A.; Srivastava, S.; Garg, S. Development of a software tool and criteria evaluation for efficient design of small interfering RNA. Biochem. Biophys. Res. Commun. 2011, 404, 313–320. [Google Scholar] [CrossRef]
- Zhong, R.; Kim, J.; Kim, H.S.; Kim, M.; Lum, L.; Levine, B.; Xiao, G.; White, M.A.; Xie, Y. Computational detection and suppression of sequence-specific off-target phenotypes from whole genome RNAi screens. Nucleic Acids Res. 2014, 42, 8214–8222. [Google Scholar] [CrossRef]
- Fedorov, Y.; Anderson, E.M.; Birmingham, A.; Reynolds, A.; Karpilow, J.; Robinson, K.; Leake, D.; Marshall, W.S.; Khvorova, A. Off-target effects by siRNA can induce toxic phenotype. RNA 2006, 12, 1188–1196. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, C.; Sharma, A.R.; Sharma, G.; Doss, C.G.P.; Lee, S.S. Therapeutic miRNA and siRNA: Moving from bench to clinic as next generation medicine. Mol. Ther. Nucleic Acids 2017, 8, 132–143. [Google Scholar] [CrossRef] [PubMed]
- Habtemariam, B.A.; Karsten, V.; Attarwala, H.; Goel, V.; Melch, M.; Clausen, V.A.; Garg, P.; Vaishnaw, A.K.; Sweetser, M.T.; Robbie, G.J.; et al. Single-Dose Pharmacokinetics and Pharmacodynamics of Transthyretin Targeting N-acetylgalactosamine-Small Interfering Ribonucleic Acid Conjugate, Vutrisiran, in Healthy Subjects. Clin. Pharmacol. Ther. 2021, 109, 372–382. [Google Scholar] [CrossRef] [PubMed]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef] [PubMed]
- Chakrabortty, A.; Patton, D.J.; Smith, B.F.; Agarwal, P. miRNAs: Potential as Biomarkers and Therapeutic Targets for Cancer. Genes 2023, 14, 1375. [Google Scholar] [CrossRef] [PubMed]
- Fabbri, M.; Paone, A.; Calore, F.; Galli, R.; Croce, C.M. A new role for microRNAs, as ligands of Toll-like receptors. RNA Biol. 2013, 10, 169–174. [Google Scholar] [CrossRef] [PubMed]
- Austin, T.B.M. First microRNA mimic enters clinic. Nat. Biotechnol. 2013, 31, 577. [Google Scholar]
- Bader, A.G. miR-34–a microRNA replacement therapy is headed to the clinic. Front. Genet. 2012, 3, 120. [Google Scholar] [CrossRef]
- Aqeilan, R.I.; Calin, G.A.; Croce, C.M. miR-15a and miR-16-1 in cancer: Discovery, function and future perspectives. Cell Death Differ. 2010, 17, 215–220. [Google Scholar] [CrossRef]
- Hayes, J.; Peruzzi, P.P.; Lawler, S. MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol. Med. 2014, 20, 460–469. [Google Scholar] [CrossRef]
- Pereira, D.M.; Rodrigues, P.M.; Borralho, P.M.; Rodrigues, C.M. Delivering the promise of miRNA cancer therapeutics. Drug Discov. Today 2013, 18, 282–289. [Google Scholar] [CrossRef] [PubMed]
- Jackson, A.L.; Bartz, S.R.; Schelter, J.; Kobayashi, S.V.; Burchard, J.; Mao, M.; Li, B.; Cavet, G.; Linsley, P.S. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 2003, 21, 635–637. [Google Scholar] [CrossRef] [PubMed]
- Chorn, G.; Klein-McDowell, M.; Zhao, L.; Saunders, M.A.; Flanagan, W.M.; Willingham, A.T.; Lim, L.P. Single-stranded microRNA mimics. RNA 2012, 10, 1796–1804. [Google Scholar] [CrossRef] [PubMed]
- Ferino, A.; Miglietta, G.; Picco, R.; Vogel, S.; Wengel, J.; Xodo, L.E. MicroRNA therapeutics: Design of single-stranded miR-216b mimics to target KRAS in pancreatic cancer cells. RNA Biol. 2018, 15, 1273–1285. [Google Scholar] [CrossRef] [PubMed]
- Piacenti, V.; Langella, E.; Autiero, I.; Nolan, J.C.; Piskareva, O.; Adamo, M.F.; Saviano, M.; Moccia, M. A combined experimental and computational study on peptide nucleic acid (PNA) analogues of tumor suppressive miRNA-34a. Bioorg. Chem. 2019, 91, 103165. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.S.; Song, Y.K.; Durinck, S.; Chen, Q.R.; Cheuk, A.T.C.; Tsang, P.; Zhang, Q.; Thiele, C.J.; Slack, A.; Shohet, J.; et al. The MYCN oncogene is a direct target of miR-34a. Oncogene 2008, 27, 5204–5213. [Google Scholar] [CrossRef] [PubMed]
- Moccia, M.; Mercurio, F.A.; Langella, E.; Piacenti, V.; Leone, M.; Adamo, M.F.; Saviano, M. Structural insights on tiny peptide nucleic acid (PNA) analogues of miRNA-34a: An in silico and experimental integrated approach. Front. Chem. 2020, 8, 568575. [Google Scholar] [CrossRef]
- Dhuri, K.; Gaddam, R.R.; Vikram, A.; Slack, F.J.; Bahal, R. Therapeutic potential of chemically modified synthetic, triplex peptide nucleic acid–based oncomiR inhibitors for cancer therapy. Cancer Res. 2021, 81, 5613–5624. [Google Scholar] [CrossRef]
- Malik, S.; Lim, J.; Slack, F.J.; Braddock, D.T.; Bahal, R. Next generation miRNA inhibition using short anti-seed PNAs encapsulated in PLGA nanoparticles. J. Control. Release 2020, 327, 406–419. [Google Scholar] [CrossRef]
- Wahane, A.; Malik, S.; Shih, K.C.; Gaddam, R.R.; Chen, C.; Liu, Y.; Nieh, M.P.; Vikram, A.; Bahal, R. Dual-modality poly-L-histidine nanoparticles to deliver peptide nucleic acids and paclitaxel for in vivo cancer therapy. ACS Appl. Mater. Interfaces 2021, 13, 45244–45258. [Google Scholar] [CrossRef]
- Malik, S.; Kumar, V.; Liu, C.H.; Shih, K.C.; Krueger, S.; Nieh, M.P.; Bahal, R. Head on Comparison of Self-and Nano-Assemblies of Gamma Peptide Nucleic Acid Amphiphiles. Adv. Funct. Mater. 2022, 32, 2109552. [Google Scholar] [CrossRef] [PubMed]
- Dhuri, K.; Pradeep, S.P.; Shi, J.; Anastasiadou, E.; Slack, F.J.; Gupta, A.; Zhong, X.B.; Bahal, R. Simultaneous targeting of multiple oncomiRs with phosphorothioate or PNA-based anti-miRs in lymphoma cell lines. Pharm. Res. 2022, 39, 2709–2720. [Google Scholar] [CrossRef]
- Milani, R.; Brognara, E.; Fabbri, E.; Manicardi, A.; Corradini, R.; Finotti, A.; Gasparello, J.; Borgatti, M.; Cosenza, L.C.; Lampronti, I.; et al. Targeting miR-155-5p and miR-221-3p by peptide nucleic acids induces caspase-3 activation and apoptosis in temozolomide-resistant T98G glioma cells. Int. J. Oncol. 2019, 55, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Malik, S.; Suh, H.W.; Xiao, Y.; Deng, Y.; Fan, R.; Huttner, A.; Bindra, R.S.; Singh, V.; Saltzman, W.M.; et al. Anti-seed PNAs targeting multiple oncomiRs for brain tumor therapy. Sci. Adv. 2023, 9, eabq7459. [Google Scholar] [CrossRef] [PubMed]
- Kubota, Y.; Nash, R.; Klungland, A.; Schar, P.; Barnes, D.; Lindahl, T. Reconstitution of DNA base excision-repair with purified human proteins: Interaction between DNA polymerase beta and the XRCC1 protein. EMBO J. 1996, 15, 6662–6670. [Google Scholar] [CrossRef] [PubMed]
- Pascucci, B.; Stucki, M.; Jònsson, Z.; Dogliotti, E.; Ulrich Hubscher, U. Long patch base excision repair with purified human proteins: DNA ligase I as patch size mediator for DNA polymerase δ and ε. J. Biol. Chem. 1999, 274, 33696–33702. [Google Scholar] [CrossRef] [PubMed]
- Pascucci, B.; Maga, G.; Hubscher, U.; Bjoras, M.; Seeberg, E.; Hickson, I.D.; Villani, G.; Giordano, C.; Cellai, L.; Dogliotti, E. Reconstitution of the base excision repair pathway for 7,8-dihydro-8-oxoguanine with purified human proteins. Nucleic Acids Res. 2002, 30, 2124–2130. [Google Scholar] [CrossRef] [PubMed]
- Chatgilialoglu, C.; Ferreri, C.; Terzidis, M.A. Purine 5′,8-cyclonucleoside lesions: Chemistry and biology. Chem. Soc. Rev. 2011, 40, 1368–1382. [Google Scholar] [CrossRef]
- Yuan, B.; Wang, J.; Cao, H.; Sun, R.; Wang, Y. High throughput analysis of the mutagenic and cytotoxic properties of DNA lesions by next-generation sequencing. Nucleic Acids Res. 2011, 39, 5945–5954. [Google Scholar] [CrossRef]
- You, C.; Dai, X.; Yuan, B.; Wang, J.; Wang, J.; Brooks, P.J.; Niedernhofer, L.J.; Wang, Y. A quantitative assay for assessing the effects of DNA lesions on transcription. Nat. Chem. Biol. 2012, 8, 817–822. [Google Scholar] [CrossRef]
- Dizdaroglu, M. Free-radical-induced formation of an 8,5′-cyclo-2′-deoxyguanosine moiety in deoxyribonucleic acid. Biochem. J. 1986, 238, 247–254. [Google Scholar] [CrossRef] [PubMed]
- Dirksen, M.L.; Blakely, W.F.; Holwitt, E.; Dizdaroglu, M. Effect of DNA conformation on the hydroxyl radical-induced formation of 8,5′-cyclopurine 2′-deoxyribonucleoside residues in DNA. Int. J. Radiat. Biol. 1988, 54, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Kuraoka, I.; Bender, C.; Romieu, A.; Cadet, J.; Wood, R.D.; Lindahl, T. DNA by the nucleotide excision-repair pathway in human cells. Proc. Natl. Acad. Sci. USA 2000, 97, 3832–3837. [Google Scholar] [CrossRef] [PubMed]
- Chatgilialoglu, C.; Ferreri, C.; Krokidis, M.G.; Masi, A.; Terzidis, M.A. On the relevance of hydroxyl radical to purine DNA damage. Free Radic. Res. 2021, 55, 384–404. [Google Scholar]
- Jaruga, P.; Dizdaroglu, M. 8,5′-Cyclopurine-2′-Deoxynucleosides in DNA: Mechanisms of Formation, Measurement, Repair and Biological Effects. DNA Repair 2008, 7, 1413–1425. [Google Scholar] [CrossRef]
- Chatgilialoglu, C.; Krokidis, M.G.; Masi, A.; Barata-Vallejo, S.; Ferreri, C.; Terzidis, M.A.; Szreder, T.; Bobrowski, K. New Insights into the Reaction Paths of Hydroxyl Radicals with Purine Moieties in DNA and Double-Stranded Oligodeoxynucleotides. Molecules 2019, 24, 3860. [Google Scholar] [CrossRef]
- Boussicault, F.; Kaloudis, P.; Caminal, C.; Mulazzani, Q.G.; Chatgilialoglu, C. The fate of C5′ radicals of purine nucleosides under oxidative conditions. J. Am. Chem. Soc. 2008, 130, 8377–8385. [Google Scholar] [CrossRef]
- Guerrero, C.R.; Wang, J.; Wang, Y. Induction of 8, 5′-cyclo-2′-deoxyadenosine and 8,5′-cyclo-2′-deoxyguanosine in isolated DNA by Fenton-type reagents. Chem. Res. Toxicol. 2013, 26, 1361–1366. [Google Scholar] [CrossRef]
- Chatgilialoglu, C.; Krokidis, M.G.; Masi, A.; Barata-Vallejo, S.; Ferreri, C.; Pascucci, B.; D’Errico, M. Assessing the Formation of Purine Lesions in Mitochondrial DNA of Cockayne Syndrome Cells. Biomolecules 2022, 12, 1630. [Google Scholar] [CrossRef]
- Terzidis, M.A.; Prisecaru, A.; Molphy, Z.; Barron, N.; Randazzo, A.; Dumont, E.; Krokidis, M.G.; Kellett, A.; Chatgilialoglu, C. Radical-induced purine lesion formation is dependent on DNA helical topology. Free Radic. Res. 2016, 50, S91–S101. [Google Scholar]
- Chatgilialoglu, C.; Ferreri, C.; Masi, A.; Sansone, A.; Terzidis, M.A.; Tsakos, M. A problem solving approach for the diastereoselective synthesis of (5′S)- and (5′R)-5′,8-cyclopurine lesions. Org. Chem. Front. 2014, 1, 698–702. [Google Scholar] [CrossRef]
- Romieu, A.; Gasparutto, D.; Molko, D.; Cadet, J. Site-Specific Introduction of (5′S)-5′,8-Cyclo-2′-deoxyadenosine into oligodeoxyribonucleotides. J. Org. Chem. 1998, 63, 5245–5249. [Google Scholar] [CrossRef]
- Romieu, A.; Gasparutto, D.; Cadet, J. Synthesis and characterization of oligonucleotides containing 5′,8-cyclopurine 2′-deoxyribonucleosides: (5′R)-5′,8-cyclo-2′-deoxyadenosine, (5′S)-5′,8-cyclo-2′-deoxyguanosine, (5′R)-5′,8-cyclo-2′-deoxyguanosine. Chem. Res. Toxicol. 1999, 12, 412–421. [Google Scholar] [CrossRef] [PubMed]
- Zaliznyak, T.; Lukin, M.; de los Santos, C. Structure and stability of duplex DNA containing (5′S)-5′,8-cyclo-2′-deoxyadenosine: An oxidatively generated lesion repaired by NER. Chem. Res. Toxicol. 2012, 25, 2103–2111. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Das, R.S.; Basu, A.K.; Stone, M.P. Structure of (5′S)-8,5′-cyclo-2′-deoxyguanosine in DNA. J. Am. Chem. Soc. 2011, 133, 20357–20368. [Google Scholar] [CrossRef]
- Masi, A.; Sabbia, A.; Ferreri, C.; Manoli, F.; Lai, Y.; Laverde, E.; Liu, Y.; Krokidis, M.G.; Chatgilialoglu, C.; Faraone Mennella, M.R. Diastereomeric Recognition of 5’,8-cyclo-2’-Deoxyadenosine Lesions by Human Poly(ADP-ribose) Polymerase 1 in a Biomimetic Model. Cells 2019, 8, 116. [Google Scholar] [CrossRef]
- Jaruga, P.; Theruvathu, J.; Dizdaroglu, M.; Brooks, P.J. Complete release of (5′S)-8,5′-cyclo-2′-deoxyadenosine from dinucleotides, oligodeoxynucleotides, and DNA, and direct comparison of its levels in cellular DNA with other oxidatively induced DNA lesions. Nucleic Acids Res. 2004, 32, e87. [Google Scholar] [CrossRef]
- Terzidis, M.A.; Chatgilialoglu, C. An ameliorative protocol for the quantification of purine 5’,8-cyclo-2’-deoxynucleoside in oxidized DNA. Front. Chem. 2015, 3, 47. [Google Scholar] [CrossRef]
- Cai, Y.; Kropachev, K.; Terzidis, M.A.; Masi, A.; Chatgilialoglu, C.; Shafirovich, V.; Geacintov, N.E.; Broyde, S. Differences in the access of lesions to the nucleotide excision repair machinery in nucleosomes. Biochemistry 2015, 54, 4181–4185. [Google Scholar] [CrossRef]
- Nyaga, S.G.; Jaruga, P.; Lohani, A.; Dizdaroglu, M.; Evans, M.K. Accumulation of oxidatively induced DNA damage in human breast cancer cell lines following treatment with hydrogen peroxide. Cell Cycle 2007, 6, 1472–1478. [Google Scholar] [CrossRef]
- Krokidis, M.G.; Prasinou, P.; Efthimiadou, E.K.; Boari, A.; Ferreri, C.; Chatgilialoglu, C. Effects of Aging and Disease Conditions in Brain of Tumor-Bearing Mice: Evaluation of Purine DNA Damages and Fatty Acid Pool Changes. Biomolecules 2022, 12, 1075. [Google Scholar] [CrossRef] [PubMed]
- Krokidis, M.G.; D’Errico, M.; Pascucci, B.; Parlanti, E.; Masi, A.; Ferreri, C.; Chatgilialoglu, C. Oxygen-Dependent Accumulation of Purine DNA Lesions in Cockayne Syndrome Cells. Cells 2020, 9, 1671. [Google Scholar] [CrossRef] [PubMed]
- Masi, A.; Fortini, P.; Krokidis, M.G.; Romeo, E.F.; Bascietto, C.; De Angelis, P.; Guglielmi, V.; Chatgilialoglu, C. Increased Levels of 5’,8-Cyclopurine DNA Lesions in Inflammatory Bowel Diseases. Redox Biol. 2020, 34, 101562. [Google Scholar] [CrossRef] [PubMed]
- Krokidis, M.G.; Parlanti, E.; D’Errico, M.; Pascucci, B.; Pino, A.; Alimonti, A.; Pietraforte, D.; Masi, A.; Ferreri, C.; Chatgilialoglu, C. Purine DNA Lesions at Different Oxygen Concentration in DNA Repair-Impaired Human Cells (EUE-siXPA). Cells 2019, 8, 1377. [Google Scholar] [CrossRef] [PubMed]
- Kirkali, G.; de Souza-Pinto, N.C.; Jaruga, P.; Bohr, V.A.; Dizdaroglu, M. Accumulation of (5′S)-8,5′-cyclo-2′-deoxyadenosine in organs of Cockayne syndrome complementation group B gene knockout mice. DNA Repair 2009, 8, 274–278. [Google Scholar] [CrossRef] [PubMed]
- Sarmini, L.; Meabed, M.; Emmanouil, E.; Atsaves, G.; Robeska, E.; Karwowski, B.T.; Campalans, A.; Gimisis, T.; Khobta, A. Requirement of transcription-coupled nucleotide excision repair for the removal of a specific type of oxidatively induced DNA damage. Nucleic Acids Res. 2023, 51, 4982–4994. [Google Scholar] [CrossRef]
- Karwowski, B.T. The Influence of (5′R)- and (5′S)-5′,8-Cyclo-2′-Deoxyadenosine on UDG and hAPE1 activity. Tandem lesions are the base excision repair system’s nightmare. Cells 2019, 8, 1303. [Google Scholar] [CrossRef]
- Boguszewska, K.; Szewczuk, M.; Kaźmierczak-Barańska, J.; Karwowski, B.T. How (5′S) and (5′R) 5′,8-cyclo-2′-deoxypurines affect base excision repair of clustered DNA damage in nuclear extracts of Xrs5 cells? A biochemical study. Cells 2021, 10, 725. [Google Scholar] [CrossRef]
- Boguszewska, K.; Kaźmierczak-Barańska, J.; Karwowski, B.T. The Influence of 5′,8-Cyclo-2′-deoxypurines on the Mitochondrial Repair of Clustered DNA Damage in Xrs5 Cells: The Preliminary Study. Molecules 2021, 26, 7042. [Google Scholar] [CrossRef]
- Boguszewska, K.; Kaźmierczak-Barańska, J.; Karwowski, B.T. Mutagenicity of a bi-stranded clustered DNA lesion containing (5′S) or (5′R) 5′, 8-cyclo-2′-deoxyAdenosine in Escherichia coli model. Acta Biochim. Pol. 2022, 69, 865–869. [Google Scholar] [CrossRef]
- Jiang, Z.; Xu, M.; Lai, Y.; Laverde, E.E.; Terzidis, M.A.; Masi, A.; Chatgilialoglu, C.; Liu, Y. Bypass of a 5′,8-cyclopurine-2′-deoxynucleoside by DNA polymerase beta during DNA replication and base excision repair leads to nucleotide misinsertions and DNA strand breaks. DNA Repair 2015, 33, 24–34. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Lai, Y.; Jiang, Z.; Terzidis, M.A.; Masi, A.; Chatgilialoglu, C.; Liu, Y. A 5′,8-cyclo-2′-deoxypurine lesion induces trinucleotide repeat deletion via a unique lesion bypass by DNA polymerase β. Nucleic Acids Res. 2014, 42, 13749–13763. [Google Scholar] [CrossRef] [PubMed]

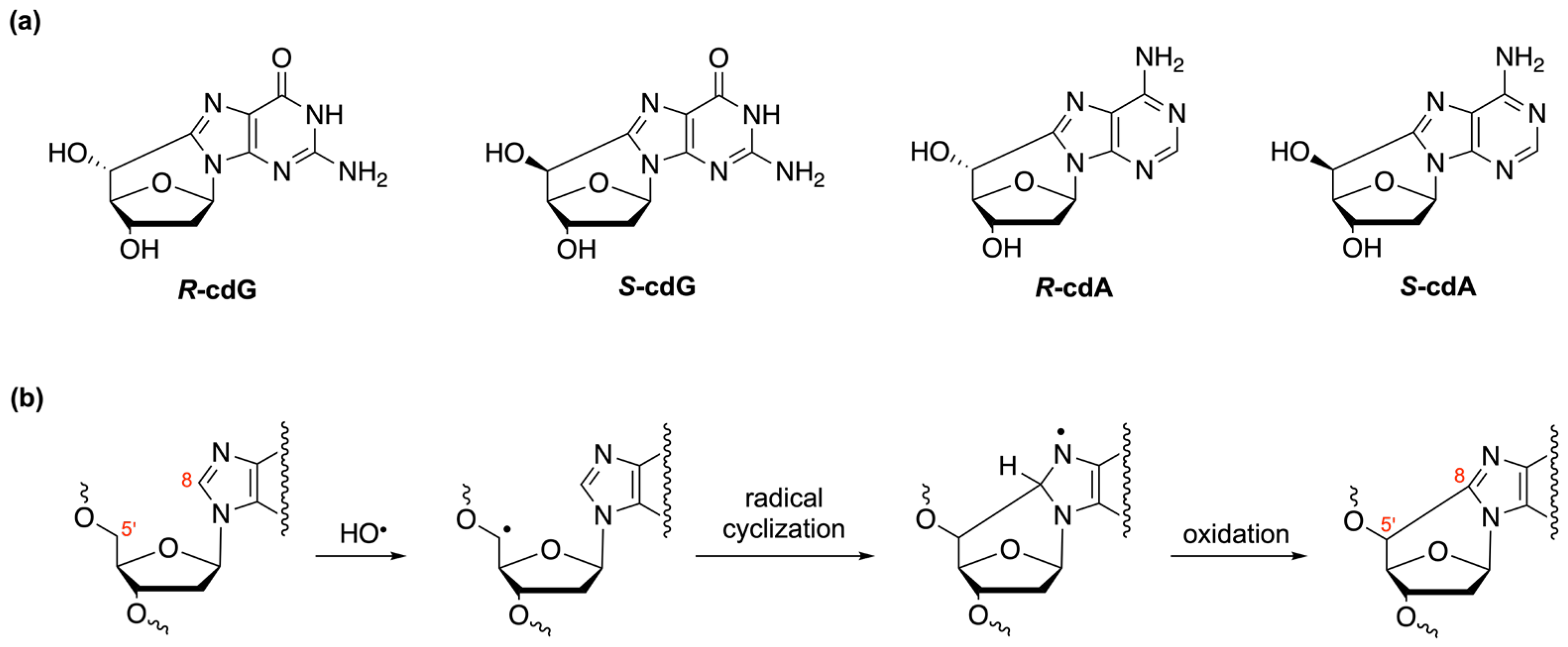
| Oligomer Lengths | Sequences (Double-Stranded) | Melting Temperatures (Tm °C) | References |
|---|---|---|---|
| 11 | 5′-d(CGT ACX CAT GC)-3′ 3′-d(GCA TGT GTA CG)-5′ | 49.5 (X = dA) 42.0 (X = 5′S-cdA) | [116] |
| 12 | 5′-d(GTG CXT GTT TGT)-3′ 3′-d(CAC GCA CAA ACA)-5′ | 55.0 (X = dG) 46.0 ± 1 (X =5′S-cdG) | [117] |
| 14 | 5′-d(ATC GTG XCT GAT CT)-3′ 3′-d(TAG CAC TGA CTA GA)-5′ | 54.0 ± 1 (X = dA) 48.0 ± 1 (X = 5′S-cdA) | [109] |
| 17 | 5′-d(CCA CCA ACX CTA CCA CC)-3′ 3′-d(GGT GGT TGT GAT GGT GG)-5′ | 65.2 ± 0.6 (X = dA) 58.9 ± 0.6 (X = 5′R-cdA) 60.5 ± 0.6 (X = 5′S-cdA) | [37] |
| 17 | 5′-d(CCA CCA ACX CTA CCA CC)-3′ 3′-d(GGT GGT TGC GAT GGT GG)-5′ | 66.2 ± 0.7 (X = dG) 63.4 ± 1.0 (X = 5′R-cdG) 63.5 ± 0.6 (X = 5′S-cdG) | [37] |
| 23 | 5′-d(GCA GAC ATA TCC TAG AGX CAT AT)-3′ 3′-d(CGT CTG TAT AGG ATC TCT GTA TA)-5′ | 60.0 ± 0.3 (X = dA) 59.0 ± 0.2 (X = 5′R-cdA) 58.0 ± 0.3 (X = 5′S-cdA) | [118] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moccia, M.; Pascucci, B.; Saviano, M.; Cerasa, M.T.; Terzidis, M.A.; Chatgilialoglu, C.; Masi, A. Advances in Nucleic Acid Research: Exploring the Potential of Oligonucleotides for Therapeutic Applications and Biological Studies. Int. J. Mol. Sci. 2024, 25, 146. https://doi.org/10.3390/ijms25010146
Moccia M, Pascucci B, Saviano M, Cerasa MT, Terzidis MA, Chatgilialoglu C, Masi A. Advances in Nucleic Acid Research: Exploring the Potential of Oligonucleotides for Therapeutic Applications and Biological Studies. International Journal of Molecular Sciences. 2024; 25(1):146. https://doi.org/10.3390/ijms25010146
Chicago/Turabian StyleMoccia, Maria, Barbara Pascucci, Michele Saviano, Maria Teresa Cerasa, Michael A. Terzidis, Chryssostomos Chatgilialoglu, and Annalisa Masi. 2024. "Advances in Nucleic Acid Research: Exploring the Potential of Oligonucleotides for Therapeutic Applications and Biological Studies" International Journal of Molecular Sciences 25, no. 1: 146. https://doi.org/10.3390/ijms25010146
APA StyleMoccia, M., Pascucci, B., Saviano, M., Cerasa, M. T., Terzidis, M. A., Chatgilialoglu, C., & Masi, A. (2024). Advances in Nucleic Acid Research: Exploring the Potential of Oligonucleotides for Therapeutic Applications and Biological Studies. International Journal of Molecular Sciences, 25(1), 146. https://doi.org/10.3390/ijms25010146










