Biological Effects of “Inflammageing” on Human Oral Cells: Insights into a Potential Confounder of Age-Related Diseases
Abstract
:1. Introduction
2. Results
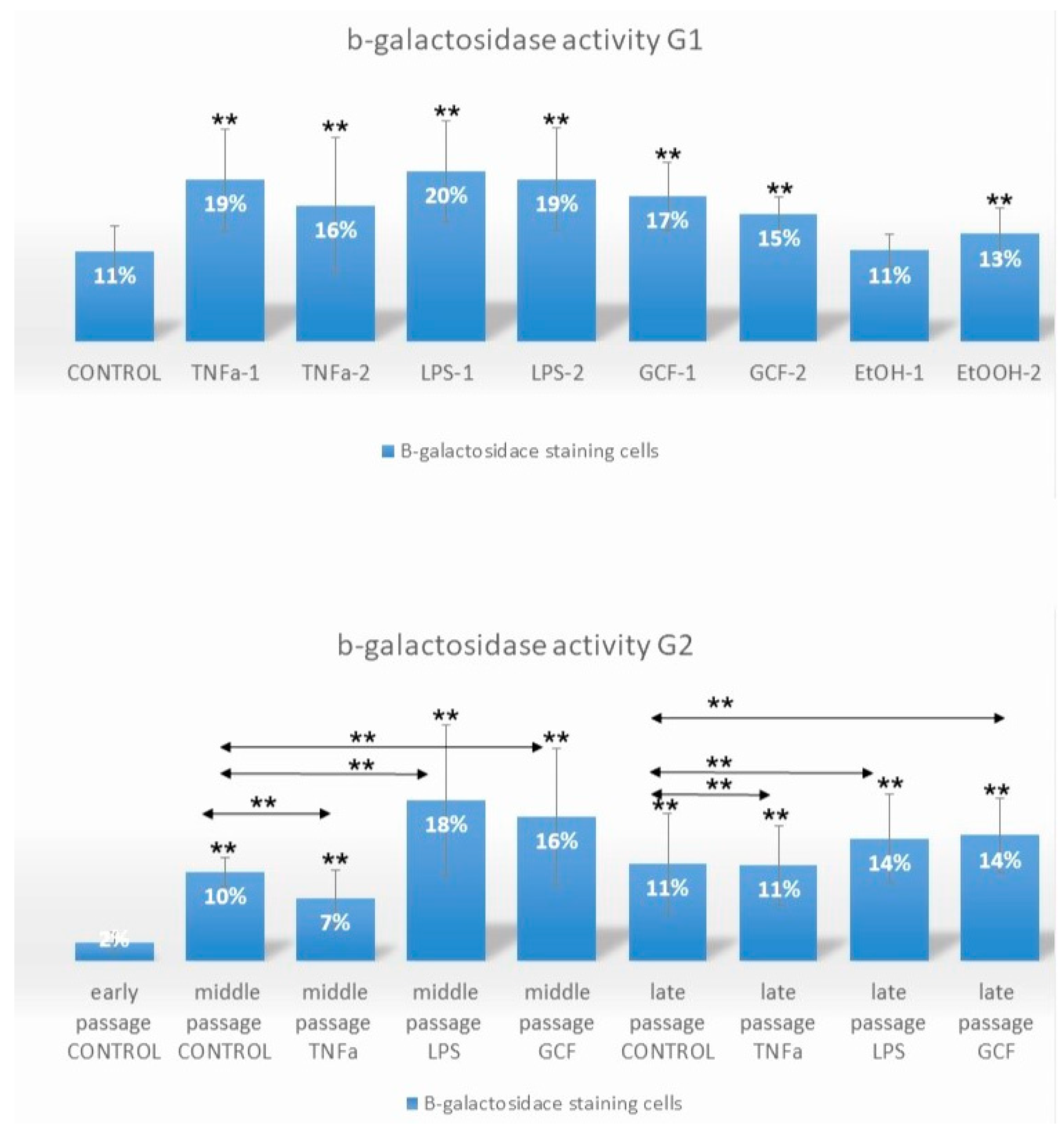
2.1. Senescence-Related Beta-Galactosidase Activity
2.2. Senescence-Related Gene Expression Patterns
2.3. SASP-Related Marker Expression Analysis
3. Discussion
4. Materials and Methods
- G1a received no treatment (negative control group).
- G1b was treated with two concentrations of TNFa (TNFa-1 = 0.1 μL/mL and TNFa-2 = 1 μL/mL).
- G1c was treated with two concentrations of LPS (LPS-1 = 0.2 μL/mL and LPS-2 = 1 μL/mL).
- G1d was treated with two concentrations/dilutions of GCF (GCF-1 = 5 μL/mL and GCF-2 = 10 μL/mL).
- G1e was treated with two concentrations of ethanol EtOH (positive control group) (EtOH-1 = 100 mM and EtOH-2 = 500 mM).
- G2a received no treatment (negative control group C).
- G2b was treated long-term with 1 μL/mL TNFa.
- G2c was treated long-term with 0.2 μL/mL LPS.
- G2d was treated long-term with 10 μL/mL GCF.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xia, S.; Zhang, X.; Zheng, S.; Khanabdali, R.; Kalionis, B.; Wu, J.; Wan, W.; Tai, X. An Update on Inflamm-Aging: Mechanisms, Prevention, and Treatment. J. Immunol. Res. 2016, 2016, 8426874. [Google Scholar] [CrossRef] [PubMed]
- Franceschi, C.; Bonafè, M.; Valensin, S.; Olivieri, F.; De Luca, M.; Ottaviani, E.; De Benedictis, G. Inflamm-aging. An evolutionary perspective on immunosenescence. Ann. N. Y. Acad. Sci. 2000, 908, 244–254. [Google Scholar] [CrossRef] [PubMed]
- Franceschi, C.; Capri, M.; Monti, D.; Giunta, S.; Olivieri, F.; Sevini, F.; Panourgia, M.P.; Invidia, L.; Celani, L.; Scurti, M.; et al. Inflammaging and Anti-Inflammaging: A Systemic Perspective on Aging and Longevity Emerged from Studies in Humans. Mech. Ageing Dev. 2007, 128, 92–105. [Google Scholar] [CrossRef] [PubMed]
- Clegg, A.; Young, J.; Iliffe, S.; Rikkert, M.O.; Rockwood, K. Frailty in elderly people. Lancet 2013, 381, 752–762. [Google Scholar] [CrossRef] [PubMed]
- Doyle, R.; Sadlier, D.M.; Godson, C. Pro-resolving lipid mediators: Agents of anti-ageing? Semin. Immunol. 2018, 40, 36–48. [Google Scholar] [CrossRef] [PubMed]
- Anastassiadou, V.; Management of Complex Conditions in Geriatric Dentistry. Integrated Dental Care for the Elderly; Electronic Repository of the Action “Hellenic Academic E-Books/Kallipos”. 2015. ISBN 978-960-603-244-8. Available online: https://repository.kallipos.gr/handle/11419/3356 (accessed on 7 June 2020).
- Barnes, V.M.; Kennedy, A.D.; Panagakos, F.; Devizio, W.; Trivedi, H.M.; Jönsson, T.; Guo, L.; Cervi, S.; Scannapieco, F.A. Global metabolomic analysis of human saliva and plasma from healthy and diabetic subjects, with and without periodontal disease. PLoS ONE 2014, 9, e105181. [Google Scholar] [CrossRef]
- Panagiotakos, D.B.; Milias, G.A.; Pitsavos, C.; Stefanadis, C. MedDietScore: A computer program that evaluates the adherence to the Mediterranean dietary pattern and its relation to cardiovascular disease risk. Comput. Methods Programs Biomed. 2006, 83, 73–77. [Google Scholar] [CrossRef]
- Lu, M.; Xuan, S.; Wang, Z. Oral microbiota: A new view of body health. Food Sci. Hum. Wellness 2019, 8, 8–15, ISSN 2213-4530. [Google Scholar] [CrossRef]
- Chrysohoou, C.; Pitsavos, C.; Panagiotakos, D.B.; Skoumas, J.; Stefanadis, C. Association between prehypertension status and inflammatory markers related to atherosclerotic disease: The ATTICA Study. Am. J. Hypertens. 2004, 17, 568–573. [Google Scholar] [CrossRef]
- Scannapieco, F.A.; Cantos, A. Oral inflammation and infection, and chronic medical diseases: Implications for the elderly. Periodontol. 2000 2016, 72, 153–175. [Google Scholar] [CrossRef]
- Franceschi, C.; Garagnani, P.; Parini, P.; Giuliani, C.; Santoro, A. Inflammaging: A new immune-metabolic viewpoint for age-related diseases. Nat. Rev. Endocrinol. 2018, 14, 576–590. [Google Scholar] [CrossRef] [PubMed]
- Fulop, T.; Larbi, A.; Witkowski, J.M.; McElhaney, J.; Loeb, M.; Mitnitski, A.; Pawelec, G. Aging, frailty and age-related diseases. Biogerontology 2010, 11, 547–563. [Google Scholar] [CrossRef] [PubMed]
- da Costa, J.P.; Vitorino, R.; Silva, G.M.; Vogel, C.; Duarte, A.C.; Rocha-Santos, T. A synopsis on aging-Theories, mechanisms and future prospects. Ageing Res. Rev. 2016, 29, 90–112. [Google Scholar] [CrossRef] [PubMed]
- Tchkonia, T.; Thomou, T.; Zhu, Y.; Karagiannides, I.; Pothoulakis, C.; Jensen, M.D.; Kirkland, J.L. Mechanisms and metabolic implications of regional differences among fat depots. Cell Metab. 2013, 17, 644–656. [Google Scholar] [CrossRef] [PubMed]
- Freund, A.; Orjalo, A.V.; Desprez, P.Y.; Campisi, J. Inflammatory networks during cellular senescence: Causes and consequences. Trends Mol. Med. 2010, 16, 238–246. [Google Scholar] [CrossRef] [PubMed]
- Xue, W.; Zender, L.; Miething, C.; Dickins, R.A.; Hernando, E.; Krizhanovsky, V.; Cordon-Cardo, C.; Lowe, S.W. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature 2007, 445, 656–660. [Google Scholar] [CrossRef]
- Krizhanovsky, V.; Yon, M.; Dickins, R.A.; Hearn, S.; Simon, J.; Miething, C.; Yee, H.; Zender, L.; Lowe, S.W. Senescence of Activated Stellate Cells Limits Liver Fibrosis. Cell 2008, 134, 657–667, ISSN 0092-8674. [Google Scholar] [CrossRef]
- Bradley, J.R. TNF-mediated inflammatory disease. J. Pathol. 2008, 214, 149–160. [Google Scholar] [CrossRef]
- Kalliolias, G.D.; Ivashkiv, L.B. TNF biology, pathogenic mechanisms and emerging therapeutic strategies. Nat. Rev. Rheumatol. 2016, 12, 49–62. [Google Scholar] [CrossRef]
- Aquino-Martinez, R.; Rowsey, J.L.; Fraser, D.G.; Eckhardt, B.A.; Khosla, S.; Farr, J.N.; Monroe, D.G. LPS-induced premature osteocyte senescence: Implications in inflammatory alveolar bone loss and periodontal disease pathogenesis. Bone 2020, 132, 115220. [Google Scholar] [CrossRef] [PubMed]
- Bozkurt, S.B.; Hakki, S.S.; Hakki, E.E.; Durak, Y.; Kantarci, A. Porphyromonas gingivalis Lipopolysaccharide Induces a Pro-inflammatory Human Gingival Fibroblast Phenotype. Inflammation 2017, 40, 144–153. [Google Scholar] [CrossRef]
- Apatzidou, D.A.; Nile, C.; Bakopoulou, A.; Konstantinidis, A.; Lappin, D.F. Stem cell-like populations and immunoregulatory molecules in periodontal granulation tissue. J. Periodontal. Res. 2018, 53, 610–621. [Google Scholar] [CrossRef]
- Subbarao, K.C.; Nattuthurai, G.S.; Sundararajan, S.K.; Sujith, I.; Joseph, J.; Syedshah, Y.P. Gingival Crevicular Fluid: An Overview. J. Pharm. Bioallied Sci. 2019, 11 (Suppl. S2), S135–S139. [Google Scholar] [CrossRef]
- Chen, J.R.; Lazarenko, O.P.; Haley, R.L.; Blackburn, M.L.; Badger, T.M.; Ronis, M.J. Ethanol impairs estrogen receptor signaling resulting in accelerated activation of senescence pathways, whereas estradiol attenuates the effects of ethanol in osteoblasts. J. Bone Miner. Res. 2009, 24, 221–230. [Google Scholar] [CrossRef]
- Wyganowska-Świątkowska, M.; Nowak, A.; Paszyńska, E.; Grzech-Lesniak, K. Ethanol influence on gingival fibroblasts—A real-time in vitro study. Ann. Agric. Environ. Med. 2018, 25, 647–650. [Google Scholar] [CrossRef]
- Bae, W.J.; Park, J.S.; Kang, S.K.; Kwon, I.K.; Kim, E.C. Effects of Melatonin and Its Underlying Mechanism on Ethanol-Stimulated Senescence and Osteoclastic Differentiation in Human Periodontal Ligament Cells and Cementoblasts. Int. J. Mol. Sci. 2018, 19, 1742. [Google Scholar] [CrossRef]
- Yang, H.; Hu, C.; Li, F.; Liang, L.; Liu, L. Effect of lipopolysaccharide on the biological characteristics of human skin fibroblasts and hypertrophic scar tissue formation. IUBMB Life 2013, 65, 526–532. [Google Scholar] [CrossRef]
- Mountziaris, P.M.; Tzouanas, S.N.; Mikos, A.G. Dose effect of tumor necrosis factor-alpha on in vitro osteogenic differentiation of mesenchymal stem cells on biodegradable polymeric microfiber scaffolds. Biomaterials 2010, 31, 1666–1675. [Google Scholar] [CrossRef]
- Hayflick, L.; Moorhead, P.S. The serial cultivation of human diploid cell strains. Exp. Cell. Res. 1961, 25, 585–621. [Google Scholar] [CrossRef]
- Dimri, G.P.; Lee, X.; Basile, G.; Acosta, M.; Scott, G.; Roskelley, C.; Medrano, E.E.; Linskens, M.; Rubelj, I.; Pereira-Smith, O.; et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. USA 1995, 92, 9363–9367. [Google Scholar] [CrossRef]
- Suzuki, K.; Susaki, E.A.; Nagaoka, I. Lipopolysaccharides and Cellular Senescence: Involvement in Atherosclerosis. Int. J. Mol. Sci. 2022, 23, 11148. [Google Scholar] [CrossRef] [PubMed]
- Nozu, A.; Hamano, S.; Tomokiyo, A.; Hasegawa, D.; Sugii, H.; Yoshida, S.; Mitarai, H.; Taniguchi, S.; Wada, N.; Maeda, H. Senescence and odontoblastic differentiation of dental pulp cells. J. Cell Physiol. 2018, 234, 849–859. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zeng, J.; Wang, X.; Zheng, M.; Luan, Q. P53 mediates lipopolysaccharide-induced inflammation in human gingival fibroblasts. J. Periodontol. 2018, 89, 1142–1151. [Google Scholar] [CrossRef] [PubMed]
- Feng, G.; Zheng, K.; Cao, T.; Zhang, J.; Lian, M.; Huang, D.; Wei, C.; Gu, Z.; Feng, X. Repeated stimulation by LPS promotes the senescence of DPSCs via TLR4/MyD88-NF-κB-p53/p21 signaling. Cytotechnology 2018, 70, 1023–1035. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.O.; Huh, A.J.; Han, S.H.; Kim, J.M. Analysis of cellular senescence induced by lipopolysaccharide in pulmonary alveolar epithelial cells. Arch. Gerontol. Geriatr. 2012, 54, e35–e41. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Fu, H.; Zhu, R.; Wu, X.; Ji, X.; Li, X.; Jiang, H.; Lin, Z.; Tang, X.; Sun, S.; et al. BRD4 contributes to LPS-induced macrophage senescence and promotes progression of atherosclerosis-associated lipid uptake. Aging 2020, 12, 9240–9259. [Google Scholar] [CrossRef] [PubMed]
- Dumont, P.; Balbeur, L.; Remacle, J.; Toussaint, O. Appearance of biomarkers of in vitro ageing after successive stimulation of WI-38 fibroblasts with IL-1alpha and TNFalpha: Senescence associated beta-galactosidase activity and morphotype transition. J. Anat. 2000, 197 Pt 4, 529–537. [Google Scholar] [CrossRef]
- Mavrogonatou, E.; Konstantinou, A.; Kletsas, D. Long-term exposure to TNF-α leads human skin fibroblasts to a p38 MAPK- and ROS-mediated premature senescence. Biogerontology 2018, 19, 237–249. [Google Scholar] [CrossRef]
- Wagner, W.; Horn, P.; Castoldi, M.; Diehlmann, A.; Bork, S.; Saffrich, R.; Benes, V.; Blake, J.; Pfister, S.; Eckstein, V.; et al. Replicative Senescence of Mesenchymal Stem Cells: A Continuous and Organized Process. PLoS ONE 2008, 3, e2213. [Google Scholar] [CrossRef]
- Wagner, W.; Bork, S.; Horn, P.; Krunic, D.; Walenda, T.; Diehlmann, A.; Benes, V.; Blake, J.; Huber, F.X.; Eckstein, V.; et al. Aging and Replicative Senescence Have Related Effects on Human Stem and Progenitor Cells. PLoS ONE 2009, 4, e5846. [Google Scholar] [CrossRef]
- Noren Hooten, N.; Evans, M.K. Techniques to Induce and Quantify Cellular Senescence. J. Vis. Exp. 2017, 123, 55533. [Google Scholar] [CrossRef]
- Debacq-Chainiaux, F.; Erusalimsky, J.D.; Campisi, J.; Toussaint, O. Protocols to detect senescence-associated beta-galactosidase (SA-betagal) activity, a biomarker of senescent cells in culture and in vivo. Nat. Protoc. 2009, 4, 1798–1806. [Google Scholar] [CrossRef] [PubMed]
- Campisi, J.; d’Adda di Fagagna, F. Cellular senescence: When bad things happen to good cells. Nat. Rev. Mol. Cell Biol. 2007, 8, 729–740. [Google Scholar] [CrossRef]
- GeneCards: The Human Gene Database. Available online: https://www.genecards.org/ (accessed on 6 May 2020).
- Hernandez-Segura, A.; de Jong, T.V.; Melov, S.; Guryev, V.; Campisi, J.; Demaria, M. Unmasking Transcriptional Heterogeneity in Senescent Cells. Curr. Biol. 2017, 27, 2652–2660.e4. [Google Scholar] [CrossRef]
- Hernandez-Segura, A.; Rubingh, R.; Demaria, M. Identification of stable senescence-associated reference genes. Aging Cell. 2019, 18, e12911. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.M.; Byun, H.O.; Jee, B.A.; Cho, H.; Seo, Y.H.; Kim, Y.S.; Park, M.H.; Chung, H.Y.; Woo, H.G.; Yoon, G. Implications of time-series gene expression profiles of replicative senescence. Aging Cell 2013, 12, 622–634. [Google Scholar] [CrossRef]
- Marthandan, S.; Priebe, S.; Baumgart, M.; Groth, M.; Cellerino, A.; Guthke, R.; Hemmerich, P.; Diekmann, S. Similarities in Gene Expression Profiles during In Vitro Aging of Primary Human Embryonic Lung and Foreskin Fibroblasts. Biomed Res. Int. 2015, 2015, 731938. [Google Scholar] [CrossRef] [PubMed]
- Marthandan, S.; Baumgart, M.; Priebe, S.; Groth, M.; Schaer, J.; Kaether, C.; Guthke, R.; Cellerino, A.; Platzer, M.; Diekmann, S.; et al. Conserved Senescence Associated Genes and Pathways in Primary Human Fibroblasts Detected by RNA-Seq. PLoS ONE 2016, 11, e0154531. [Google Scholar] [CrossRef]
- Fukami, J.; Anno, K.; Ueda, K.; Takahashi, T.; Ide, T. Enhanced expression of cyclin D1 in senescent human fibroblasts. Mech. Ageing Dev. 1995, 81, 139–157. [Google Scholar] [CrossRef]
- Leontieva, O.V.; Demidenko, Z.N.; Blagosklonny, M.V. MEK drives cyclin D1 hyperelevation during geroconversion. Cell Death Differ. 2013, 20, 1241–1249. [Google Scholar] [CrossRef]
- Huang, C.J.; Yang, S.H.; Huang, S.M.; Lin, C.M.; Chien, C.C.; Chen, Y.C.; Lee, C.L.; Wu, H.H.; Chang, C.C. A predicted protein, KIAA0247, is a cell cycle modulator in colorectal cancer cells under 5-FU treatment. J. Transl. Med. 2011, 9, 82. [Google Scholar] [CrossRef] [PubMed]
- Polato, F.; Rusconi, P.; Zangrossi, S.; Morelli, F.; Boeri, M.; Musi, A.; Marchini, S.; Castiglioni, V.; Scanziani, E.; Torri, V.; et al. DRAGO (KIAA0247), a new DNA damage-responsive, p53-inducible gene that cooperates with p53 as oncosuppressor. [Corrected]. J. Natl. Cancer Inst. 2014, 106, dju053. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Dodig-Crnković, T.; Schwenk, J.M.; Tao, S.C. Current applications of antibody microarrays. Clin. Proteomics 2018, 15, 7. [Google Scholar] [CrossRef]
- Bostanci, N.; Belibasakis, G. Gingival crevicular fluid and its immune mediators in the proteomic era. Periodontology 2000 2017, 76, 68–84. [Google Scholar] [CrossRef] [PubMed]
- Maggio, M.; Guralnik, J.M.; Longo, D.L.; Ferrucci, L. Interleukin-6 in aging and chronic disease: A magnificent pathway. J. Gerontol. A Biol. Sci. Med. Sci. 2006, 61, 575–584. [Google Scholar] [CrossRef]
- Rea, I.M.; Gibson, D.S.; McGilligan, V.; McNerlan, S.E.; Alexander, H.D.; Ross, O.A. Age and Age-Related Diseases: Role of Inflammation Triggers and Cytokines. Front. Immunol. 2018, 9, 586. [Google Scholar] [CrossRef]
- Morley, J.E.; Baumgartner, R.N. Cytokine-Related Aging Process. J. Gerontol. 2004, 59, M924–M929. [Google Scholar] [CrossRef]
- Gong, X.; Gong, W.; Kuhns, D.B.; Ben-Baruch, A.; Howard, O.M.; Wang, J.M. Monocyte chemotactic protein-2 (MCP-2) uses CCR1 and CCR2B as its functional receptors. J. Biol. Chem. 1997, 272, 11682–11685. [Google Scholar] [CrossRef]
- Yousefzadeh, M.J.; Schafer, M.J.; Noren Hooten, N.; Atkinson, E.J.; Evans, M.K.; Baker, D.J.; Quarles, E.K.; Robbins, P.D.; Ladiges, W.C.; LeBrasseur, N.K.; et al. Circulating levels of monocyte chemoattractant protein-1 as a potential measure of biological age in mice and frailty in humans. Aging Cell 2018, 17, e12706. [Google Scholar] [CrossRef]
- Kamei, N.; Tobe, K.; Suzuki, R.; Ohsugi, M.; Watanabe, T.; Kubota, N.; Ohtsuka-Kowatari, N.; Kumagai, K.; Sakamoto, K.; Kobayashi, M.; et al. Overexpression of monocyte chemoattractant protein-1 in adipose tissues causes macrophage recruitment and insulin resistance. J. Biol. Chem. 2006, 281, 26602–26614. [Google Scholar] [CrossRef]
- El Khoury, J.; Toft, M.; Hickman, S.E.; Means, T.K.; Terada, K.; Geula, C.; Luster, A.D. Ccr2 deficiency impairs microglial accumulation and accelerates progression of Alzheimer-like disease. Nat. Med. 2007, 13, 432–438. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Anshita, D.; Ravichandiran, V. MCP-1: Function, regulation, and involvement in disease. Int. Immunopharmacol. 2021, 101 Pt B, 107598. [Google Scholar] [CrossRef]
- Bettcher, B.M.; Neuhaus, J.; Wynn, M.J.; Elahi, F.M.; Casaletto, K.B.; Saloner, R.; Fitch, R.; Karydas, A.; Kramer, J.H. Increases in a Pro-inflammatory Chemokine, MCP-1, Are Related to Decreases in Memory Over Time. Front. Aging Neurosci. 2019, 11, 25. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Gao, Y.; Ding, P.; Wu, T.; Ji, G. The role of CXCL family members in different diseases. Cell Death Discov. 2023, 9, 212. [Google Scholar] [CrossRef] [PubMed]
- Silva, T.A.; Garlet, G.P.; Fukada, S.Y.; Silva, J.S.; Cunha, F.Q. Chemokines in oral inflammatory diseases: Apical periodontitis and periodontal disease. J. Dent. Res. 2007, 86, 306–319. [Google Scholar] [CrossRef] [PubMed]
- Prasad, G.; McCullough, M. Chemokines and cytokines as salivary biomarkers for the early diagnosis of oral cancer. Int. J. Dent. 2013, 2013, 813756. [Google Scholar] [CrossRef]
- Sahingur, S.E.; Yeudall, W.A. Chemokine function in periodontal disease and oral cavity cancer. Front. Immunol. 2015, 6, 214. [Google Scholar] [CrossRef]
- Sarode, G.; Sarode, S.; Patil, S.; Yadahalli, R. Significance of CC Group of Chemokines in Oral Squamous Cell Carcinoma and Oral Potential Malignant Disorders: A Review. World J. Dent. 2021, 12, 160–165. [Google Scholar] [CrossRef]
- Bakopoulou, A.; Leyhausen, G.; Volk, J.; Tsiftsoglou, A.; Garefis, P.; Koidis, P.; Geurtsen, W. Assessment of the impact of two different isolation methods on the osteo/odontogenic differentiation potential of human dental stem cells derived from deciduous teeth. Calcif. Tissue Int. 2011, 88, 130–141. [Google Scholar] [CrossRef]
- Bakopoulou, A.; Apatzidou, D.; Aggelidou, E.; Gousopoulou, E.; Leyhausen, G.; Volk, J.; Kritis, A.; Koidis, P.; Geurtsen, W. Isolation and prolonged expansion of oral mesenchymal stem cells under clinical-grade, GMP-compliant conditions differentially affects “stemness” properties. Stem. Cell Res. Ther. 2017, 8, 247. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- UniProt Consortium. UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023, 51, D523–D531. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef] [PubMed]





| Gene Symbol | Forward (5′-3′) | Reverse (5′-3′) | Amplicon Size (bp) |
|---|---|---|---|
| CCND1 | AGCTGTGCATCTACACCGAC | GAAATCGTGCGGGGTCATTG | 113 |
| SUSD6 | TTAGCTGCCGTCTCAACGAG | CTGGTCACGCCTGCTATGAT | 170 |
| STAG1 | GATTGCAGCTCCGTTGAAGG | GCCGACCATCGACCTAGTTT | 125 |
| GAPDH | GACAGTCAGCCGCATCTTCT | GCGCCCAATACGACCAAATC | 104 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alexakou, E.; Bakopoulou, A.; Apatzidou, D.A.; Kritis, A.; Malousi, A.; Anastassiadou, V. Biological Effects of “Inflammageing” on Human Oral Cells: Insights into a Potential Confounder of Age-Related Diseases. Int. J. Mol. Sci. 2024, 25, 5. https://doi.org/10.3390/ijms25010005
Alexakou E, Bakopoulou A, Apatzidou DA, Kritis A, Malousi A, Anastassiadou V. Biological Effects of “Inflammageing” on Human Oral Cells: Insights into a Potential Confounder of Age-Related Diseases. International Journal of Molecular Sciences. 2024; 25(1):5. https://doi.org/10.3390/ijms25010005
Chicago/Turabian StyleAlexakou, Elli, Athina Bakopoulou, Danae A. Apatzidou, Aristeidis Kritis, Andigoni Malousi, and Vassiliki Anastassiadou. 2024. "Biological Effects of “Inflammageing” on Human Oral Cells: Insights into a Potential Confounder of Age-Related Diseases" International Journal of Molecular Sciences 25, no. 1: 5. https://doi.org/10.3390/ijms25010005







