Neuroprotective Effect of Antiapoptotic URG7 Protein on Human Neuroblastoma Cell Line SH-SY5Y
Abstract
:1. Introduction
2. Results
2.1. URG7 Expression in Mouse Brain Tissues and Primary Cells
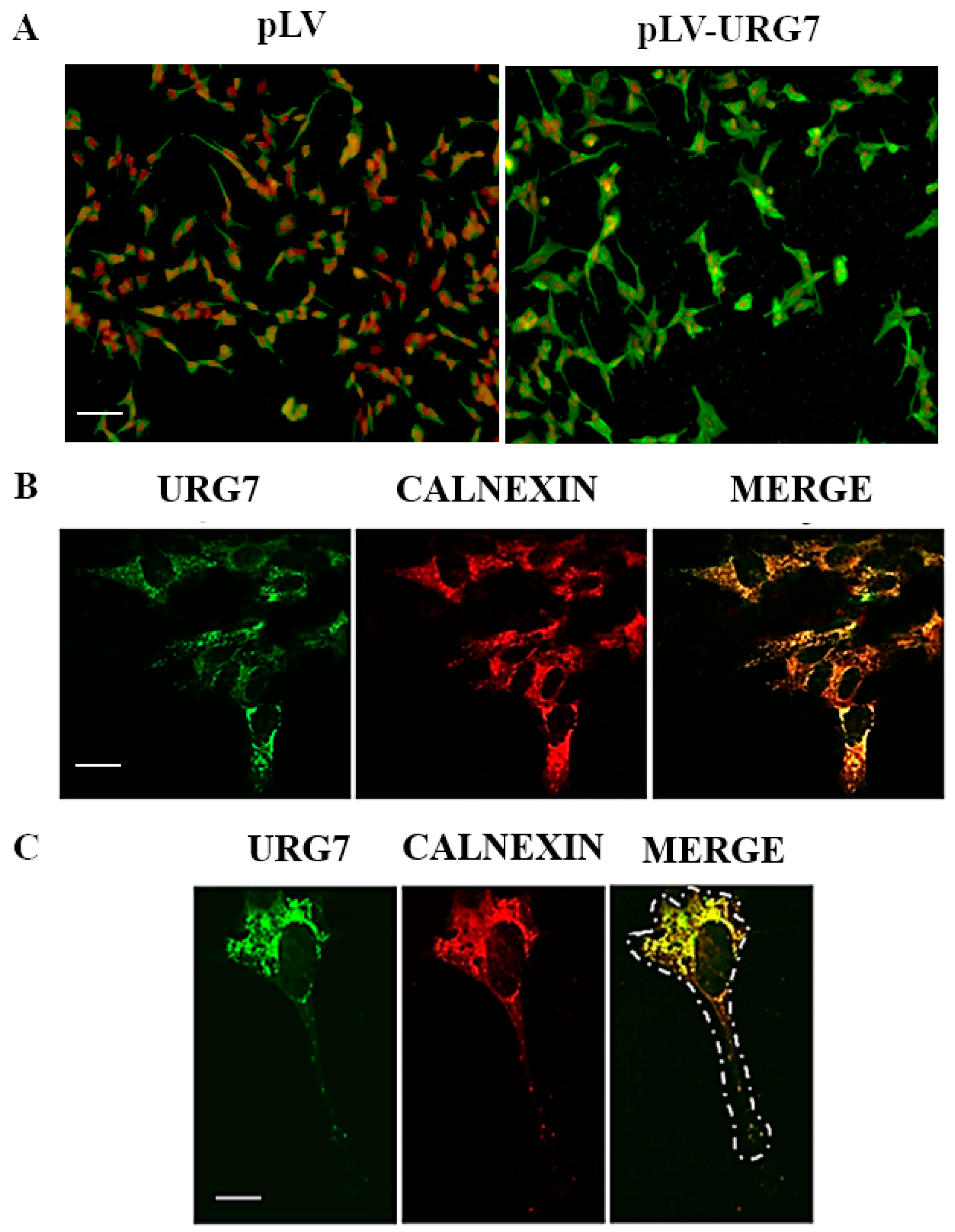
2.2. Stable Clones’ Generation and Intracellular Localization of URG7 in SH-SY5Y Cells
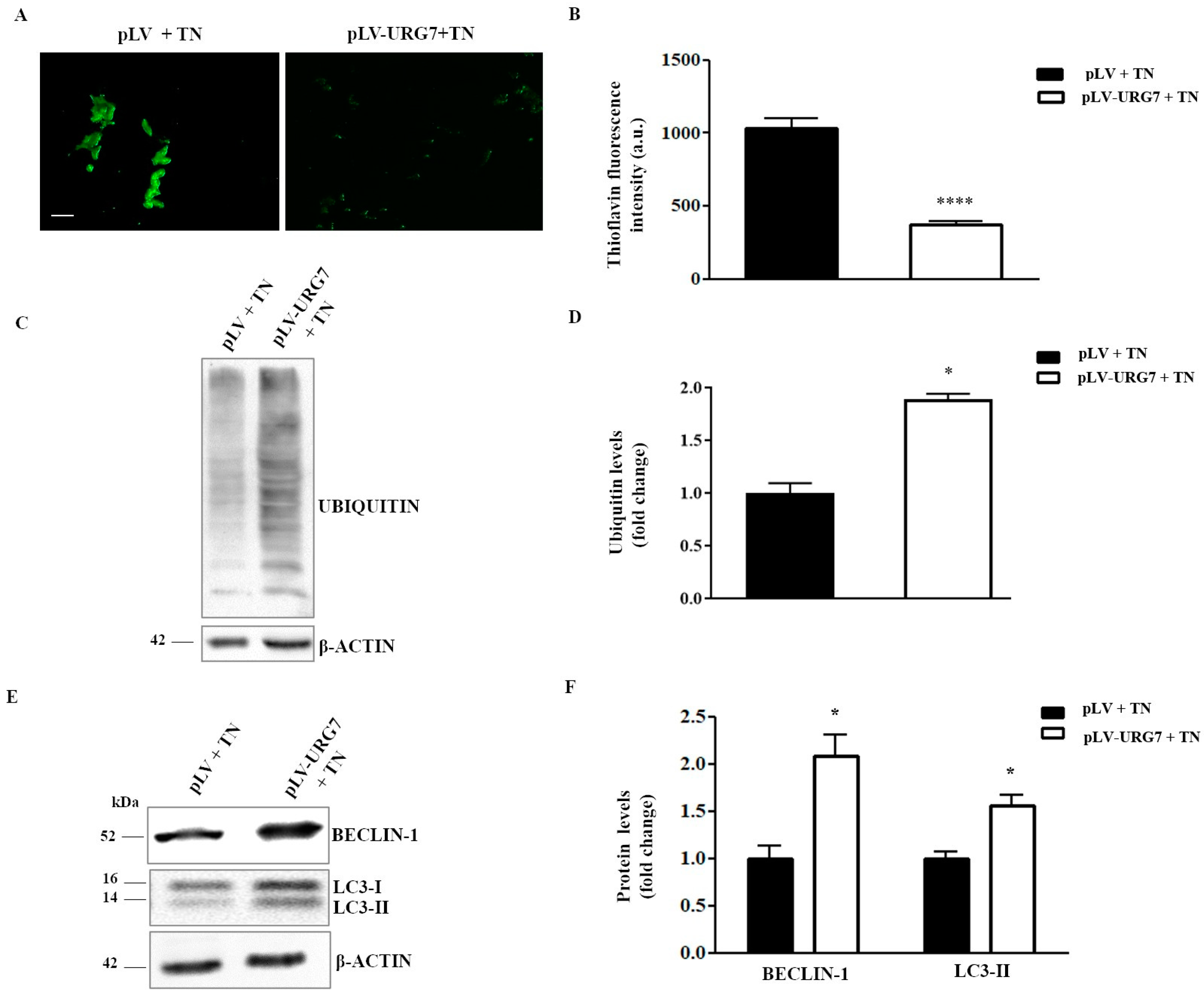
2.3. Mitigation of ER Stress in URG7-SH-SY5Y Stable Clones
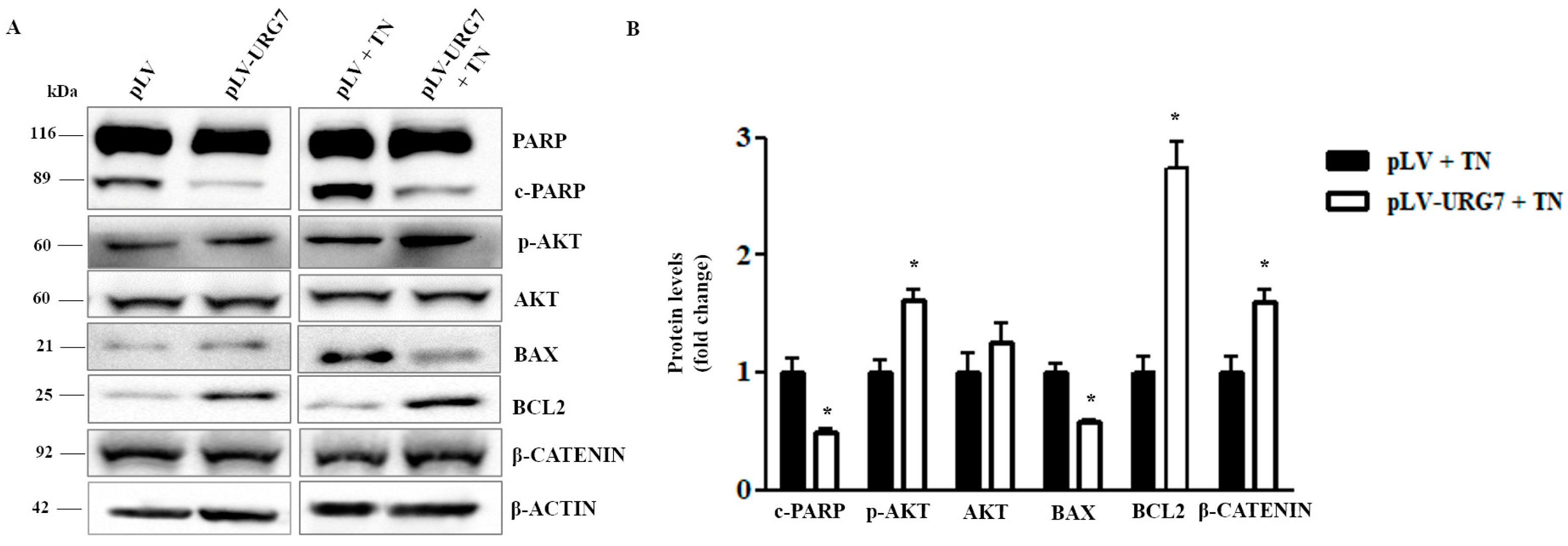
2.4. URG7 Favors Cell Survival and Inhibits Apoptosis in ER Stressed Cells
2.5. URG7 Affects Protein Catabolism in ER Stressed Cells
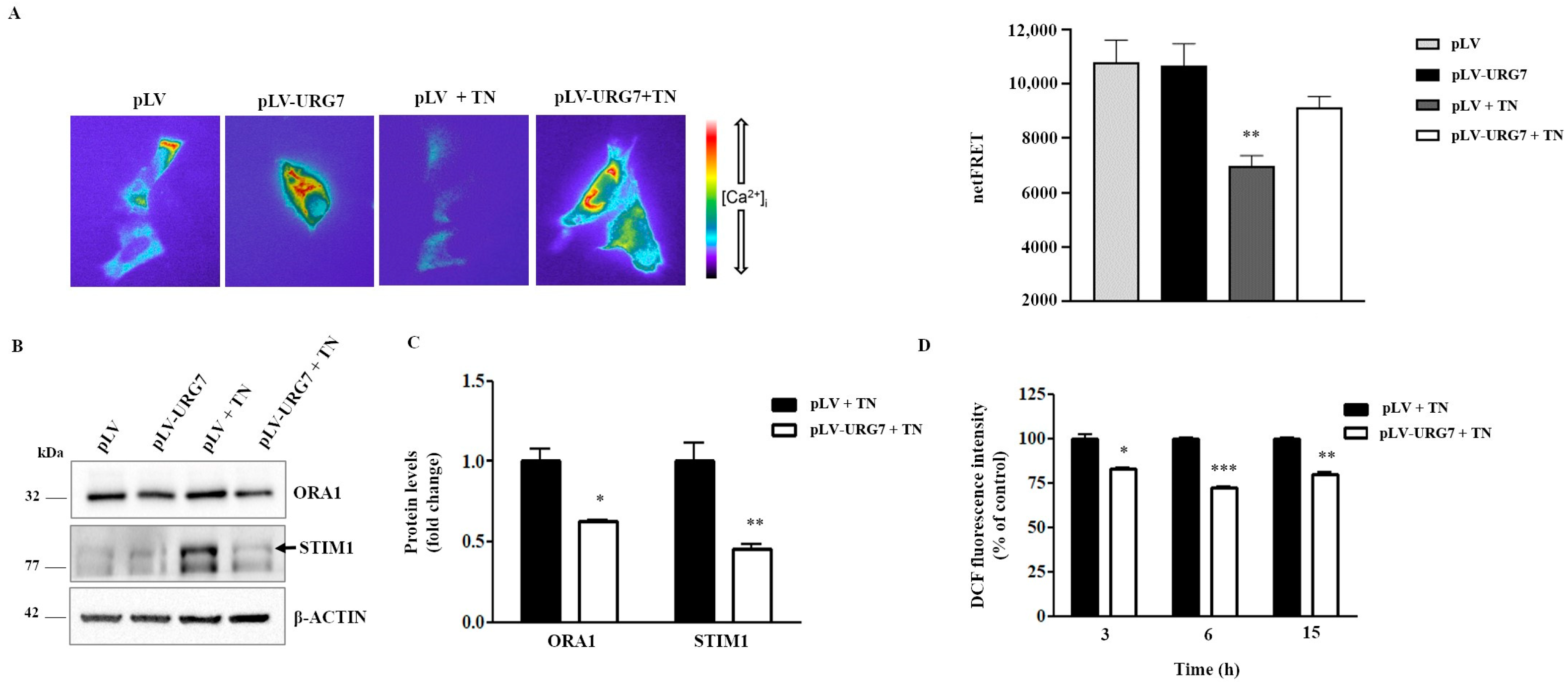
2.6. URG7 Affects ER Calcium Depletion and Reduces ROS Production in SH-SY5Y Stressed Cells
3. Discussion
4. Materials and Methods
4.1. Reagents and Antibodies
4.2. Animals, Brain Lysis and Primary Cultures
4.3. Cell Culture and Treatments
4.4. URG7 Stable Cell Line Generation
4.5. Immunofluorescence and Confocal Microscopy
4.6. Western Blotting Analysis
4.7. Determination of Intracellular Reactive Oxygen Species (ROS)
4.8. Measurement of Intraluminal ER Ca2+
4.9. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Klaips, C.L.; Jayaraj, G.G.; Hartl, F.U. Pathways of cellular proteostasis in aging and disease. J. Cell Biol. 2018, 217, 51–63. [Google Scholar] [CrossRef] [PubMed]
- Hetz, C.; Zhang, K.; Kaufman, R.J. Mechanisms, regulation and functions of the unfolded protein response. Nat. Rev. Mol. Cell Biol. 2020, 21, 421–438. [Google Scholar] [CrossRef] [PubMed]
- Ventura, S. Protein misfolding diseases. Futur. Sci. OA 2015, 1, FSO38. [Google Scholar] [CrossRef] [PubMed]
- Hetz, C.; Saxena, S. ER stress and the unfolded protein response in neurodegeneration. Nat. Rev. Neurol. 2017, 13, 477–491. [Google Scholar] [CrossRef] [PubMed]
- Ostuni, A.; Lara, P.; Armentano, M.F.; Miglionico, R.; Salvia, A.M.; Mönnich, M.; Carmosino, M.; Lasorsa, F.M.; Monné, M.; Nilsson, I.; et al. The hepatitis B x antigen anti-apoptotic effector URG7 is localized to the endoplasmic reticulum membrane. FEBS Lett. 2013, 587, 3058–3062. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Lara, P.; Ostuni, A.; Presto, J.; Johansson, J.; Nilsson, I.; Kim, H. Live-cell topology assessment of URG7, MRP6102 and SP-C using glycosylatable green fluorescent protein in mammalian cells. Biochem. Biophys. Res. Commun. 2014, 450, 1587–1592. [Google Scholar] [CrossRef]
- Lian, Z.; Liu, J.; Pan, J.; Satiroglu Tufan, N.L.; Zhu, M.; Arbuthnot, P.; Kew, M.; Clayton, M.M.; Feitelson, M.A. A cellular gene up-regulated by hepatitis B virus—Encoded X antigen promotes hepatocellular growth and survival. Hepatology 2001, 34, 146–157. [Google Scholar] [CrossRef]
- Feitelson, M.A. Mechanisms of HBx Mediated Liver Cancer: Multiple Pathways and Opportunities. In Hepatocellular Carcinoma—Basic Research; Lau, J.W.Y., Ed.; InTech: London, UK, 2012. [Google Scholar]
- Armentano, M.F.; Caterino, M.; Miglionico, R.; Ostuni, A.; Pace, M.C.; Cozzolino, F.; Monti, M.; Milella, L.; Carmosino, M.; Pucci, P.; et al. New insights on the functional role of URG7 in the cellular response to ER stress. Biol. Cell 2018, 110, 147–158. [Google Scholar] [CrossRef]
- Dufey, E.; Sepúlveda, D.; Rojas-Rivera, D.; Hetz, C. Cellular mechanisms of endoplasmic reticulum stress signaling in health and disease. 1. An overview. Am. J. Physiol. Cell Physiol. 2014, 307, C582–C594. [Google Scholar] [CrossRef]
- Oda, T.; Kosuge, Y.; Arakawa, M.; Ishige, K.; Ito, Y. Distinct mechanism of cell death is responsible for tunicamycin-induced ER stress in SK-N-SH and SH-SY5Y cells. Neurosci. Res. 2008, 60, 29–39. [Google Scholar] [CrossRef]
- Wei, J.; Zhao, F.; Shang, Y.; Liu, X.; Huang, H.; Lao, F. Protective Effect of Zeaxanthin against Tunicamycin-induced Cell Damage in SH-SY5Y Cell. Food Sci. Technol. Res. 2018, 24, 1101–1109. [Google Scholar] [CrossRef]
- Beriault, D.R.; Werstuck, G.H. Detection and quantification of endoplasmic reticulum stress in living cells using the fluorescent compound, Thioflavin T. Biochim. Biophys. Acta Mol. Cell Res. 2013, 1833, 2293–2301. [Google Scholar] [CrossRef] [PubMed]
- Lebeau, P.F.; Platko, K.; Byun, J.H.; Austin, R.C. Calcium as a reliable marker for the quantitative assessment of endoplasmic reticulum stress in live cells. J. Biol. Chem. 2021, 296, 100779. [Google Scholar] [CrossRef] [PubMed]
- Sattler, R.; Tymianski, M. Molecular mechanisms of calcium-dependent excitotoxicity. J. Mol. Med. 2000, 78, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Wegierski, T.; Kuznicki, J. Neuronal calcium signaling via store-operated channels in health and disease. Cell Calcium 2018, 74, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Joseph, S.K.; Booth, D.M.; Young, M.P.; Hajnóczky, G. Redox regulation of ER and mitochondrial Ca2+ signaling in cell survival and death. Cell Calcium 2019, 79, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Remondelli, P.; Renna, M. The endoplasmic reticulum unfolded protein response in neurodegenerative disorders and its potential therapeutic significance. Front. Mol. Neurosci. 2017, 10, 187. [Google Scholar] [CrossRef]
- Dandurand, J.; Ostuni, A.; Armentano, M.F.; Crudele, M.A.; Dolce, V.; Marra, F.; Samouillan, V.; Bisaccia, F.; Dandurand, J.; Ostuni, A.; et al. Calorimetry and FTIR reveal the ability of URG7 protein to modify the aggregation state of both cell lysate and amylogenic α-synuclein. AIMS Biophys. 2020, 7, 189–203. [Google Scholar] [CrossRef]
- Kovalevich, J.; Langford, D. Neuronal Cell Culture. Neuronal Cell Cult. Methods Protoc. 2013, 1078, 35–44. [Google Scholar] [CrossRef]
- Xicoy, H.; Wieringa, B.; Martens, G.J.M. The SH-SY5Y cell line in Parkinson’s disease research: A systematic review. Mol. Neurodegener. 2017, 12, 10. [Google Scholar] [CrossRef]
- Zhang, L.H.; Zhang, X. Roles of GRP78 in physiology and cancer. J. Cell. Biochem. 2010, 110, 1299–1305. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, I.M.; Abdelmalek, D.H.; Elfiky, A.A. GRP78: A cell’s response to stress. Life Sci. 2019, 226, 156–163. [Google Scholar] [CrossRef] [PubMed]
- Voronin, M.V.; Abramova, E.V.; Verbovaya, E.R.; Vakhitova, Y.V.; Seredenin, S.B. Chaperone-Dependent Mechanisms as a Pharmacological Target for Neuroprotection. Int. J. Mol. Sci. 2023, 24, 823. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Tirasophon, W.; Shen, X.; Michalak, M.; Prywes, R.; Okada, T.; Yoshida, H.; Mori, K.; Kaufman, R.J. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 2002, 16, 452–466. [Google Scholar] [CrossRef] [PubMed]
- Okada, T.; Yoshida, H.; Akazawa, R.; Negishi, M.; Mori, K. Distinct roles of activating transcription factor 6 (ATF6) and double-stranded RNA-activated protein kinase-like endoplasmic reticulum kinase (PERK) in transcription during the mammalian unfolded protein response. Biochem. J. 2002, 366, 585–594. [Google Scholar] [CrossRef] [PubMed]
- Pitale, P.; Gorbatyuk, O.; Gorbatyuk, M. Neurodegeneration: Keeping ATF4 on a tight leash. Front. Cell. Neurosci. 2017, 11, 410. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.Z.; Lawson, B.; Brewer, J.W.; Zinszner, H.; Sanjay, A.; Mi, L.J.; Boorstein, R.; Kreibich, G.; Hendershot, L.M.; Ron, D. Signals from the stressed endoplasmic reticulum induce C/EBP-homologous protein (CHOP/GADD153). Mol. Cell. Biol. 1996, 16, 4273–4280. [Google Scholar] [CrossRef]
- Zinszner, H.; Kuroda, M.; Wang, X.Z.; Batchvarova, N.; Lightfoot, R.T.; Remotti, H.; Stevens, J.L.; Ron, D. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 1998, 12, 982–995. [Google Scholar] [CrossRef]
- Oyadomari, S.; Araki, E.; Mori, M. Endoplasmic reticulum stress-mediated apoptosis in pancreatic β-cells. Apoptosis 2002, 7, 335–345. [Google Scholar] [CrossRef]
- Distelhorst, C.W.; McCormick, T.S. Bcl-2 acts subsequent to and independent of Ca2+ fluxes to inhibit apoptosis in thapsigargin and glucocorticoid treated mouse lymphoma cells. Cell Calcium 1996, 19, 473–483. [Google Scholar] [CrossRef]
- Wei, M.C.; Zong, W.X.; Cheng, E.H.Y.; Lindsten, T.; Panoutsakopoulou, V.; Ross, A.J.; Roth, K.A.; Macgregor, G.R.; Thompson, C.B.; Korsmeyer, S.J. Proapoptotic BAX and BAK: A requisite gateway to mitochondrial dysfunction and death. Science 2001, 292, 727–730. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Yankner, B.A. Apoptosis in the nervous system. Nature 2000, 407, 802–809. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Lian, Z.; Wallet, S.; Feitelson, M.A. The hepatitis B x antigen effector, URG7, blocks tumour necrosis factor α-mediated apoptosis by activation of phosphoinositol 3-kinase and β-catenin. J. Gen. Virol. 2007, 88, 3275–3285. [Google Scholar] [CrossRef] [PubMed]
- Salvesen, G.S.; Dixit, V.M. Caspases: Intracellular Signaling by Proteolysis. Cell 1997, 91, 443–446. [Google Scholar] [CrossRef] [PubMed]
- Ogata, M.; Hino, S.; Saito, A.; Morikawa, K.; Kondo, S.; Kanemoto, S.; Murakami, T.; Taniguchi, M.; Tanii, I.; Yoshinaga, K.; et al. Autophagy Is Activated for Cell Survival after Endoplasmic ReticulumStress. Mol. Cell. Biol. 2006, 26, 9220–9231. [Google Scholar] [CrossRef] [PubMed]
- Rashid, H.-O.; Yadav, R.K.; Kim, H.-R.; Chae, H.-J. ER stress: Autophagy induction, inhibition and selection. Autophagy 2015, 11, 1956–1977. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Arikkath, J.; Yang, L.; Guo, M.L.; Periyasamy, P.; Buch, S. Interplay of endoplasmic reticulum stress and autophagy in neurodegenerative disorders. Autophagy 2016, 12, 225–244. [Google Scholar] [CrossRef]
- Hosoi, T.; Nomura, J.; Tanaka, K.; Ozawa, K.; Nishi, A.; Nomura, Y. Link between endoplasmic reticulum stress and autophagy in neurodegenerative diseases. Endoplasmic Reticulum Stress Dis. 2017, 4, 37–45. [Google Scholar] [CrossRef]
- B’Chir, W.; Maurin, A.C.; Carraro, V.; Averous, J.; Jousse, C.; Muranishi, Y.; Parry, L.; Stepien, G.; Fafournoux, P.; Bruhat, A. The eIF2α/ATF4 pathway is essential for stress-induced autophagy gene expression. Nucleic Acids Res. 2013, 41, 7683–7699. [Google Scholar] [CrossRef]
- Calì, T.; Ottolini, D.; Brini, M. Calcium and endoplasmic reticulum-mitochondria tethering in neurodegeneration. DNA Cell Biol. 2013, 32, 140–146. [Google Scholar] [CrossRef]
- Görlach, A.; Bertram, K.; Hudecova, S.; Krizanova, O. Calcium and ROS: A mutual interplay. Redox Biol. 2015, 6, 260–271. [Google Scholar] [CrossRef] [PubMed]
- Bahar, E.; Kim, H.; Yoon, H. ER stress-mediated signaling: Action potential and Ca2+ as key players. Int. J. Mol. Sci. 2016, 17, 1558. [Google Scholar] [CrossRef] [PubMed]
- Pietrafesa, G.; De Zio, R.; Scorza, S.I.; Armentano, M.F.; Pepe, M.; Forleo, C.; Procino, G.; Gerbino, A.; Svelto, M.; Carmosino, M. Targeting unfolded protein response reverts ER stress and ER Ca2+ homeostasis in cardiomyocytes expressing the pathogenic variant of Lamin A/C R321X. J. Transl. Med. 2023, 21, 340. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.S.; Kaufman, R.J. Endoplasmic Reticulum Stress and Oxidative Stress in Cell Fate Decision and Human Disease. Antioxid. Redox Signal. 2014, 21, 396. [Google Scholar] [CrossRef] [PubMed]
- Martinez, A.; Lopez, N.; Gonzalez, C.; Hetz, C. Targeting of the unfolded protein response (UPR) as therapy for Parkinson’s disease. Biol. Cell 2019, 111, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Siafaka, P.I.; Okur, M.E.; Erim, P.D.; Çağlar, E.Ş.; Özgenç, E.; Gündoğdu, E.; Köprülü, R.E.P.; Karantas, I.D.; Üstündağ Okur, N. Protein and Gene Delivery Systems for Neurodegenerative Disorders: Where Do We Stand Today? Pharmaceutics 2022, 14, 2425. [Google Scholar] [CrossRef]
- Emmi, A.; Sandre, M.; Russo, F.P.; Tombesi, G.; Garrì, F.; Campagnolo, M.; Carecchio, M.; Biundo, R.; Spolverato, G.; Macchi, V.; et al. Duodenal alpha-Synuclein Pathology and Enteric Gliosis in Advanced Parkinson’s Disease. Mov. Disord. 2023, 38, 885–894. [Google Scholar] [CrossRef]
- Iannotta, L.; Biosa, A.; Kluss, J.H.; Tombesi, G.; Kaganovich, A.; Cogo, S.; Plotegher, N.; Civiero, L.; Lobbestael, E.; Baekelandt, V.; et al. Divergent Effects of G2019S and R1441C LRRK2 Mutations on LRRK2 and Rab10 Phosphorylations in Mouse Tissues. Cells 2020, 9, 2344. [Google Scholar] [CrossRef]
- Pike, A.F.; Longhena, F.; Faustini, G.; van Eik, J.M.; Gombert, I.; Herrebout, M.A.C.; Fayed, M.M.H.E.; Sandre, M.; Varanita, T.; Teunissen, C.E.; et al. Dopamine signaling modulates microglial NLRP3 inflammasome activation: Implications for Parkinson’s disease. J. Neuroinflamm. 2022, 19, 50. [Google Scholar] [CrossRef]
- Masato, A.; Plotegher, N.; Terrin, F.; Sandre, M.; Faustini, G.; Thor, A.; Adams, S.; Berti, G.; Cogo, S.; De Lazzari, F.; et al. DOPAL initiates αSynuclein-dependent impaired proteostasis and degeneration of neuronal projections in Parkinson’s disease. NPJ Park. Dis. 2023, 9, 42. [Google Scholar] [CrossRef]
- Ostuni, A.; Cuviello, F.; Salvia, A. Immunochemical Characterization of the Specific Sequence of URG7 Protein. J. Biomol. Res. Ther. 2016, 5, 3–7. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Grynkiewicz, G.; Poenie, M.; Tsien, R.Y. A new generation of Ca2+ indicators with greatly improved fluorescence properties. J. Biol. Chem. 1985, 260, 3440–3450. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nigro, I.; Miglionico, R.; Carmosino, M.; Gerbino, A.; Masato, A.; Sandre, M.; Bubacco, L.; Antonini, A.; Rinaldi, R.; Bisaccia, F.; et al. Neuroprotective Effect of Antiapoptotic URG7 Protein on Human Neuroblastoma Cell Line SH-SY5Y. Int. J. Mol. Sci. 2024, 25, 481. https://doi.org/10.3390/ijms25010481
Nigro I, Miglionico R, Carmosino M, Gerbino A, Masato A, Sandre M, Bubacco L, Antonini A, Rinaldi R, Bisaccia F, et al. Neuroprotective Effect of Antiapoptotic URG7 Protein on Human Neuroblastoma Cell Line SH-SY5Y. International Journal of Molecular Sciences. 2024; 25(1):481. https://doi.org/10.3390/ijms25010481
Chicago/Turabian StyleNigro, Ilaria, Rocchina Miglionico, Monica Carmosino, Andrea Gerbino, Anna Masato, Michele Sandre, Luigi Bubacco, Angelo Antonini, Roberta Rinaldi, Faustino Bisaccia, and et al. 2024. "Neuroprotective Effect of Antiapoptotic URG7 Protein on Human Neuroblastoma Cell Line SH-SY5Y" International Journal of Molecular Sciences 25, no. 1: 481. https://doi.org/10.3390/ijms25010481
APA StyleNigro, I., Miglionico, R., Carmosino, M., Gerbino, A., Masato, A., Sandre, M., Bubacco, L., Antonini, A., Rinaldi, R., Bisaccia, F., & Armentano, M. F. (2024). Neuroprotective Effect of Antiapoptotic URG7 Protein on Human Neuroblastoma Cell Line SH-SY5Y. International Journal of Molecular Sciences, 25(1), 481. https://doi.org/10.3390/ijms25010481








