Comparative Analysis of Cyclization Techniques in Stapled Peptides: Structural Insights into Protein–Protein Interactions in a SARS-CoV-2 Spike RBD/hACE2 Model System
Abstract
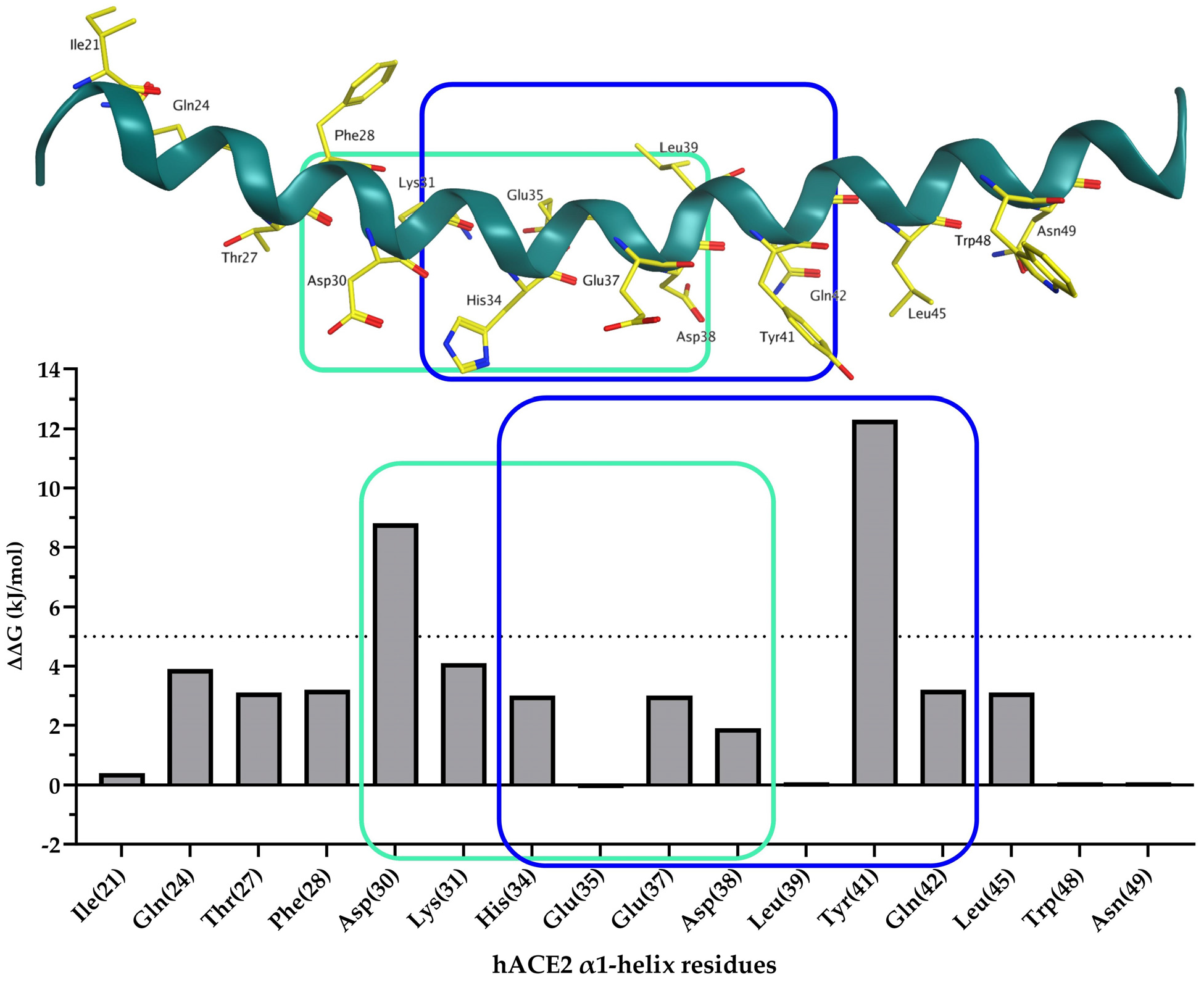
1. Introduction
2. Results and Discussion
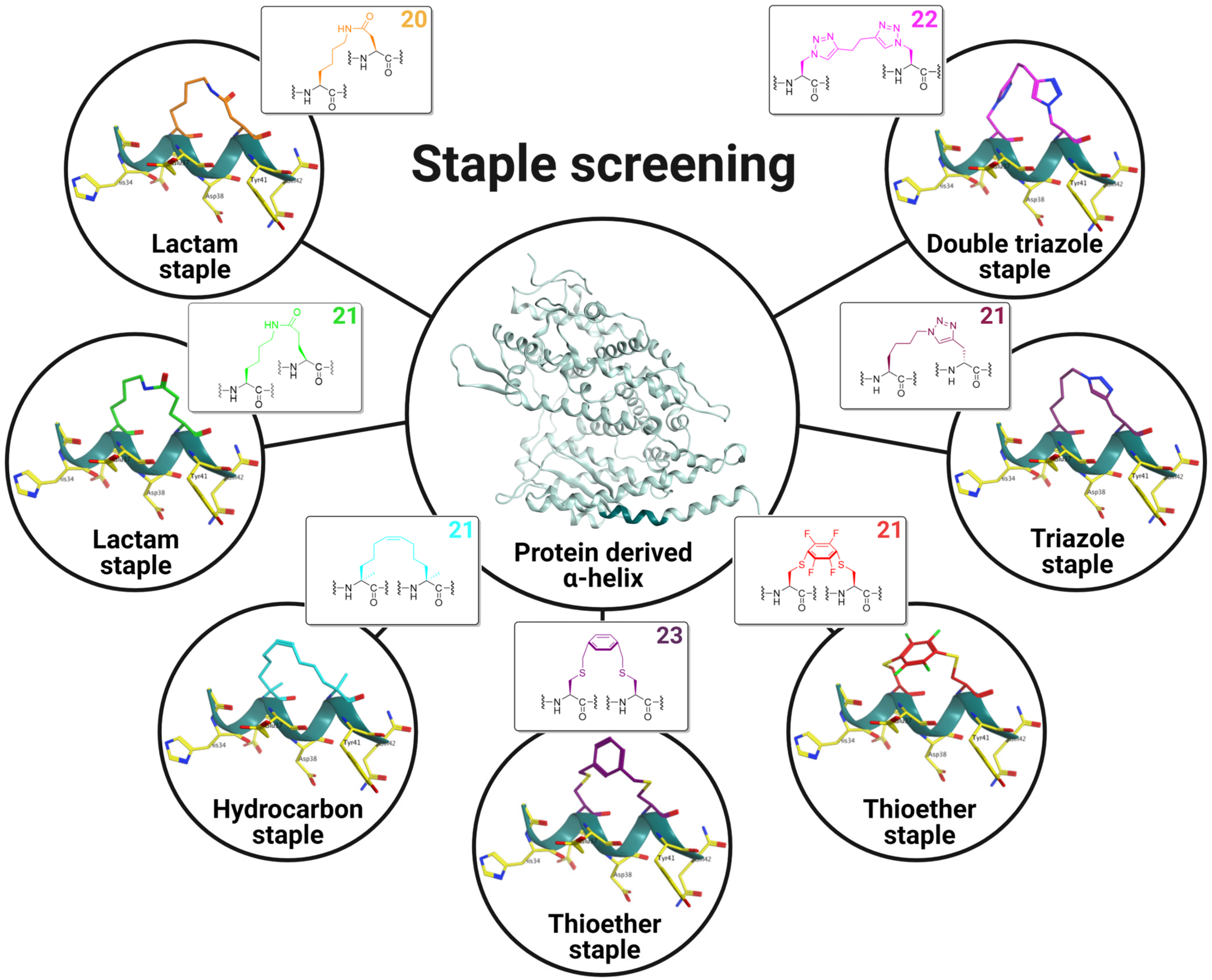
2.1. Experimental Design and Synthesis
2.2. Circular Dichroism
2.3. Nuclear Magnetic Resonance
2.3.1. 1H Resonance Assignments
2.3.2. NMR-Based Peptide Structural Comparisons and Hydrocarbon Staple Isomeric Investigations
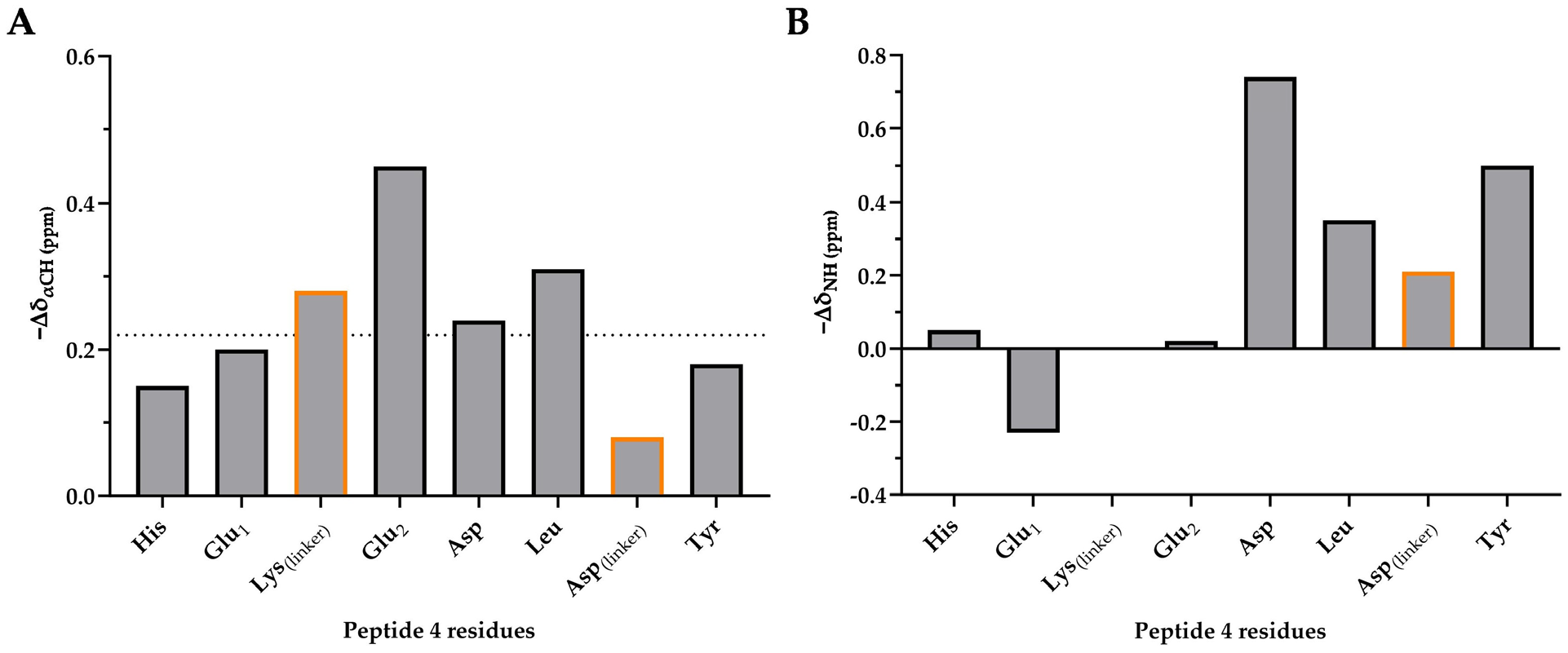
2.3.3. Secondary Chemical Shifts of Peptides 4, 9, 10 and 11
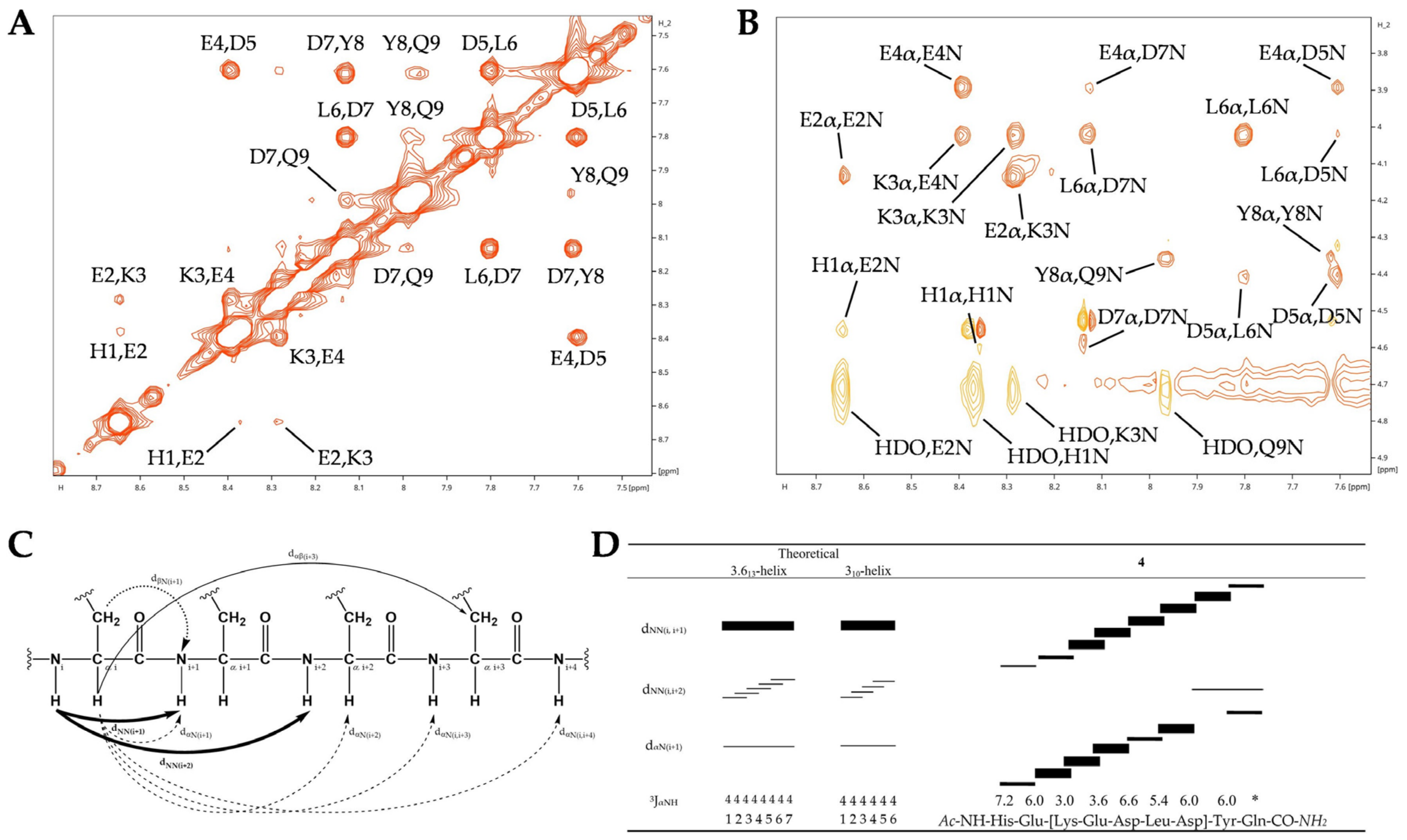
2.3.4. α-Helical NOESY Connectivities of Peptide 4
2.3.5. Comparative NOESY Connectivities for RCM-Based Staple Peptides
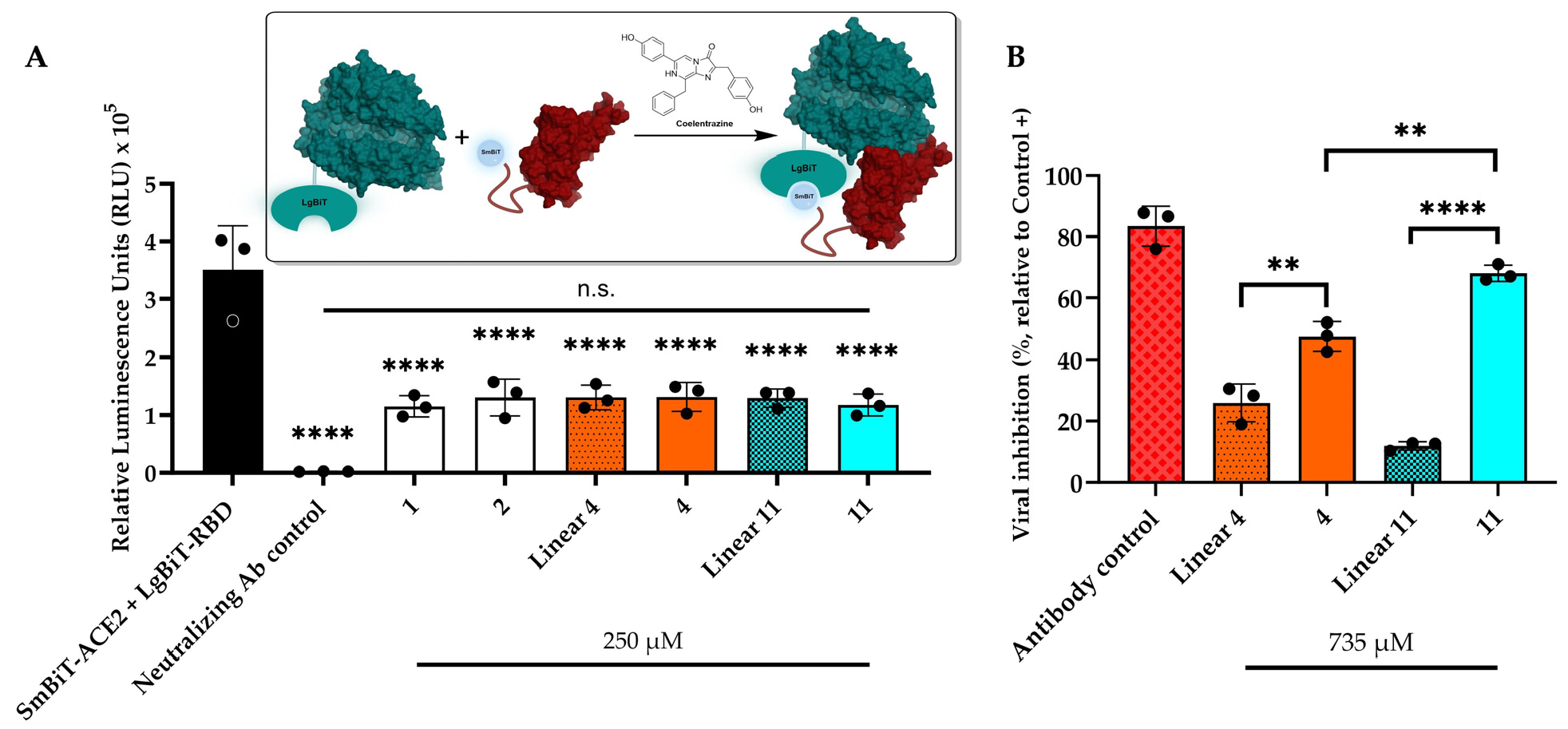
2.4. Inhibition Assays
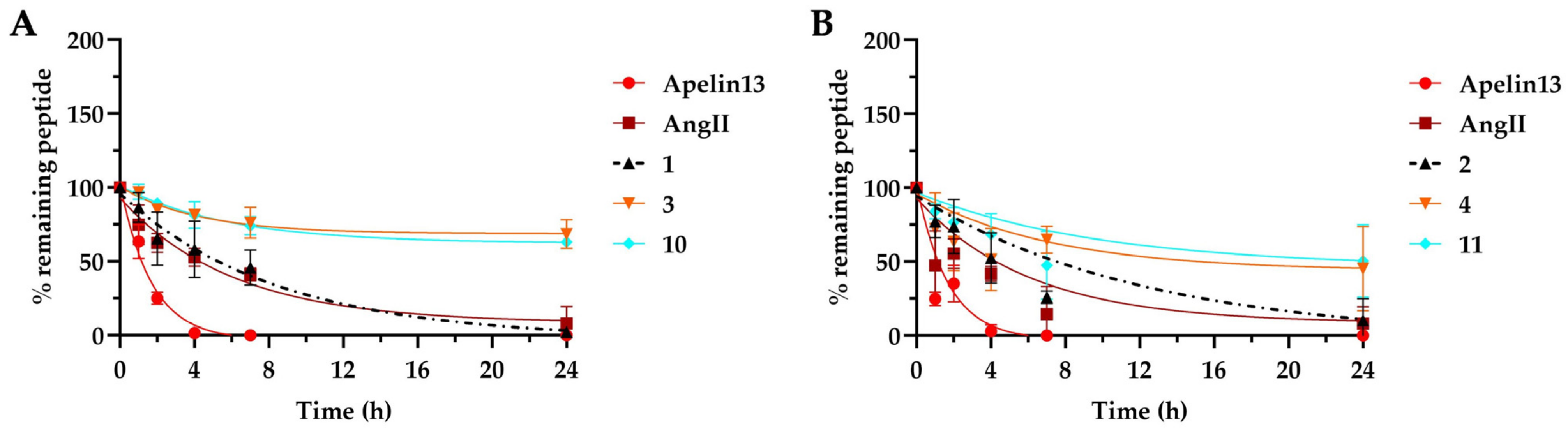
2.5. Plasma Stability
3. Materials and Methods
3.1. Reagents
3.2. Peptide Synthesis
3.2.1. Solid-Phase Peptide Synthesis
3.2.2. N-Terminal Acetylation
3.2.3. Lactam Staple Formation
3.2.4. Hydrocarbon Staple Formation
3.2.5. Triazole Staple Formation
3.2.6. Double Triazole Staple Formation
3.2.7. Thioether Staple Formation (S-Alkylation)
3.2.8. Thioether Staple Formation (S-Arylation)
3.2.9. Peptide Cleavage and Side Chain Deprotection
3.2.10. Peptide Purification
3.3. General Procedures
3.3.1. Peptide Characterization and Data Analysis
UPLC-MS Analysis
HRMS Analysis
3.3.2. Peptide Plasma Stability
3.3.3. Circular Dichroism Spectroscopy
Circular Dichroism Measurements
Quantification of Helicity
3.3.4. Nuclear Magnetic Resonance Measurements and Data Analysis
3.3.5. Bioreporter-Based Neutralization Assay
Cell Maintenance
Generation of Biosensors
Biosensor Assay
Bioreporter-Based Inhibition Assay Data Analysis
3.3.6. Neutralization Assay
Lentivirus Vector Design
Cell Maintenance
Pseudovirus-Based Inhibition Assay
Pseudovirus-Based Inhibition Assay Analysis
3.3.7. Peptide Design and Visualization
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 Spike Receptor-Binding Domain Bound to the ACE2 Receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhang, Y.; Wu, L.; Niu, S.; Song, C.; Zhang, Z.; Lu, G.; Qiao, C.; Hu, Y.; Yuen, K.Y.; et al. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell 2020, 181, 894–904.e9. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, M.; Gao, J. Enhanced Receptor Binding of SARS-CoV-2 through networks of Hydrogen-Bonding and hydrophobic Interactions. Proc. Natl. Acad. Sci. USA 2020, 117, 13967–13974. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.; Thornton, J.M. Review Principles of Protein-Protein Interactions. Proc. Natl. Acad. Sci. USA 1996, 93, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.C.; Gestwicki, J.E. Features of Protein-Protein Interactions That Translate into Potent Inhibitors: Topology, Surface Area and Affinity. Expert Rev. Mol. Med. 2012, 14, e16. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and Computational Approaches to Estimate Solubility and Permeability in Drug Discovery and Development Settings. Adv. Drug Deliv. Rev. 2001, 46, 4–17. [Google Scholar] [CrossRef] [PubMed]
- Buchwald, P. Small-Molecule Protein-Protein Interaction Inhibitors: Therapeutic Potential in Light of Molecular Size, Chemical Space, and Ligand Binding Efficiency Considerations. IUBMB Life 2010, 62, 724–731. [Google Scholar] [CrossRef] [PubMed]
- Bogan, A.A.; Thorn, K.S. Anatomy of Hot Spots in Protein Interfaces. J. Mol. Biol. 1998, 280, 1–9. [Google Scholar] [CrossRef]
- Zhang, G.; Zhang, J.; Gao, Y.; Li, Y.; Li, Y. Strategies for Targeting Undruggable Targets. Expert Opin. Drug Discov. 2022, 17, 55–69. [Google Scholar] [CrossRef]
- Dougherty, P.G.; Qian, Z.; Pei, D. Macrocycles as Protein-Protein Interaction Inhibitors. Biochem. J. 2017, 474, 1109–1125. [Google Scholar] [CrossRef]
- Hansen, J.; Baum, A.; Pascal, K.E.; Russo, V.; Giordano, S.; Wloga, E.; Fulton, B.O.; Yan, Y.; Koon, K.; Patel, K.; et al. Studies in Humanized Mice and Convalescent Humans Yield a SARS-CoV-2 Antibody Cocktail. Science 2020, 369, 1010–1014. [Google Scholar] [CrossRef] [PubMed]
- Weinreich, D.M.; Sivapalasingam, S.; Norton, T.; Ali, S.; Gao, H.; Bhore, R.; Musser, B.J.; Soo, Y.; Rofail, D.; Im, J.; et al. REGN-COV2, a Neutralizing Antibody Cocktail, in Outpatients with Covid-19. N. Engl. J. Med. 2021, 384, 238–251. [Google Scholar] [CrossRef] [PubMed]
- Horne, W.S.; Grossmann, T.N. Proteomimetics as Protein-Inspired Scaffolds with Defined Tertiary Folding Patterns. Nat. Chem. 2020, 12, 331–337. [Google Scholar] [CrossRef] [PubMed]
- Mabonga, L.; Kappo, A.P. Peptidomimetics: A Synthetic Tool for Inhibiting Protein–Protein Interactions in Cancer. Int. J. Pept. Res. Ther. 2020, 26, 225–241. [Google Scholar] [CrossRef]
- Bullock, B.N.; Jochim, A.L.; Arora, P.S. Assessing Helical Protein Interfaces for Inhibitor Design. J. Am. Chem. Soc. 2011, 133, 14220–14223. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wu, M.; Chang, Q.; Zhao, X. Stapling Strategy Enables Improvement of Antitumor Activity and Proteolytic Stability of Host-Defense Peptide Hymenochirin-1B. RSC Adv. 2018, 8, 22268–22275. [Google Scholar] [CrossRef] [PubMed]
- De Araujo, A.D.; Lim, J.; Wu, K.C.; Hoang, H.N.; Nguyen, H.T.; Fairlie, D.P. Landscaping Macrocyclic Peptides: Stapling HDM2-Binding Peptides for Helicity, Protein Affinity, Proteolytic Stability and Cell Uptake. RSC Chem. Biol. 2022, 3, 895–904. [Google Scholar] [CrossRef] [PubMed]
- Curreli, F.; Victor, S.M.B.; Ahmed, S.; Drelich, A.; Tong, X.; Tseng, C.-T.K.; Hillyer, C.D.; Debnath, A.K. Stapled Peptides Based on Human Angiotensin-Converting Enzyme 2 (ACE2) Potently Inhibit SARS-CoV-2 Infection In Vitro. MBio 2020, 11, 10–1128. [Google Scholar] [CrossRef]
- Maas, M.N.; Hintzen, J.C.J.; Löffler, P.M.G.; Mecinović, J. Targeting SARS-CoV-2 Spike Protein by Stapled HACE2 Peptides. Chem. Commun. 2021, 57, 3283–3286. [Google Scholar] [CrossRef]
- Karoyan, P.; Vieillard, V.; Gómez-Morales, L.; Odile, E.; Guihot, A.; Luyt, C.E.; Denis, A.; Grondin, P.; Lequin, O. Human ACE2 Peptide-Mimics Block SARS-CoV-2 Pulmonary Cells Infection. Commun. Biol. 2021, 4, 197. [Google Scholar] [CrossRef]
- Quagliata, M.; Stincarelli, M.A.; Papini, A.M.; Giannecchini, S.; Rovero, P. Antiviral Activity against SARS-CoV-2 of Conformationally Constrained Helical Peptides Derived from Angiotensin-Converting Enzyme 2. ACS Omega 2023, 8, 22665–22672. [Google Scholar] [CrossRef] [PubMed]
- Larue, R.C.; Xing, E.; Kenney, A.D.; Zhang, Y.; Tuazon, J.A.; Li, J.; Yount, J.S.; Li, P.K.; Sharma, A. Rationally Designed ACE2-Derived Peptides Inhibit SARS-CoV-2. Bioconjug Chem. 2021, 32, 215–223. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Goreshnik, I.; Coventry, B.; Case, J.B.; Miller, L.; Kozodoy, L.; Chen, R.E.; Carter, L.; Walls, A.C.; Park, Y.-J.; et al. De Novo Design of Picomolar SARS-CoV-2 Miniprotein Inhibitors. Science 2020, 370, 426–431. [Google Scholar] [CrossRef] [PubMed]
- Sommese, R.F.; Sivaramakrishnan, S.; Baldwin, R.L.; Spudich, J.A. Helicity of Short E-R/K Peptides. Protein Sci. 2010, 19, 2001–2005. [Google Scholar] [CrossRef] [PubMed]
- Marqusee, S.; Baldwin, R.L. Helix Stabilization by Glu-W0Lys+ Salt Bridges in Short Peptides of de Novo Design (Model a-Helix/Protein Folding/Protein Electrostatics). Proc. Natl. Acad. Sci. USA 1987, 84, 8898–8902. [Google Scholar] [CrossRef] [PubMed]
- Houston, M.E., Jr.; Gannon, C.L.; Kayand, C.M.; Hodges, R.S. Lactam Bridge Stabilization of A-Helical Peptides: Ring Size, Orientation and Positional Effects. J. Pept. Sci. Off. Publ. Eur. Pept. Soc. 1995, 1, 274–282. [Google Scholar] [CrossRef] [PubMed]
- Felix, A.M.; Wang, C.T.; Campbell, R.M.; Toome, V.; Fry, D.C.; Madison, V.S. Biologically Active Cyclic (Lactam) Analogs of Growth Hormone-Releasing Factor: Effect of Ring Size and Location on Conformation and Biological Activity. In Peptides Chemistry and Biology; Smith, J.A., Rivier, J.E., Eds.; Springer: Leiden, The Netherlands, 1992; pp. 77–79. [Google Scholar]
- Shim, S.Y.; Kim, Y.W.; Verdine, G.L. A New i, i + 3 Peptide Stapling System for α-Helix Stabilization. Chem. Biol. Drug Des. 2013, 82, 635–642. [Google Scholar] [CrossRef] [PubMed]
- Bird, G.H.; Madani, N.; Perry, A.F.; Princiotto, A.M.; Supko, J.G.; He, X.; Gavathiotis, E.; Sodroski, J.G.; Walensky, L.D.; Snyder, S.H. Hydrocarbon Double-Stapling Remedies the Proteolytic Instability of a Lengthy Peptide Therapeutic. Proc. Natl. Acad. Sci. USA 2010, 107, 14093–14098. [Google Scholar] [CrossRef]
- Bernal, F.; Wade, M.; Godes, M.; Davis, T.N.; Whitehead, D.G.; Kung, A.L.; Wahl, G.M.; Walensky, L.D. A Stapled P53 Helix Overcomes HDMX-Mediated Suppression of P53. Cancer Cell 2010, 18, 411–422. [Google Scholar] [CrossRef]
- Schafmeister, C.E.; Po, J.; Verdine, G.L. An All-Hydrocarbon Cross-Linking System for Enhancing the Helicity and Metabolic Stability of Peptides. J. Am. Chem. Soc. 2000, 122, 5891–5892. [Google Scholar] [CrossRef]
- Moellering, R.E.; Cornejo, M.; Davis, T.N.; Del Bianco, C.; Aster, J.C.; Blacklow, S.C.; Kung, A.L.; Gilliland, D.G.; Verdine, G.L.; Bradner, J.E. Direct Inhibition of the NOTCH Transcription Factor Complex. Nature 2009, 462, 182–188. [Google Scholar] [CrossRef] [PubMed]
- Williams, R.M.; IM, M.-N. Asymmetric Synthesis of Monosubstituted and α,α-Disubstituted α-Amino Acids via Diastereoselective Glycine Enolate Alkylations. ChemInform 1991, 23, 255. [Google Scholar]
- Ueda, A.; Makura, Y.; Kakazu, S.; Kato, T.; Umeno, T.; Hirayama, K.; Doi, M.; Oba, M.; Tanaka, M. E-Selective Ring-Closing Metathesis in α-Helical Stapled Peptides Using Carbocyclic α,α-Disubstituted α-Amino Acids. Org. Lett. 2022, 24, 1049–1054. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chou, D.H.-C. A Thiol-Ene Coupling Approach to Native Peptide Stapling and Macrocyclization. Angew. Chem. 2015, 127, 11081–11084. [Google Scholar] [CrossRef]
- Paterson, D.L.; Flanagan, J.U.; Shepherd, P.R.; Harris, P.W.R.; Brimble, M.A. Variable-Length Ester-Based Staples for α-Helical Peptides by Using A Double Thiol-Ene Reaction. Chem.—A Eur. J. 2020, 26, 10826–10833. [Google Scholar] [CrossRef]
- Cantel, S.; Isaad, A.L.C.; Scrima, M.; Levy, J.J.; DiMarchi, R.D.; Rovero, P.; Halperin, J.A.; D’Ursi, A.M.; Papini, A.M.; Chorev, M. Synthesis and Conformational Analysis of a Cyclic Peptide Obtained via i to i + 4 Intramolecular Side-Chain to Side-Chain Azide-Alkyne 1,3-Dipolar Cycloaddition. J. Org. Chem. 2008, 73, 5663–5674. [Google Scholar] [CrossRef]
- Kawamoto, S.A.; Coleska, A.; Ran, X.; Yi, H.; Yang, C.Y.; Wang, S. Design of Triazole-Stapled BCL9 α-Helical Peptides to Target the β-Catenin/B-Cell CLL/Lymphoma 9 (BCL9) Protein-Protein Interaction. J. Med. Chem. 2012, 55, 1137–1146. [Google Scholar] [CrossRef]
- Torres, O.; Yüksel, D.; Bernardina, M.; Kumar, K.; Bong, D. Peptide Tertiary Structure Nucleation by Side-Chain Crosslinking with Metal Complexation and Double “Click” Cycloaddition. ChemBioChem 2008, 9, 1701–1705. [Google Scholar] [CrossRef]
- Scrima, M.; Le Chevalier-Isaad, A.; Rovero, P.; Papini, A.M.; Chorev, M.; D’Ursi, A.M. Cui Catalyzed Azide-Alkyne Intramolecular i-to-(i + 4) Side-Chain-to-SideChain Cyclization Promotes the Formation of Helix-Like Secondary Structures. Eur. J. Org. Chem. 2010, 2010, 446–457. [Google Scholar] [CrossRef]
- Goddard-Borger, E.D.; Stick, R.V. An Efficient, Inexpensive, and Shelf-Stable Diazotransfer Reagent: Imidazole-1-Sulfonyl Azide Hydrochloride. Org. Lett. 2007, 9, 3797–3800. [Google Scholar] [CrossRef]
- Lau, Y.H.; Spring, D.R. Efficient Synthesis of Fmoc-Protected Azido Amino Acids. Synlett 2011, 2011, 1917–1919. [Google Scholar] [CrossRef][Green Version]
- Spokoyny, A.M.; Zou, Y.; Ling, J.J.; Yu, H.; Lin, Y.-S.; Pentelute, B.L. A Perfluoroaryl-Cysteine SNAr Chemistry Approach to Unprotected Peptide Stapling Supporting Information S1. J. Am. Chem. Soc. 2013, 135, 5946–5949. [Google Scholar] [CrossRef] [PubMed]
- Dognini, P.; Killoran, P.M.; Hanson, G.S.; Halsall, L.; Chaudhry, T.; Islam, Z.; Giuntini, F.; Coxon, C.R. Using 19 F NMR and 2-Level Factorial Design to Explore Thiol-Fluoride Substitution in Hexafluorobenzene and Its Application in Peptide Stapling & Cyclisation Supporting Information Contents. Chem.–A Eur. J. 2022, 28, e202103305. [Google Scholar]
- Holmes, E.C. Severe Acute Respiratory Syndrome Coronavirus 2 Isolate Wuhan-Hu1, Complete Genome; National Library of Medicine: Bethesda, MD, USA, 2019. [Google Scholar]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.-L.; Abiona, O.; Graham, B.S.; Mclellan, J.S. Cryo-EM Structure of the 2019-NCoV Spike in the Prefusion Conformation. Science 2019, 367, 1260–1263. [Google Scholar] [CrossRef] [PubMed]
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292.e6. [Google Scholar] [CrossRef] [PubMed]
- Shang, J.; Ye, G.; Shi, K.; Wan, Y.; Luo, C.; Aihara, H.; Geng, Q.; Auerbach, A.; Li, F. Structural Basis of Receptor Recognition by SARS-CoV-2. Nature 2020, 581, 221–224. [Google Scholar] [CrossRef] [PubMed]
- Yan, R.; Zhang, Y.; Li, Y.; Xia, L.; Guo, Y.; Zhou, Q. Structural Basis for the Recognition of SARS-CoV-2 by Full-Length Human ACE2. Science 2020, 367, 1444–1448. [Google Scholar] [CrossRef]
- Yuan, M.; Wu, N.C.; Zhu, X.; Lee, C.C.D.; So, R.T.Y.; Lv, H.; Mok, C.K.P.; Wilson, I.A. A Highly Conserved Cryptic Epitope in the Receptor Binding Domains of SARS-CoV-2 and SARS-CoV. Science 2020, 368, 630–633. [Google Scholar] [CrossRef]
- Huang, X.; Pearce, R.; Zhang, Y. De Novo Design of Protein Peptides to Block Association of the SARSCoV-2 Spike Protein with Human ACE2. Aging 2020, 12, 11263. [Google Scholar] [CrossRef]
- Ali, F.; Elserafy, M.; Alkordi, M.H.; Amin, M. ACE2 Coding Variants in Different Populations and Their Potential Impact on SARS-CoV-2 Binding Affinity. Biochem. Biophys. Rep. 2020, 24, 100798. [Google Scholar] [CrossRef]
- Rohl, C.A.; Baldwin, R.L. [1] Deciphering Rules of Helix Stability in Peptides. Methods Enzymol. 1998, 295, 1–26. [Google Scholar] [PubMed]
- Huyghues-Despointes, B.M.P.; Klingler, T.M.; Baldwin, R.L. Measuring the Strength of Side-Chain Hydrogen Bonds in Peptide Helices: The Gln*Asp (i, i + 4) Interaction? Biochemistry 1995, 34, 13267–13271. [Google Scholar] [CrossRef] [PubMed]
- Tornøe, C.W.; Christensen, C.; Meldal, M. Peptidotriazoles on Solid Phase: [1,2,3]-Triazoles by Regiospecific Copper(I)-Catalyzed 1,3-Dipolar Cycloadditions of Terminal Alkynes to Azides. J. Org. Chem. 2002, 67, 3057–3064. [Google Scholar] [CrossRef] [PubMed]
- Rostovtsev, V.V.; Green, L.G.; Fokin, V.V.; Barry Sharpless, K.; Coolen, H.K.A.C.; van Leeuwen, P.W.N.M.; Nolte, R.J.M. Harrowfield in Calixarenes; Kluwer Academic Publishers: Dordrecht, The Netherlands, 2002; Volume 114. [Google Scholar]
- Hang, H.C.; Yu, C.; Kato, D.L.; Bertozzi, C.R.; Simons, K. A Metabolic Labeling Approach toward Proteomic Analysis of Mucin-Type O-Linked Glycosylation. Proc. Natl. Acad. Sci. USA 2003, 100, 14846–14851. [Google Scholar] [CrossRef] [PubMed]
- Lacroix, E.; Viguera, A.R.; Serrano, L. Elucidating the Folding Problem of α-Helices: Local Motifs, Long-Range Electrostatics, Ionic-Strength Dependence and Prediction of NMR Parameters. J. Mol. Biol. 1998, 284, 173–191. [Google Scholar] [CrossRef] [PubMed]
- Daragane, V.A.; Mayo, K.H. 13C-{1H} NMR/NOE and Multiplet Relaxation Data in Modeling Protein Dynamics of a Collagen 13C-Enriched Glycine GXX Repeat Motif Hexadecapeptide. J. Am. Chem. Soc. 1992, 114, 4326–4331. [Google Scholar] [CrossRef]
- Bakshi, K.; Liyanage, M.R.; Volkin, D.B.; Middaugh, C.R. Circular Dichroism of Peptides. Methods Mol. Biol. 2014, 1088, 247–253. [Google Scholar] [CrossRef]
- Micsonai, A.; Wien, F.; Kernya, L.; Lee, Y.H.; Goto, Y.; Réfrégiers, M.; Kardos, J. Accurate Secondary Structure Prediction and Fold Recognition for Circular Dichroism Spectroscopy. Proc. Natl. Acad. Sci. USA 2015, 112, E3095–E3103. [Google Scholar] [CrossRef]
- Wishart, D.S.; Bigam, C.G.; Sykes, B.D. 1H, 13C and Random Coil NMR Chemical Shifts of the Common Amino Acids. I. Investigations of Nearest-Neighbor Effects. J. Biomol. NMR 1995, 5, 67–81. [Google Scholar] [CrossRef]
- Jeremy, N.S. Evans 4. Protein Structure. In Biomolecular NMR Spectroscopy; Oxford University Press Inc.: New York, NY, USA, 1995; pp. 147–207. [Google Scholar]
- Houston, M.E.; Campbell, A.P.; Lix, B.; Kay, C.M.; Sykes, B.D.; Hodges, R.S. Lactam Bridge Stabilization of R-Helices: The Role of Hydrophobicity in Controlling Dimeric versus Monomeric R-Helices. Biochemistry 1996, 35, 10041–10050. [Google Scholar] [CrossRef]
- Gratias, R.; Konat, R.; Kessler, H.; Crisma, M.; Valle, G.; Polese, A.; Formaggio, F.; Toniolo, C.; Broxterman, Q.B.; Kamphuis, J. First Step Toward the Quantitative Identification of Peptide 310-Helix Conformation with NMR Spectroscopy: NMR and X-Ray Diffraction Structural Analysis of a Fully-Developed 310-Helical Peptide Standard. J. Am. Chem. Soc. 1997, 120, 4763–4770. [Google Scholar] [CrossRef]
- Azad, T.; Singaravelu, R.; Taha, Z.; Jamieson, T.R.; Boulton, S.; Crupi, M.J.F.; Martin, N.T.; Fekete, E.E.F.; Poutou, J.; Ghahremani, M.; et al. Nanoluciferase Complementation-Based Bioreporter Reveals the Importance of N-Linked Glycosylation of SARS-CoV-2 S for Viral Entry. Mol. Ther. 2021, 29, 1984–2000. [Google Scholar] [CrossRef] [PubMed]
- Murza, A.; Belleville, K.; Longpré, J.M.; Sarret, P.; Marsault, É. Stability and Degradation Patterns of Chemically Modified Analogs of Apelin-13 in Plasma and Cerebrospinal Fluid. Biopolym.—Pept. Sci. Sect. 2014, 102, 297–303. [Google Scholar] [CrossRef] [PubMed]
- Caputo, I.; Caroccia, B.; Frasson, I.; Poggio, E.; Zamberlan, S.; Morpurgo, M.; Seccia, T.M.; Calì, T.; Brini, M.; Richter, S.N.; et al. Angiotensin II Promotes SARS-CoV-2 Infection via Upregulation of ACE2 in Human Bronchial Cells. Int. J. Mol. Sci. 2022, 23, 5125. [Google Scholar] [CrossRef]
- Kaiser, E.; Colescott, R.; Bossinger, C.; Cook, P. Color Test for Detection of Free Terminal Amino Groups in the Solid-Phase Synthesis of Peptides. Anal. Biochem. 1970, 34, 595–598. [Google Scholar] [CrossRef]
- Wallimann, P.; Kennedy, R.J.; Kemp, D.S. Large Circular Dichroism Ellipticities for N-Templated Helical Polypeptides Are Inconsistent with Currently Accepted Helicity Algorithms. Angew. Chem. Int. Ed. 1999, 38, 1290–1292. [Google Scholar] [CrossRef]
- Johnson, M.C.; Lyddon, T.D.; Suarez, R.; Salcedo, B.; LePique, M.; Graham, M.; Ricana, C.; Robinson, C.; Ritter, D.G. Optimized Pseudotyping Conditions for the SARS-COV-2 Spike Glycoprotein. J. Virol. 2020, 94, 10–1128. [Google Scholar] [CrossRef]

| Residue | NH | CαH | CβH | CγH | CδH | CεH |
|---|---|---|---|---|---|---|
| N-terminus | ||||||
| His | 8.37 | 4.58 | 3.03, 3.1 | - | 7.1, * | 2, * |
| Glu2 | 8.65 | 4.15 | 1.87, 2.01 | 2.19, * | - | - |
| Lys(linker) | 8.29 | 4.04 | 1.72, 1.80 | 1.30, 1.47 | 1.12, * | *, * |
| Glu1 | 8.40 | 3.90 | 1.92 | 2.21, * | - | - |
| Asp | 7.60 | 4.4 | 2.58, 263 | - | - | - |
| Leu | 7.81 | 4.03 | 1.64, 1.76 | 1.71 | 0.74, 0.77 | - |
| Asp(linker) | 8.13 | 4.56 | 2.50, 2.72 | - | - | - |
| Tyr | 7.62 | 4.37 | 2.97, 3.01 | - | *, * | *, * |
| Gln | * | * | *, * | *, * | - | *, * |
| C-terminus |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferková, S.; Froehlich, U.; Nepveu-Traversy, M.-É.; Murza, A.; Azad, T.; Grandbois, M.; Sarret, P.; Lavigne, P.; Boudreault, P.-L. Comparative Analysis of Cyclization Techniques in Stapled Peptides: Structural Insights into Protein–Protein Interactions in a SARS-CoV-2 Spike RBD/hACE2 Model System. Int. J. Mol. Sci. 2024, 25, 166. https://doi.org/10.3390/ijms25010166
Ferková S, Froehlich U, Nepveu-Traversy M-É, Murza A, Azad T, Grandbois M, Sarret P, Lavigne P, Boudreault P-L. Comparative Analysis of Cyclization Techniques in Stapled Peptides: Structural Insights into Protein–Protein Interactions in a SARS-CoV-2 Spike RBD/hACE2 Model System. International Journal of Molecular Sciences. 2024; 25(1):166. https://doi.org/10.3390/ijms25010166
Chicago/Turabian StyleFerková, Sára, Ulrike Froehlich, Marie-Édith Nepveu-Traversy, Alexandre Murza, Taha Azad, Michel Grandbois, Philippe Sarret, Pierre Lavigne, and Pierre-Luc Boudreault. 2024. "Comparative Analysis of Cyclization Techniques in Stapled Peptides: Structural Insights into Protein–Protein Interactions in a SARS-CoV-2 Spike RBD/hACE2 Model System" International Journal of Molecular Sciences 25, no. 1: 166. https://doi.org/10.3390/ijms25010166
APA StyleFerková, S., Froehlich, U., Nepveu-Traversy, M.-É., Murza, A., Azad, T., Grandbois, M., Sarret, P., Lavigne, P., & Boudreault, P.-L. (2024). Comparative Analysis of Cyclization Techniques in Stapled Peptides: Structural Insights into Protein–Protein Interactions in a SARS-CoV-2 Spike RBD/hACE2 Model System. International Journal of Molecular Sciences, 25(1), 166. https://doi.org/10.3390/ijms25010166







