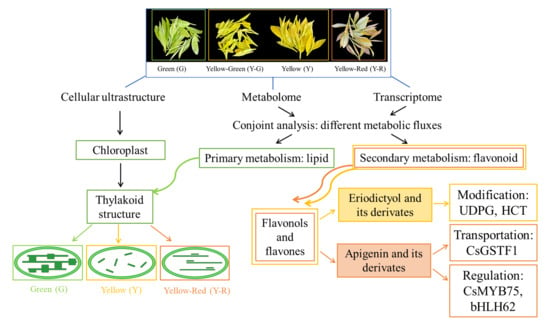
Integrated Analysis of Metabolome and Transcriptome Revealed Different Regulatory Networks of Metabolic Flux in Tea Plants [Camellia sinensis (L.) O. Kuntze] with Varied Leaf Colors
Abstract
:1. Introduction
2. Results
2.1. The Phenotype and Relative Chlorophyll Concentration of Tea Leaves Varied Coordinately
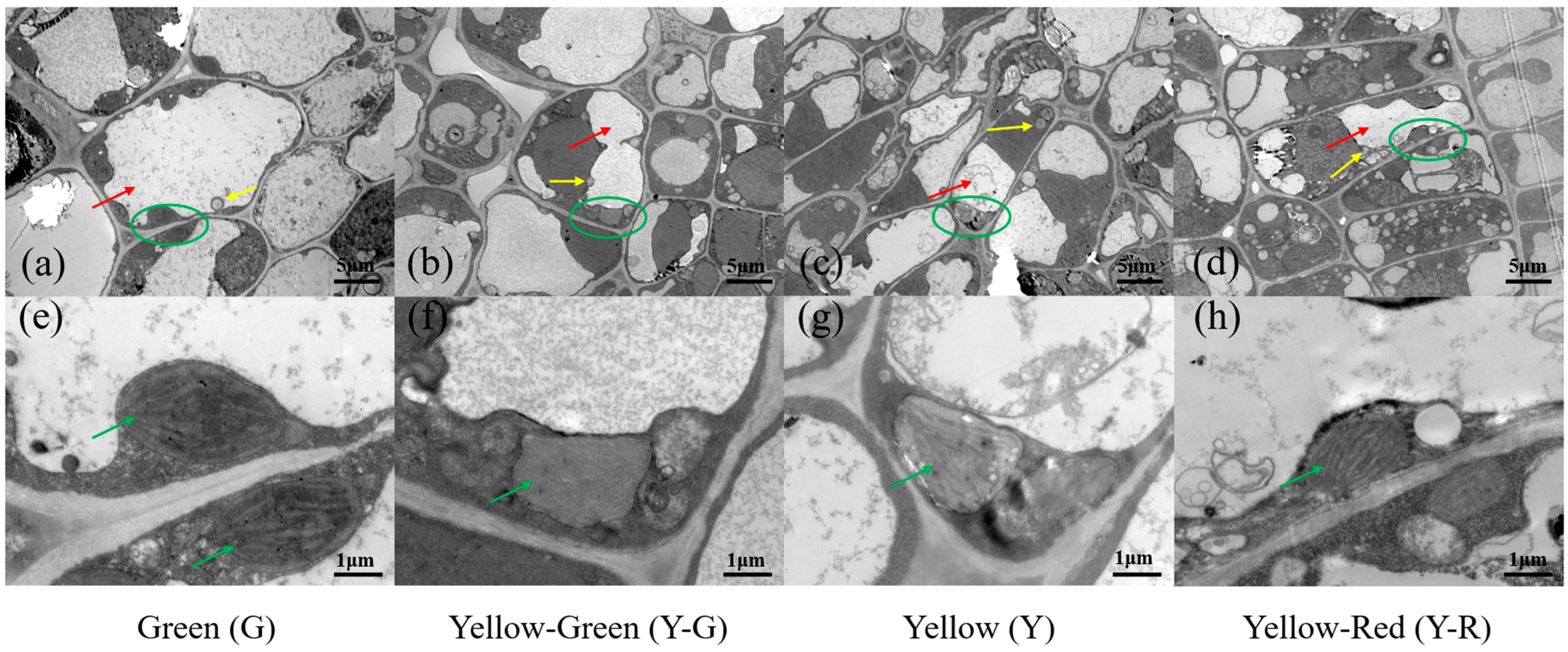
2.2. The Cellular Ultrastructure of Tea Leaves with Different Colors
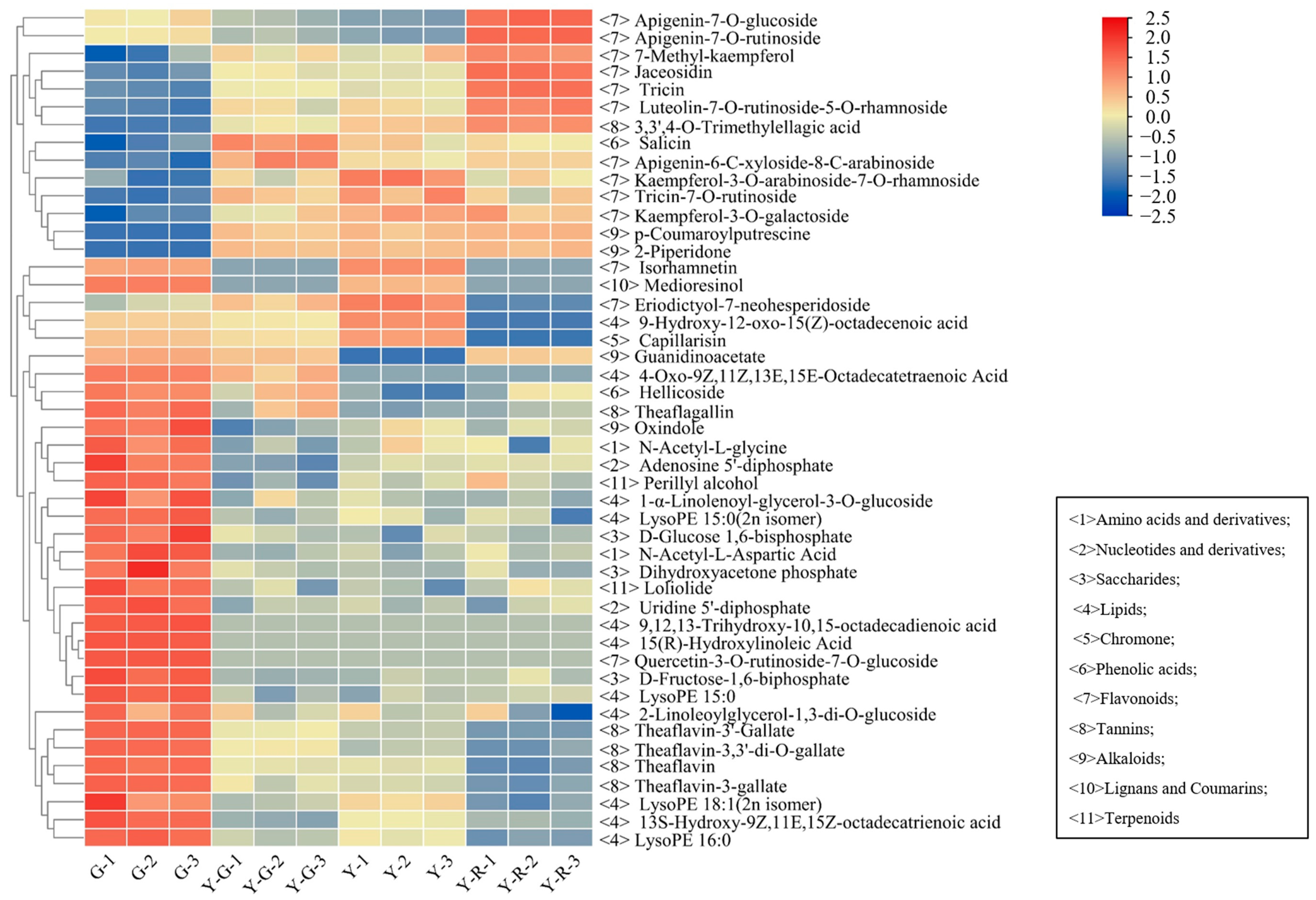
2.3. Differentially Accumulated Metabolites (DAMs) in Green and Yellow Leaves
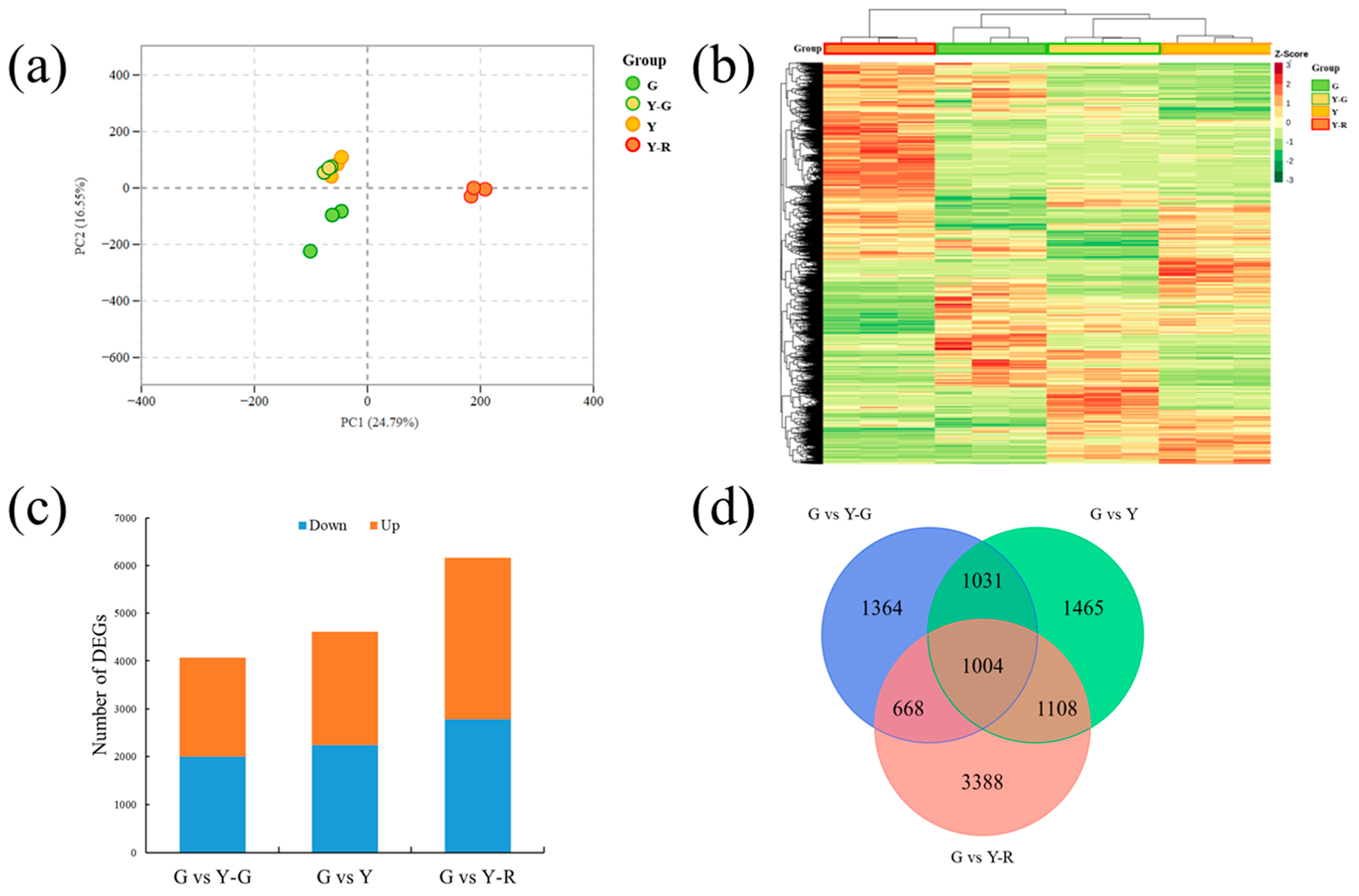
2.4. Differentially Expressed Genes (DEGs) in Green and Yellow Leaves
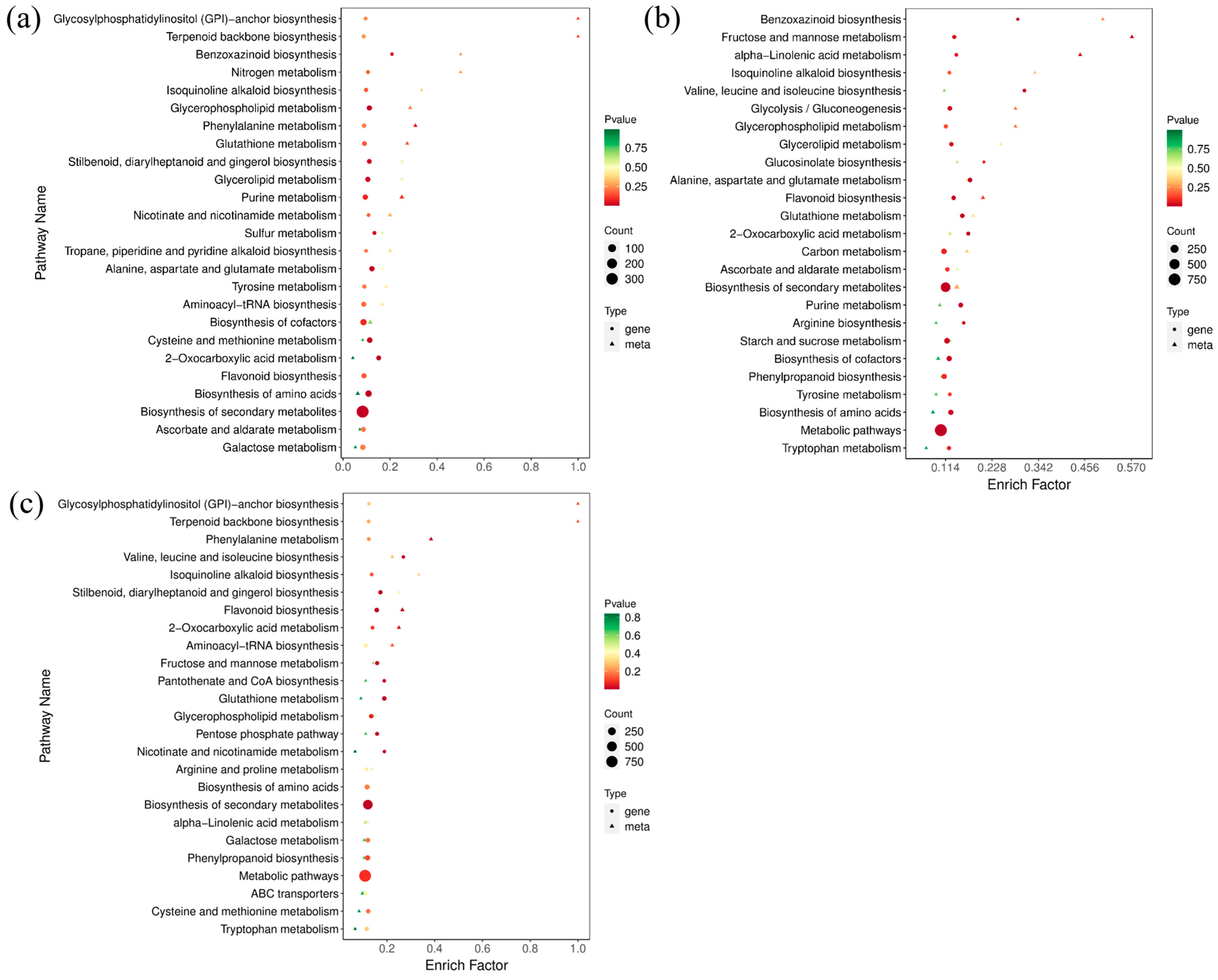
2.5. DAMs and DEGs Were Enriched in Consistent Metabolic Pathways
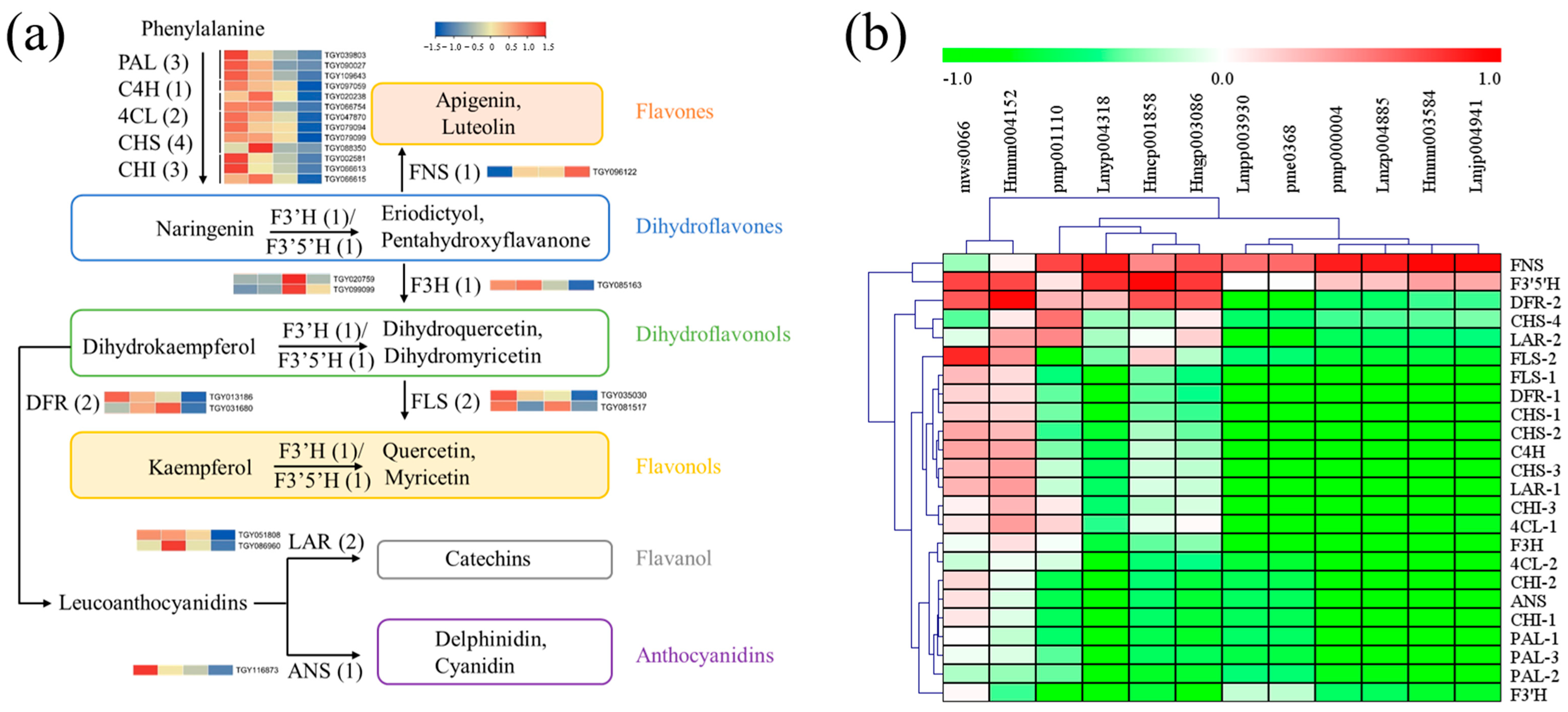
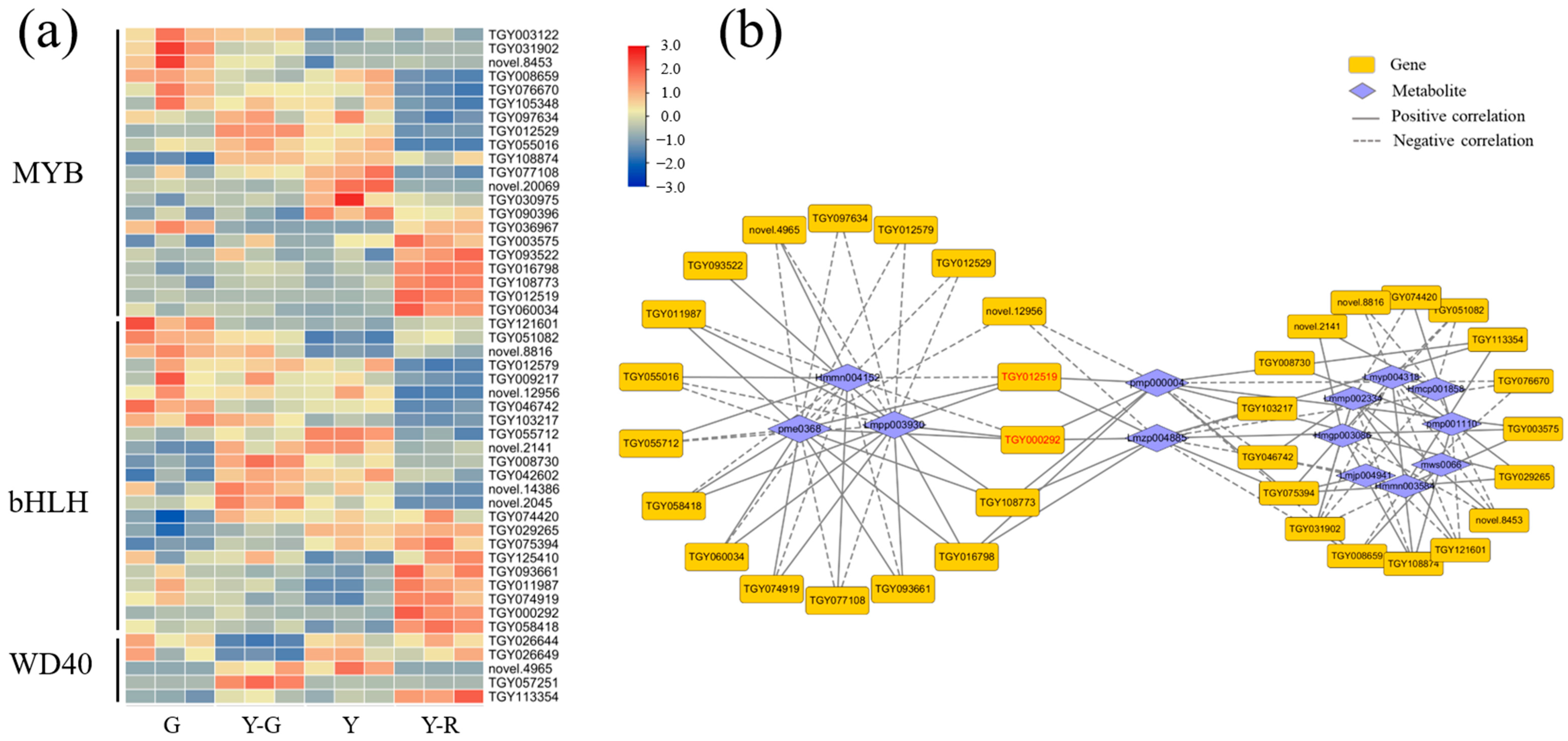
2.6. Flavonoid Pathway Was Regulated Differentially in Green and Yellow Leaves
3. Discussion
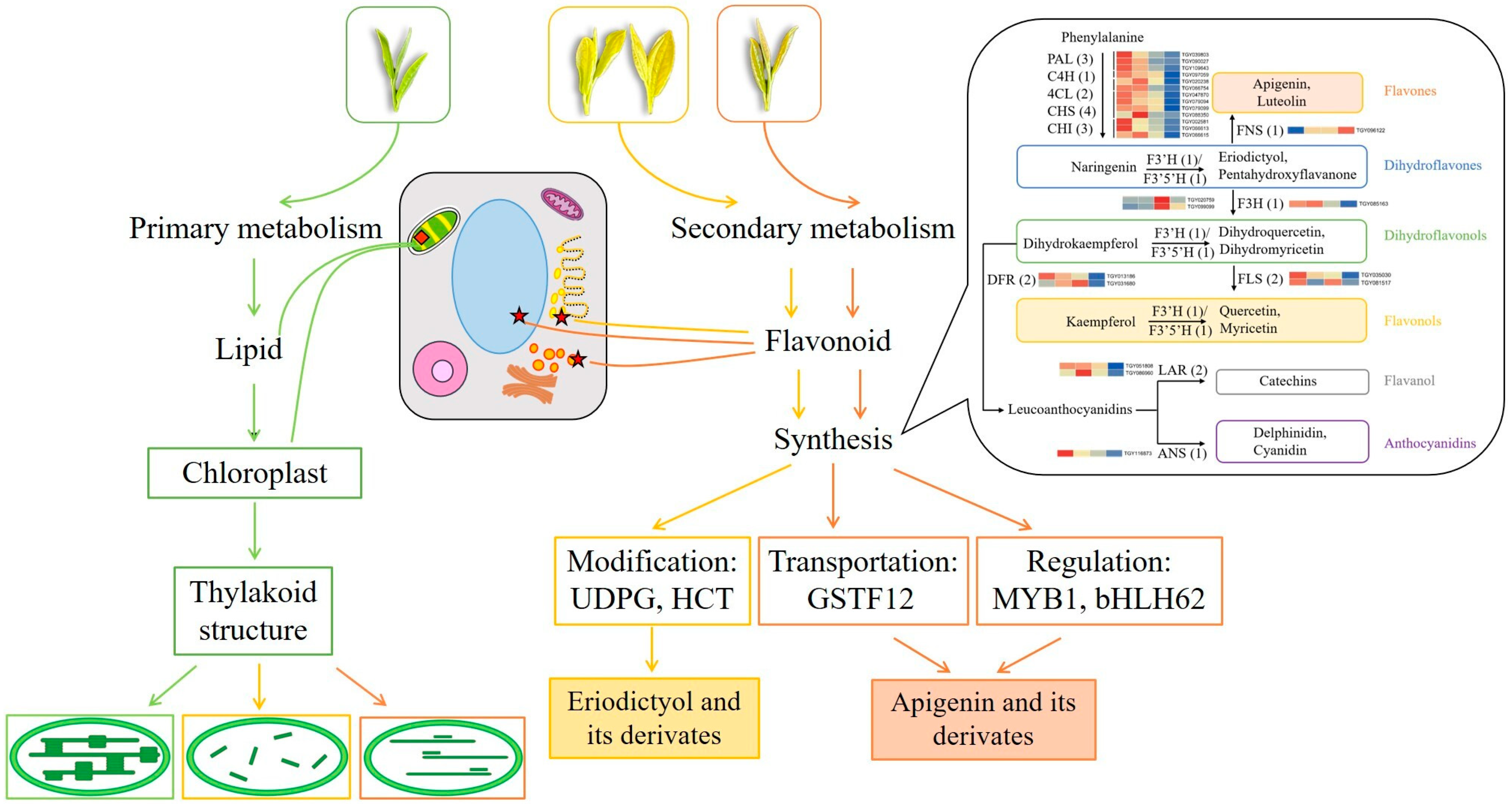
3.1. There Are Differences in Primary and Secondary Metabolic Flux in Green and Yellow Leaves
3.2. Decrease in Lipid Metabolism Leads to the Disruption of Chloroplast Structure in Tea Leaves
3.3. Increase in Flavones and Flavonols Metabolism Are Responsible for the Yellow Color Formation of Tea Leaves
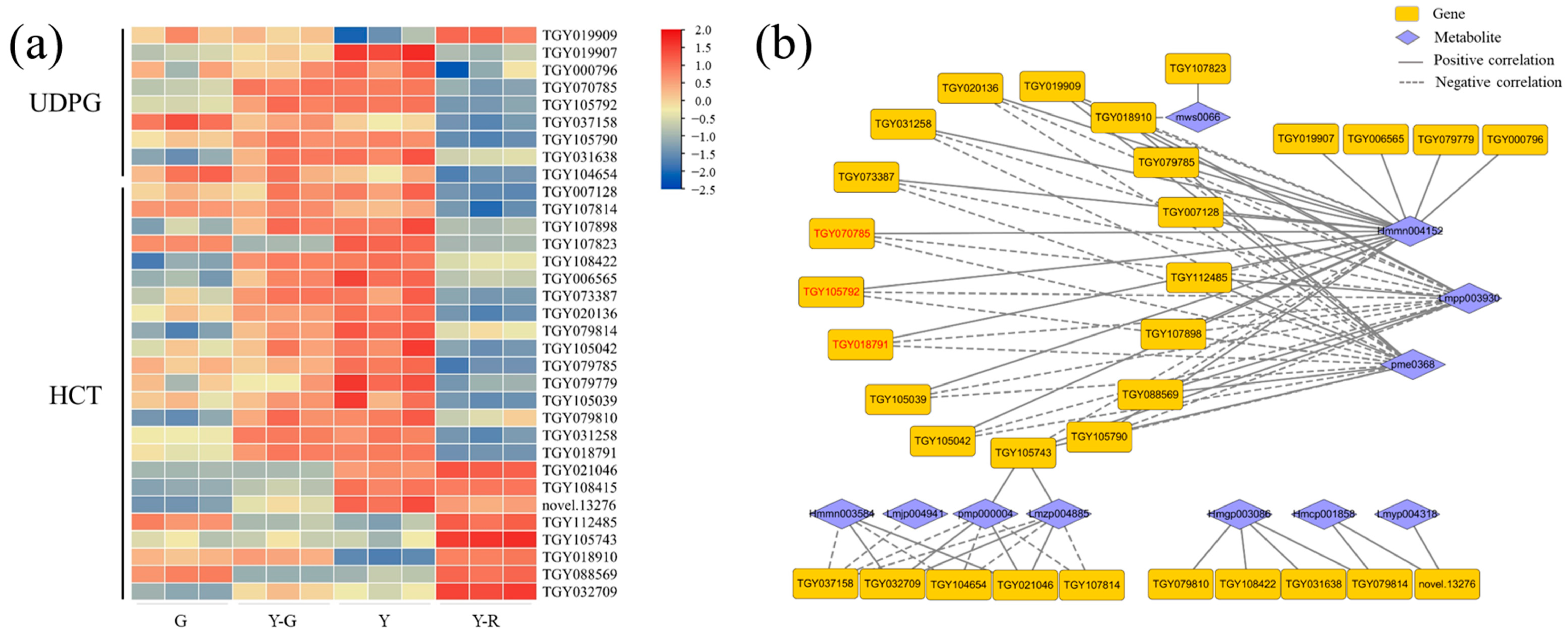
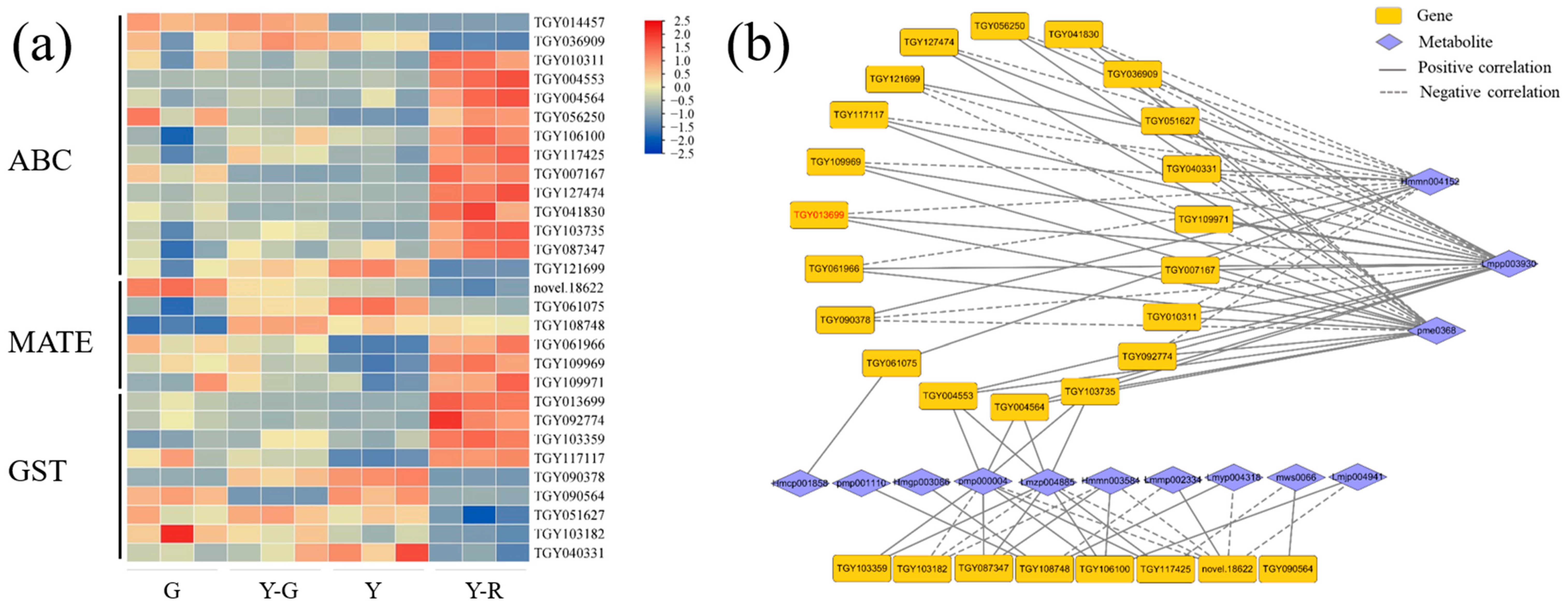
3.4. Modification, Transportation, and Transcription Factor Regulation of Flavones and Flavonols Also Affect Leaf Color in Different Yellow Tea Leaves
4. Materials and Methods
4.1. Plant Materials
4.2. Determination of Leaf SPAD Value
4.3. Transmission Electron Microscopy Analysis of Tea Leaves
4.4. Metabolomic Profiling Analysis
4.5. Transcriptomic Profiling Analysis
4.6. Conjoint Analysis of Metabolome and Transcriptome
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pan, S.Y.; Nie, Q.; Tai, H.C.; Song, X.L.; Tong, Y.F.; Zhang, L.J.; Wu, X.W.; Lin, Z.H.; Zhang, Y.Y.; Ye, D.Y.; et al. Tea and tea drinking: China’s outstanding contributions to the mankind. Chin. Med. 2022, 17, 27. [Google Scholar] [CrossRef]
- Chen, Y.; Li, Y.; Shen, C.; Xiao, L. Topics and trends in fresh tea (Camellia sinensis) leaf research: A comprehensive bibliometric study. Front. Plant Sci. 2023, 14, 1092511. [Google Scholar] [CrossRef]
- Yue, C.; Wang, Z.; Yang, P. Review: The effect of light on the key pigment compounds of photosensitive etiolated tea plant. Bot. Stud. 2021, 62, 21. [Google Scholar] [CrossRef]
- Zhu, M.Z.; Zhou, F.; Ran, L.S.; Li, Y.L.; Tan, B.; Wang, K.B.; Huang, J.A.; Liu, Z.H. Metabolic profiling and gene expression analyses of purple-leaf formation in tea cultivars (Camellia sinensis var. sinensis and var. assamica). Front. Plant Sci. 2021, 12, 606962. [Google Scholar] [CrossRef]
- Hao, X.; Zhang, W.; Liu, Y.; Zhang, H.; Ren, H.; Chen, Y.; Wang, L.; Zeng, J.; Yang, Y.; Wang, X. Pale green mutant analyses reveal the importance of CsGLKs in chloroplast developmental regulation and their effects on flavonoids biosynthesis in tea plant. Plant Physiol. Biochem. 2020, 146, 392–402. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, M.; Gao, X.; Zhou, F.; Shen, C.; Liu, Z. Multi-omics research in albino tea plants: Past, present, and future. Sci. Hortic. 2020, 261, 108943. [Google Scholar] [CrossRef]
- Xu, P.; Su, H.; Jin, R.; Mao, Y.; Xu, A.; Cheng, H.; Wang, Y.; Meng, Q. Shading Effects on Leaf Color Conversion and Biosynthesis of the Major Secondary Metabolites in the Albino Tea Cultivar ‘Yujinxiang’. J. Agric. Food Chem. 2020, 68, 2528–2538. [Google Scholar] [CrossRef]
- Zheng, Y.; Wang, P.; Chen, X.; Yue, C.; Guo, Y.; Yang, J.; Sun, Y.; Ye, N. Integrated transcriptomics and metabolomics provide novel insight into changes in specialized metabolites in an albino tea cultivar (Camellia sinensis (L.) O. Kuntz). Plant Physiol. Biochem. 2021, 160, 27–36. [Google Scholar] [CrossRef]
- Chen, X.; Wang, P.; Zheng, Y.; Gu, M.; Lin, X.; Wang, S.; Jin, S.; Ye, N. Comparison of metabolome and transcriptome of flavonoid biosynthesis pathway in a puple-leaf tea germplasm Jinmingzao and a green-leaf tea germplasm Huangdan reveals their relationship with genetic mechanisms of color formation. Int. J. Mol. Sci. 2020, 21, 4167. [Google Scholar] [CrossRef]
- Huang, R.; Wang, J.Y.; Yao, M.Z.; Ma, C.L.; Chen, L. Quantitative trait loci mapping for free amino acid content using an albino population and SNP markers provides insight into the genetic improvement of tea plants. Hortic. Res. 2023, 9, uhab029. [Google Scholar] [CrossRef]
- Wu, Q.; Chen, Z.; Sun, W.; Deng, T.; Chen, M. De novo sequencing of the leaf transcriptome reveals complex light-responsive regulatory networks in Camellia sinensis cv. Baijiguan. Front. Plant. Sci. 2016, 7, 332. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Chen, M.; Wu, Q.; Zeng, W.; Chen, Z.; Sun, W. Combined analysis of lipidomics and trancriptomics revealed the key pathways and genes of lipids in light-sensitive albino tea plant (Camellia sinensis cv. Baijiguan). Front. Plant Sci. 2022, 13, 1035119. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Kong, X.; Zhang, Y.; Wang, S.; Zhou, H.; Fang, D.; Yue, W.; Chen, C. Metabolomic Profiling in Combination with Data Association Analysis Provide Insights about Potential Metabolic Regulation Networks among Non-Volatile and Volatile Metabolites in Camellia sinensis cv. Baijiguan. Plants 2022, 11, 2557. [Google Scholar] [CrossRef]
- Scarano, A.; Chieppa, M.; Santino, A. Looking at flavonoid biodiversity in horticultural crops: A colored mine with nutritional benefits. Plants 2018, 7, 98. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Dixon, R.A. The ‘ins’ and ‘outs’ of flavonoid transport. Trends Plant Sci. 2010, 15, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Hichri, I.; Barrieu, F.; Bogs, J.; Kappel, C.; Delrot, S.; Lauvergeat, V. Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J. Exp. Bot. 2011, 62, 2465. [Google Scholar] [CrossRef] [PubMed]
- Dong, F.; Shi, Y.; Liu, M.; Fan, K.; Zhang, Q.; Ruan, J. iTRAQ-based quantitative proteomics analysis reveals the mechanism underlying the weakening of carbon metabolism in chlorotic tea leaves. Int. J. Mol. Sci. 2018, 19, 3943. [Google Scholar] [CrossRef]
- Kumari, M.; Thakur, S.; Kumar, A.; Joshi, R.; Kumar, P.; Shankar, R.; Kumar, R. Regulation of color transition in purple tea (Camellia sinensis). Planta 2020, 251, 35. [Google Scholar] [CrossRef]
- Li, N.N.; Lu, J.L.; Li, Q.S.; Zheng, X.Q.; Wang, X.C.; Wang, L.; Wang, Y.C.; Ding, C.Q.; Liang, Y.R.; Yang, Y.J. Dissection of chemical composition and associated gene expression in the pigment-deficient tea cultivar ‘Xiaoxueya’ reveals an albino phenotype and metabolite formation. Front. Plant Sci. 2019, 10, 1543. [Google Scholar] [CrossRef]
- Havaux, M.; Kloppstech, K. The protective functions of carotenoid and flavonoid pigments against excess visible radiation at chilling temperature investigated in Arabidopsis npq and tt mutants. Planta 2001, 213, 953–966. [Google Scholar] [CrossRef]
- Shitan, N.; Yazaki, K. Dynamism of vacuoles toward survival strategy in plants. Biochim. Biophys. Acta-Biomembr. 2019, 1862, 183127. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhou, B.; Li, J.; Tang, H.; Tang, J.; Yang, Z. Formation and Change of Chloroplast-Located Plant Metabolites in Response to Light Conditions. Int. J. Mol. Sci. 2018, 19, 654. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, K.; Fujii, S.; Sato, M.; Toyooka, K.; Wada, H. Specific role of phosphatidylglycerol and functional overlaps with other thylakoid lipids in Arabidopsis chloroplast biogenesis. Plant Cell Rep. 2014, 34, 631–642. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Li, P.; Xia, T.; Wan, X. Exploring plant metabolic genomics: Chemical diversity, metabolic complexity in the biosynthesis and transport of specialized metabolites with the tea plant as a model. Crit. Rev. Biotechnol. 2020, 40, 667–688. [Google Scholar] [CrossRef] [PubMed]
- Shoji, J.; Kikuma, T.; Kitamoto, K. Vesicle trafficking, organelle functions, and unconventional secretion in fungal physiology and pathogenicity. Curr. Opin. Microbiol. 2014, 20, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Liu, M.; Ruan, J. Integrated transcriptome and metabolic analyses reveals novel insights into free amino acid metabolism in Huangjinya tea cultivar. Front. Plant Sci. 2017, 8, 291. [Google Scholar] [CrossRef]
- Zhang, Q.; Liu, M.; Ruan, J. Metabolomics analysis reveals the metabolic and functional roles of flavonoids in light-sensitive tea leaves. BMC Plant Biol. 2017, 17, 64. [Google Scholar] [CrossRef]
- Wei, K.; Wang, L.; Zhang, Y.; Ruan, L.; Li, H.; Wu, L.; Xu, L.; Zhang, C.; Zhou, X.; Cheng, H.; et al. A coupled role for CsMYB75 and CsGSTF1 in anthocyanin hyperaccumulation in purple tea. Plant J. 2019, 97, 5. [Google Scholar] [CrossRef]
- Liu, G.F.; Han, Z.X.; Feng, L.; Gao, L.P.; Gao, M.J.; Gruber, M.; Zhang, Z.L.; Xia, T.; Wan, X.C.; Wei, S. Metabolic flux redirection and transcriptomic reprogramming in the albino tea cultivar ‘Yu-Jin-Xiang’ with an emphasis on catechin production. Sci. Rep. 2017, 7, 45062. [Google Scholar] [CrossRef]
- Mei, X.; Zhang, K.; Lin, Y.; Su, H.; Lin, C.; Chen, B.; Yang, H. Metabolic and transcriptomic profiling reveals etiolated mechanism in Huangyu tea (Camellia sinensis) leaves. Int. J. Mol. Sci. 2022, 23, 15044. [Google Scholar] [CrossRef]
- Xuan, L.; Zhang, C.; Yan, T.; Wu, D.; Hussain, N.; Li, Z.; Chen, M.; Pan, J. TRANSPARENT TESTA 4-mediated flavonoids negatively affect embryonic fatty acid biosynthesis in Arabidopsis. Plant Cell Environ. 2018, 41, 2773–2790. [Google Scholar] [CrossRef] [PubMed]
- Tattini, M.; Landi, M.; Brunetti, C.; Giordano, C.; Remorini, D.; Gould, K.S.; Guidi, L. Epidermal coumaroyl anthocyanins protect sweet basil against excess light stress: Multiple consequences of light attenuation. Physiol. Plant. 2014, 152, 585–598. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.Y.; Burgos, A.; Ma, L.; Zhang, Q.; Tang, D.; Ruan, J. Lipidomics analysis unravels the effect of nitrogen fertilization on lipid metabolism in tea plant (Camellia sinensis L.). BMC Plant Biol. 2017, 17, 165. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.T.; Chen, L.J.; Herrfurth, C.; Feussner, I.; Li, H. Reduced biosynthesis of digalactosyldiacylglycerol, a major chloroplast membrane lipid, leads to oxylipin overproduction and phloem cap lignification in Arabidopsis. Plant Cell 2016, 28, 219–232. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Lam, S.M.; Zuo, J.; Yuan, S.; Lv, J.; Shi, J.; Gao, L.; Chen, B.; Sui, Y.; Shui, G.; et al. Lipidomics reveals the difference of membrane lipid catabolism between chilling injury sensitive and non-sensitive green bell pepper in response to chilling. Postharvest Biol. Technol. 2021, 182, 111714. [Google Scholar] [CrossRef]
- Bejaoui, F.; Salas, J.J.; Nouairi, I.; Smaoui, A.; Abdelly, C.; Martínez-Force, E.; Youssef, N.B. Changes in chloroplast lipid contents and chloroplast ultrastructure in Sulla carnosa and Sulla coronaria leaves under salt stress. J. Plant Physiol. 2016, 198, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Q.; Wu, C.; Cheng, T.T.; Yan, Y.; Zhang, L.; Wan, Y.L.; Wang, J.W.; Liu, Q.Z.; Feng, Z.; Liu, Y. Comparative Analysis of the Metabolome and Transcriptome between the Green and Yellow-Green Regions of Variegated Leaves in a Mutant Variety of the Tree Species Pteroceltis tatarinowii. Int. J. Mol. Sci. 2022, 23, 4950. [Google Scholar] [CrossRef]
- Kobayashi, K. Role of membrane glycerolipids in photosynthesis, thylakoid biogenesis and chloroplast development. J. Plant Res. 2016, 129, 565–580. [Google Scholar] [CrossRef]
- Amaro, A.L.; Almeida, D.P.F. Lysophosphatidylethanolamine effects on horticultural commodities: A review. Postharvest Biol. Technol. 2013, 78, 92–102. [Google Scholar] [CrossRef]
- Tan, X.; Wang, Q.; Tian, B.; Zhang, H.; Lu, D.; Zhou, J. A Brassica napus Lipase Locates at the Membrane Contact Sites Involved in Chloroplast Development. PLoS ONE 2011, 6, e26831. [Google Scholar] [CrossRef]
- Yaeno, T.; Matsuda, O.; Iba, K. Role of chloroplast trienoic fatty acids in plant disease defense responses. Plant J. 2004, 40, 931–941. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Zheng, Y.; Guo, Y.; Liu, B.; Jin, S.; Liu, S.; Zhao, F.; Chen, X.; Sun, Y.; Yang, J.; et al. Widely Targeted Metabolomic and Transcriptomic Analyses of a Novel Albino Tea Mutant of “Rougui”. Forests 2020, 11, 229. [Google Scholar] [CrossRef]
- Shin, Y.H.; Yang, R.; Shi, Y.L.; Li, X.M.; Fu, Q.Y.; Lu, J.L.; Ye, J.H. Light-sensitive albino tea plants and their characterization. HortScience 2018, 53, 144–147. [Google Scholar] [CrossRef]
- Zhang, Q.; Li, C.; Jiao, Z.; Ruan, J.; Liu, M. Integration of Metabolomics and Transcriptomics Reveal the Mechanism Underlying Accumulation of Flavonols in Albino Tea Leaves. Molecules 2022, 27, 5792. [Google Scholar] [CrossRef] [PubMed]
- Herna’ndez, I.; Alegre, L.; Breusege, F.V.; Munne’-Bosch, S. How relevant are flavonoids as antioxidants in plants? Trends Plant Sci. 2009, 14, 125–132. [Google Scholar] [CrossRef]
- Agati, G.; Azzarello, E.; Pollastri, S.; Tattini, M. Flavonoids as antioxidants in plants: Location and functional significance. Plant Sci. 2012, 196, 67–76. [Google Scholar] [CrossRef]
- Tanaka, Y.; Brugliera, F.; Kalc, G.; Senior, M.; Dyson, B.; Nakamura, N.; Katsumoto, Y.; Chandler, S. Flower Color Modification by Engineering of the Flavonoid Biosynthetic Pathway: Practical Perspectives. Biosci. Biotechnol. Biochem. 2010, 74, 1760–1769. [Google Scholar] [CrossRef]
- Hoffmann, L.; Besseau, S.; Geoffroy, P.; Ritzenthaler, C.; Meyer, D.; Lapierre, C.; Pollet, B.; Legrand, M. Silencing of hydroxycinnamoyl-coenzyme A shikimate/quinate hydroxycinnamoyltransferase affects phenylpropanoid biosynthesis. Plant Cell 2004, 16, 1446–1465. [Google Scholar] [CrossRef]
- Rinaldo, A.R.; Cavallini, E.; Jia, Y.; Moss, S.M.A.; McDavid, D.A.J.; Hooper, L.C.; Robinson, S.P.; Tornielli, G.B.; Zenoni, S.; Ford, C.M.; et al. A grapevine anthocyanin acyltransferase, transcriptionally regulated by VvMYBA, can produce most acylated anthocyanins present in grape skins. Plant Physiol. 2015, 169, 1897–1916. [Google Scholar] [CrossRef]
- Lim, E.K.; Ashford, D.A.; Hou, B.; Jackson, R.G.; Bowles, D.J. Arabidopsis Glycosyltransferases as Biocatalysts in Fermentation for Regioselective Synthesis of Diverse Quercetin Glucosides. Biotechnol. Bioeng. 2004, 87, 623–631. [Google Scholar] [CrossRef]
- Griesser, M.; Vitzthum, F.; Fink, B.; Bellido, M.L.; Raasch, C.; Munoz-Blanco, J.; Schwab, W. Multi-substrate flavonol O-glucosyltransferases from strawberry (Fragaria × ananassa) achene and receptacle. J. Exp. Bot. 2008, 59, 2611–2625. [Google Scholar] [CrossRef] [PubMed]
- Montefiori, M.; Espley, R.V.; Stevenson, D.; Cooney, J.; Datson, P.M.; Saiz, A.; Atkinson, R.G.; Hellens, R.P.; Allan, A.C. Identification and characterisation of F3GT1 and F3GGT1, two glycosyltransferases responsible for anthocyanin biosynthesis in red-fleshed kiwifruit (Actinidia chinensis). Plant J. 2011, 65, 106–118. [Google Scholar] [CrossRef] [PubMed]
- Di, S.; Yan, F.; Rodas, F.R.; Rodriguez, T.O.; Murai, Y.; Iwashina, T.; Sugawara, S.; Mori, T.; Nakabayashi, R.; Yonekura-Sakakibara, K.; et al. Linkage mapping, molecular cloning and functional analysis of soybean gene Fg3 encoding flavonol 3-O-glucoside/galactoside (1→ 2) glucosyltransferase. BMC Plant Biol. 2015, 15, 126. [Google Scholar] [CrossRef] [PubMed]
- Morita, Y.; Hoshino, A.; Kikuchi, Y.; Okuhara, H.; Ono, E.; Tanaka, Y.; Fukui, Y.; Saito, N.; Nitasaka, E.; Noguchi, H.; et al. Japanese morning glory dusky mutants displaying reddish-brown or purplish-gray flowers are deficient in a novel glycosylation enzyme for anthocyanin biosynthesis, UDP-glucose:anthocyanidin 3-O-glucoside-2″-O-glucosyltransferase, due to 4-bp insertions in the gene. Plant J. 2005, 42, 353–363. [Google Scholar] [CrossRef] [PubMed]
- Levsh, O.; Chiang, Y.C.; Tung, C.F.; Noel, J.P.; Wang, Y.; Weng, J.K. Dynamic Conformational States Dictate Selectivity toward the Native Substrate in a Substrate-Permissive Acyltransferase. Biochemistry 2016, 55, 6314–6326. [Google Scholar] [CrossRef] [PubMed]
- Eudes, A.; Pereira, J.H.; Yogiswara, S.; Wang, G.; Benites, V.T.; Baidoo, E.E.; Lee, T.S.; Adams, P.D.; Keasling, J.D.; Loqué, D. Exploiting the Substrate Promiscuity of Hydroxycinnamoyl-CoA:Shikimate Hydroxycinnamoyl Transferase to Reduce Lignin. Plant Cell Physiol. 2016, 57, 568–579. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, L.; Maury, S.; Martz, F.; Geoffroy, P.; Legrand, M. Purification, cloning, and properties of an acyltransferase controlling shikimate and quinate ester intermediates in phenylpropanoid metabolism. J. Biol. Chem. 2003, 278, 95–103. [Google Scholar] [CrossRef]
- Appelhagen, I.; Jahns, O.; Bartelniewoehner, L.; Sagasser, M.; Weisshaar, B.; Stracke, R. Leucoanthocyanidin Dioxygenase in Arabidopsis thaliana: Characterization of mutant alleles and regulation by MYB-BHLH-TTG1 transcription factor complexes. Gene 2011, 484, 61–68. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, G.; Chen, J.; Xie, N.; Huang, J.; Shen, C. Translational landscape and metabolic characteristics of the etiolated tea plant (Camellia sinensis). Sci. Hortic. 2022, 303, 111193. [Google Scholar] [CrossRef]
- Li, X.; Gao, P.; Cui, D.; Wu, L.; Parkin, I.; Saberianfar, R.; Menassa, R.; Pan, H.; Westcott, N.; Gruber, M.Y. The Arabidopsis tt19-4 mutant differentially accumulates proanthocyanidin and anthocyanin through a 3′ amino acid substitution in glutathione S-transferase. Plant Cell Environ. 2011, 34, 374–388. [Google Scholar] [CrossRef]
- Kitamura, S.; Matsuda, F.; Tohge, T.; Yonekura-Sakakibara, K.; Yamazaki, M.; Saito, K.; Narumi, I. Metabolic profiling and cytological analysis of proanthocyanidins in immature seeds of Arabidopsis thaliana flavonoid accumulation mutants. Plant J. 2010, 62, 549–559. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, S.; Shikazono, N.; Tanaka, A. TRANSPARENT TESTA 19 is involved in the accumulation of both anthocyanins and proanthocyanidins in Arabidopsis. Plant J. 2004, 37, 104–114. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Lin, Y.; Zhang, K.; Rothenberg, D.O.; Zhang, H.; Zhou, H.; Su, H.; Zhang, L. The Chemical Composition and Transcriptome Analysis Reveal the Mechanism of Color Formation in Tea (Camellia sinensis) Pericarp. Int. J. Mol. Sci. 2023, 24, 13198. [Google Scholar] [CrossRef] [PubMed]
- Man, Y.P.; Wang, Y.C.; Li, Z.Z.; Jiang, Z.W.; Yang, H.L.; Gong, J.J.; He, S.S.; Wu, S.Q.; Yang, Z.Q.; Zheng, J.; et al. High-temperature inhibition of biosynthesis and transportation of anthocyanins results in the poor red coloration in red-fleshed Actinidia chinensis. Physiol. Plant. 2015, 153, 565–583. [Google Scholar] [CrossRef] [PubMed]
- Ampomah-Dwamena, C.; Thrimawithana, A.H.; Dejnoprat, S.; Lewis, D.; Espley, R.V.; Allan, A.C. A kiwifruit (Actinidia deliciosa) R2R3-MYB transcription factor modulates chlorophyll and carotenoid accumulation. New Phytol. 2019, 221, 309–325. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Tang, W.; Hu, Y.; Zhang, Y.; Sun, J.; Guo, X.; Lu, H.; Yang, Y.; Fang, C.; Niu, X.; et al. A MYB/bHLH complex regulates tissue-specific anthocyanin biosynthesis in the inner pericarp of red-centered kiwifruit Actinidia chinensis cv. Hongyang. Plant J. 2019, 99, 359–378. [Google Scholar] [CrossRef]
- Sun, B.; Zhu, Z.; Cao, P.; Chen, H.; Chen, C.; Zhou, X.; Mao, Y.; Lei, J.; Jiang, Y.; Meng, W.; et al. Purple foliage coloration in tea (Camellia sinensis L.) arises from activation of the R2R3-MYB transcription factor CsAN1. Sci. Rep. 2016, 6, 32534. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, S.; Shi, L.; Gong, D.; Zhang, S.; Zhao, Q.; Zhan, D.; Vasseur, L.; Wang, Y.; Yu, J.; et al. Haplotype-resolved genome assembly provides insights into evolutionary history of the tea plant. Nat. Genet. 2021, 53, 1250–1259. [Google Scholar] [CrossRef]
- Livaka, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using realtime quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Wang, L.; Kong, X.; Chen, Z.; Zhong, S.; Li, X.; Shan, R.; You, X.; Wei, K.; Chen, C. Integrated Analysis of Metabolome and Transcriptome Revealed Different Regulatory Networks of Metabolic Flux in Tea Plants [Camellia sinensis (L.) O. Kuntze] with Varied Leaf Colors. Int. J. Mol. Sci. 2024, 25, 242. https://doi.org/10.3390/ijms25010242
Zhang Y, Wang L, Kong X, Chen Z, Zhong S, Li X, Shan R, You X, Wei K, Chen C. Integrated Analysis of Metabolome and Transcriptome Revealed Different Regulatory Networks of Metabolic Flux in Tea Plants [Camellia sinensis (L.) O. Kuntze] with Varied Leaf Colors. International Journal of Molecular Sciences. 2024; 25(1):242. https://doi.org/10.3390/ijms25010242
Chicago/Turabian StyleZhang, Yazhen, Liyuan Wang, Xiangrui Kong, Zhihui Chen, Sitong Zhong, Xinlei Li, Ruiyang Shan, Xiaomei You, Kang Wei, and Changsong Chen. 2024. "Integrated Analysis of Metabolome and Transcriptome Revealed Different Regulatory Networks of Metabolic Flux in Tea Plants [Camellia sinensis (L.) O. Kuntze] with Varied Leaf Colors" International Journal of Molecular Sciences 25, no. 1: 242. https://doi.org/10.3390/ijms25010242
APA StyleZhang, Y., Wang, L., Kong, X., Chen, Z., Zhong, S., Li, X., Shan, R., You, X., Wei, K., & Chen, C. (2024). Integrated Analysis of Metabolome and Transcriptome Revealed Different Regulatory Networks of Metabolic Flux in Tea Plants [Camellia sinensis (L.) O. Kuntze] with Varied Leaf Colors. International Journal of Molecular Sciences, 25(1), 242. https://doi.org/10.3390/ijms25010242