QTL Detection and Candidate Gene Identification for Eating and Cooking Quality Traits in Rice (Oryza sativa L.) via a Genome-Wide Association Study
Abstract
:1. Introduction
2. Results
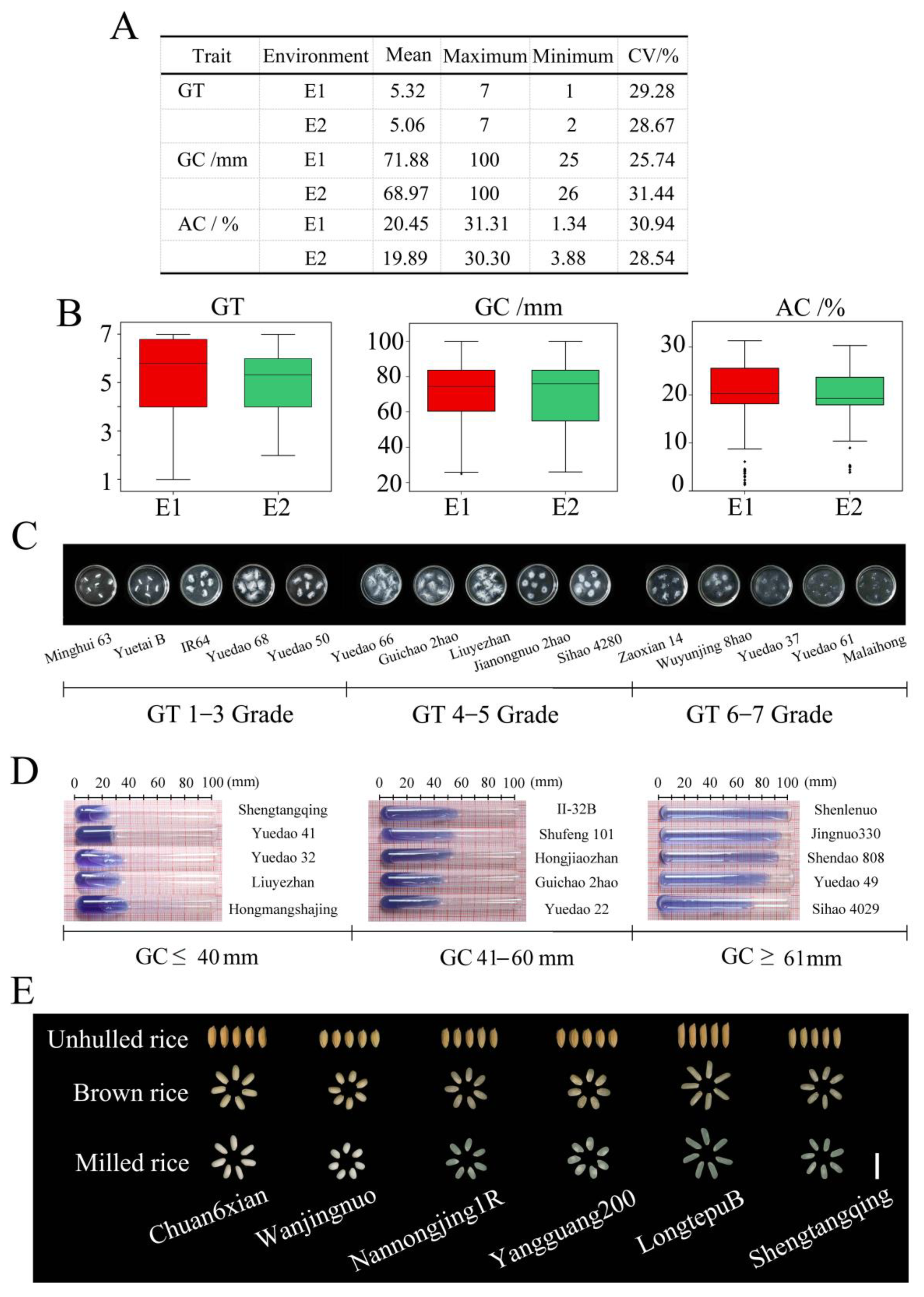
2.1. Phenotypic Variation in ECQ-Related Traits
2.2. Identification of QTLs for ECQ-Related Traits Using GWAS
2.3. Identification of Candidate Genes
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Measurement of Rice GT, GC, and AC
4.3. Genome-Wide Association Study
4.4. Candidate Gene Analysis
4.5. Quantitative Reverse Transcription PCR Analysis of Candidate Genes
4.6. Haplotype Analysis
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Juliano, B.O. The chemical basis of rice grain quality. In Proceedings on the Workshop on Chemical Aspects of Rice Grain Quality; Brady, N.C., Ed.; IRRI Philippines: Laguna, Philippines, 1979; pp. 69–90. [Google Scholar]
- Xu, C.W.; Mo, H.D. Qualitative-quantitative analysis for Inheritance of gelatinization temperature in Indica rice (Oryza sativa subsp. indica). Acta Agron. Sin. 1996, 22, 385–391. Available online: https://zwxb.chinacrops.org/CN/Y1996/V22/104/385 (accessed on 5 September 2023).
- Umemoto, T.; Yano, M.; Satoh, H.; Shomura, A.; Nakamura, Y. Mapping of a gene responsible for the difference in amylopectin structure between japonica-type and indica-type rice varieties. Theor. Appl. Genet. 2002, 104, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.Y.; Zeng, D.L.; Cheng, F.M.; Tian, Z.X.; Guo, L.B.; Su, Y.; Yan, M.X.; Jiang, H.; Dong, G.J.; Huang, Y.C.; et al. ALK, the key gene for gelatinization temperature, is a modifier gene for gel consistency in rice. J. Integr. Plant Biol. 2011, 53, 756–765. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.Y.; Cheng, Z.J.; Zhang, X.; Guo, X.P.; Su, N.; Jiang, L.; Mao, L.; Wan, J.M. Double repression of soluble starch synthase genes SSIIa and SSIIIa in rice (Oryza sativa L.) uncovers interactive effects on the physicochemical properties of starch. Genome 2011, 54, 448–459. [Google Scholar] [CrossRef] [PubMed]
- Umemoto, T.; Aoki, N. Single-nucleotide polymorphisms in rice starch synthase IIa that alter starch gelatinisation and starch association of the enzyme. Funct. Plant Biol. 2005, 32, 763–768. [Google Scholar] [CrossRef] [PubMed]
- Fan, M.Y.; Wang, X.J.; Wang, X.H.; Tang, L.; Xu, Q.; Xu, Z.J. Research advances of amylopectin structure in rice. Chin. J. Rice Sci. 2017, 31, 124–132. [Google Scholar] [CrossRef]
- Bao, J.S.; Corke, H.; Sun, M. Nucleotide diversity in Starch synthase IIa and validation of single nucleotide polymorphisms in relation to starch gelatinization temperature and other physicochemical properties in rice (Oryza sativa L.). Theor. Appl. Genet. 2006, 113, 1171–1183. [Google Scholar] [CrossRef]
- Waters, D.; Henry, R.J.; Reinke, R.F.; Fitzgeraid, M.A. Gelatinization temperature of rice explained by polymorphisms in starch synthase. Plant Biotechnol. J. 2006, 4, 115–122. [Google Scholar] [CrossRef]
- Fujita, N.; Yoshida, M.; Asakura, N.; Ohdan, T.; Miyao, A.; Hirochika, H.; Nakamyra, Y. Functional and characterization of starch synthase I using mutants in rice. Plant Physiol. 2006, 140, 1070–1084. [Google Scholar] [CrossRef]
- Yang, B.W.; Xiang, X.C.; Xu, S.J.; Xu, L.; Wang, Q. Effects for interaction of Wx and SSIII-2 on rice eating and cooking qualities. Acta Bot. Boreali-Occident Sin. 2017, 37, 879–884. [Google Scholar] [CrossRef]
- Vandeputte, G.E.; Delcour, J.A. From sucrose to starch granule to starch physical behaviour: A focus on rice starch. Carbohyd. Polym. 2004, 58, 245–266. [Google Scholar] [CrossRef]
- Tian, Z.X.; Qian, Q.; Liu, Q.Q.; Yan, M.; Liu, X.; Yan, C.J.; Liu, G.; Gao, Z.; Tang, S.; Zeng, D.; et al. Allele diversities in rice starch biosynthesis lead to a diverse array of rice eating and cooking qualities. Proc. Natl. Acad. Sci. USA 2009, 106, 21760–21765. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.H.; Zhang, C.Q.; Gu, M.H.; Liu, Q.Q. Progress in the allelic variation of Wx gene and it’s application in rice breeding. Chin. J. Rice Sci. 2015, 29, 431–438. [Google Scholar]
- Zhang, C.Q.; Zhao, D.S.; Li, Q.F.; Gu, M.H.; Liu, Q.Q. Progresses in research on cloning and functional analysis of key genes involving in rice grain quality. Sci. Agric. Sin. 2016, 49, 4267–4283. [Google Scholar] [CrossRef]
- Zhang, C.Q.; Zhu, J.H.; Chen, S.J.; Fan, X.L.; Lu, Y.; Wang, M.; Yu, H.X.; Yi, C.D.; Tang, S.Z.; Gu, M.H.; et al. Wxlv, the ancestral allele of rice Waxy gene. Mol. Plant 2019, 12, 1157–1166. [Google Scholar] [CrossRef]
- Wanchana, S.; Toojinda, T.; Tragoonrung, S.; Vanavichit, A. Duplicated coding sequence in the waxy allele of tropical glutinous rice (Oryza sativa L.). Plant Sci. 2003, 165, 1193–1199. [Google Scholar] [CrossRef]
- Wang, Z.Y.; Zheng, F.Q.; Shen, G.Z.; Gao, J.P.; Snustad, D.P.; Li, M.G.; Zhang, J.L.; Hong, M.M. The amylose content in rice endosperm is related to the post-transcriptional regulation of the waxy gene. Plant J. 1995, 7, 613–622. [Google Scholar] [CrossRef]
- Mikami, I.; Uwatoko, N.; Ikeda, Y.; Yamaguchi, J.; Hirano, H.Y.; Suzuki, Y.; Sano, Y. Allele diversification at the wx locus in landraces of Asian rice. Theor. Appl. Genet. 2008, 116, 979–989. [Google Scholar] [CrossRef]
- Chen, Z.Z.; Yang, Y.; Feng, L.H.; Sun, Y.; Zhang, C.Q.; Fan, X.L.; Li, Q.F.; Liu, Q.Q. Effect of different combinations of Wx and ALK main alleles on rice grain quality. Chin. J. Rice Sci. 2020, 34, 228–236. [Google Scholar] [CrossRef]
- Yan, C.J.; Tian, S.; Zhang, Z.Q.; Han, Y.P.; Chen, F.; Li, X.; Gu, M.H. The source of genes related to rice grain starch synthesis among cultivated varieties and its contribution to quality. Sci. Agric. Sin. 2005, 39, 865–871. [Google Scholar] [CrossRef]
- Zhu, L.J.; Gu, M.H.; Meng, X.L.; Cheung, C.K.; Yu, H.X.; Huang, J.; Sun, Y.; Shi, Y.C.; Liu, Q.Q. High-amylose rice improves indices of animal health in normal and diabetic rats. Plant Biotechnol. J. 2012, 10, 353–362. [Google Scholar] [CrossRef]
- Zhao, H.; Wang, J.M.; Zhang, Q.F.; Zhao, Q.; Mei, S.F.; Liu, X.L.; Cheng, F.M. Activities of several starch synthesis enzymes in filling grains for rice sugary endosperm mutant (Sug-11) and it’s relation to starch quality. Chin. J. Rice Sci. 2015, 29, 73–81. [Google Scholar] [CrossRef]
- Satoh, H.; Omura, T. New endosperm mutations induced by chemical mutagens in rice Oryza sativa L. Jpn. J. Breeding 1981, 31, 316–326. [Google Scholar] [CrossRef]
- She, K.C.; Kusano, H.; Koizumi, K.; Yamakawa, H.; Hakata, M.; Imamura, T.; Fukuda, M.; Naito, N.; Tsurumaki, Y.; Yaeshima, M.; et al. A novel factor FLOURY ENDOSPERM 2 is involved in regulation of rice grain size and starch quality. Plant Cell 2010, 22, 3280–3294. [Google Scholar] [CrossRef] [PubMed]
- Nishio, T.; Iida, S. Mutants having a low content of 16-kDa allergenic protein in rice (Oryza sativa L.). Theor. Appl. Genet. 1993, 86, 317–321. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.G.; Park, S.; Matsuoka, M.; An, G. White-core endosperm floury endosperm 4 in rice is generated by knockout mutations in the C4-type pyruvate orthophosphate dikinase gene (OsPPDKB). Plant J. 2005, 42, 901–911. [Google Scholar] [CrossRef]
- Fujita, N.; Yoshida, M.; Kondo, T.; Saito, K.; Utsumi, Y.; Tokunage, T.; Nishi, A.; Satoh, H.; Park, J.; Jane, J.; et al. Characterization of SSIIIa-deficient mutants of rice: The function of SSIIIa and pleiotropic effects by SSIIIa deficiency in the rice endosperm. Plant Physiol. 2007, 144, 2009–2023. [Google Scholar] [CrossRef] [PubMed]
- Ryoo, N.; Yu, C.; Park, C.S.; Baik, M.Y.; Park, I.M.; Cho, M.H.; Bhoo, S.H.; An, G.; Hahn, T.; Jeon, J.S. Knockout of a atarch synthase gene OsSSIIIa/Flo5 causes white-core floury endosperm in rice (Oryza sativa L.). Plant Cell Rep. 2007, 26, 1083–1095. [Google Scholar] [CrossRef]
- Cheng, P.; Wang, Y.H.; Liu, F.; Ren, Y.L.; Zhou, K.N.; Lv, J.; Zheng, M.; Zhao, S.L.; Zhang, L.; Wang, C.M.; et al. FLOURY ENDOSPERM 6 encodes a CBM48 domain-containing protein involved in compound granule formation and starch synthesis in rice endosperm. Plant J. 2014, 77, 917–930. [Google Scholar] [CrossRef]
- Fang, P.F.; Li, S.F.; Jiao, G.A.; Xie, L.H.; Hu, P.S.; Wei, X.J.; Tang, S.Q. Physicochemical property analysis and gene mapping of a floury endosperm mutant flo7 in rice. Chin. J. Rice Sci. 2014, 28, 447–457. [Google Scholar] [CrossRef]
- Sheng, Z.H.; Fang, P.F.; Li, S.F.; Jiao, G.A.; Xie, L.H.; Hu, P.S.; Tang, S.Q.; Wei, X.J. Phenotype of rice floury endosperm mutant flo7 and fine mapping of mutated gene. Rice Sci. 2015, 22, 162–170. [Google Scholar] [CrossRef]
- Silver, D.M.; Kötting, O.; Moorhead, G.B.G. Phosphoglucan phosphatase function sheds light on starch degradation. Trends Plant Sci. 2014, 19, 471–478. [Google Scholar] [CrossRef]
- Zhu, Y.; Cai, X.L.; Wang, Z.Y.; Hong, M.M. An interaction between a MYC protein and an EREBP protein is involved in transciptional regulation of the rice Wx gene. J. Biol. Chem. 2003, 278, 47803–47811. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.R.; Huang, W.X.; Cai, X.L. Oligomerization of rice granule-bound starch synthase 1 modulates its activity regulation. Plant Sci. 2013, 210, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Duan, L.; Dai, J.S.; Zhang, C.Q.; Li, J.; Gu, M.H.; Liu, Q.Q.; Zhu, Y. Major QTLs reduce the deleterious effects of high temperature on rice amylose content by increasing splicing efficiency of Wx pre-mRNA. Theor. Appl. Genet. 2013, 127, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Fu, F.F.; Xue, H.W. Coexpression analysis identifies rice starch regulator1, a rice AP2/EREBP family transcription factor, as a novel rice starch biosynthesis regulator. Plant Physiol. 2010, 154, 927–938. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.C.; Xu, H.; Zhu, Y.; Liu, Q.Q.; Cai, X.L. OsbZIP58, a basic leucine zipper transcription factor, regulates starch biosynthesis in rice endosperm. J. Exp. Bot. 2013, 64, 3453–3466. [Google Scholar] [CrossRef]
- Wu, J.H.; Zhu, C.F.; Pang, J.H.; Zhang, X.R.; Yang, C.L.; Xia, G.X.; Tian, Y.C.; He, C.Z. OsLOL1, a C2C2-type zinc finger protein, interacts with OsbZIP58 to promote seed germination through the modulation of gibberellin biosynthesis in Oryza sativa. Plant J. 2014, 80, 1118–1130. [Google Scholar] [CrossRef]
- Hu, C.M.; Jiang, J.H.; Li, Y.L.; Song, S.J.; Zou, Y.; Jing, C.Y.; Zhang, Y.; Wang, D.Z.; He, Q.; Dang, X.J. QTL mapping and identification of candidate genes using a genome-wide association study for heat tolerance at anthesis in rice (Oryza sativa L.). Front. Genet. 2022, 13, 983525. [Google Scholar] [CrossRef]
- Gao, Z.Y.; Zeng, D.L.; Cui, X.; Zhou, Y.H.; Yan, M.X.; Huang, D.N.; Li, J.Y.; Qian, Q. Map-based cloning of ALK gene, which controls the gelatinization temperature of rice. Sci. Chi. (Ser. C) 2003, 46, 661–668. [Google Scholar] [CrossRef]
- Zhang, C.Q.; Yang, Y.; Chen, S.J.; Liu, X.J.; Zhu, J.H.; Zhou, L.H.; Lu, Y.; Li, Q.F.; Fan, X.L.; Tang, S.Z.; et al. A rare Waxy allele coordinately improves rice eating and cooking quality and grain transparency. J. Integr. Plant Biol. 2021, 63, 889–901. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.R.; Su, F.; Huang, S.; Mei, F.T.; Niu, X.M.; Ma, C.L.; Zhang, H.; Zhu, X.G.; Zhu, J.K.; Zhang, J.S. Novel Wx alleles generated by base editing for improvement of rice grain quality. J. Integr. Plant Biol. 2021, 63, 1632–1638. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, Y.; Kamiya, N.; Morinaka, Y.; Matsuoka, M.; Sazuka, T. Auxin biosynthesis by the YUCCA genes in rice. Plant Physiol. 2007, 143, 1362–1371. [Google Scholar] [CrossRef]
- Zhang, T.; Li, R.N.; Xing, J.L.; Yan, L.; Wang, R.C.; Zhao, Y.D. The YUCCA-Auxin-WOX11 module controls crown root development in rice. Front. Plant Sci. 2018, 9, 523. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.Y.; E, Z.G.; Zhang, D.P.; Yun, Q.B.; Zhou, Y.; Niu, B.X.; Chen, C. OsYUC11-mediated auxin biosynthesis is essential for endosperm development of rice. Plant Physiol. 2021, 185, 934–950. [Google Scholar] [CrossRef]
- Cheng, Y.F.; Dai, X.H.; Zhao, Y.D. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 2006, 20, 1790–1799. [Google Scholar] [CrossRef]
- Cheng, Y.F.; Dai, X.H.; Zhao, Y.D. Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 2007, 19, 2430–2439. [Google Scholar] [CrossRef]
- Abu-Zaitoon, Y.M.; Bennett, K.; Normanly, J.; Nonhebel, H.M. A large increase in IAA during development of rice grains correlates with the expression of trytophan aminotransferase OsTAR1 and a grain specific YUCCA. Physiol. Plant 2012, 146, 487–499. [Google Scholar] [CrossRef]
- Chen, Q.G.; Dai, X.H.; De-Paoli, H.; Cheng, Y.; Takebayashi, Y.; Kasahara, H.; Kamiya, Y.; Zhao, Y.D. Auxin overproduction in shoots cannot rescue auxin deficiencies in Arabidopsis roots. Plant Cell Physiol. 2014, 55, 1072–1079. [Google Scholar] [CrossRef]
- Zhang, X.F.; Tong, J.H.; Bai, A.N.; Liu, C.M.; Xiao, L.T.; Xue, H.W. Phytohormone dynamics in developing endosperm influence rice grain shape and quality. J. Integr. Plant Biol. 2020, 62, 1625–1637. [Google Scholar] [CrossRef]
- Bernardi, J.; Lanubile, A.; Li, Q.B.; Kumar, D.; Kladnik, A.; Cook, S.D.; Ross, J.J.; Marocco, A.; Chourey, P.S. Impaired auxin biosynthesis in the defective endosperm 18 mutant is due to mutational loss of expression in the ZmYuc 1 gene encoding endosperm-specific YUCCA1 protein in maize. Plant Physiol. 2012, 160, 1317–1328. [Google Scholar] [CrossRef] [PubMed]
- Bernardi, J.; Battaglia, R.; Bagnaresi, P.; Lucini, L.; Marocco, A. Transcriptomic and metabolomic analysis of ZmYUC1 mutant reveals the role of auxin during early endosperm formation in maize. Plant Sci. 2019, 281, 133–145. [Google Scholar] [CrossRef]
- Champagne, E.T.; Bett-Garber, K.L.; Fitzgerald, M.A.; Grimm, C.C.; Lea, J.; Ohtsubo, K.I.; Jongdee, S.; Xie, L.H.; Bassinello, P.Z.; Resurreccion, A.; et al. Important sensory properties differentiating premium rice varieties. Rice 2010, 3, 270–281. [Google Scholar] [CrossRef]
- Xu, Y.J.; Ying, Y.N.; Ouyang, S.H.; Duan, X.L.; Sun, H.; Jiang, S.K.; Sun, S.C.; Bao, J.S. Factors affecting sensory quality of cooked japonica rice. Rice Sci. 2018, 2, 330–339. [Google Scholar] [CrossRef]
- Sreenivasulu, N.; Butardo, V.M.J.; Misra, G.; Cuevas, R.P.; Anacleto, R.; Kavi Kishor, P.B. Desinging climate-resilient rice with ideal grain quality suited for high-temperature stress. J. Exp. Bot. 2015, 66, 1737–1748. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.Q.; Chen, S.J.; Ren, X.Y.; Lu, Y.; Liu, D.R.; Cai, X.L.; Li, Q.F.; Gao, J.P.; Liu, Q.Q. Molecular structure and physicochemical properties of starches from rice with different amylose contents resulting from modification of OsGBSSI activity. J. Agric. Food Chem. 2017, 65, 2222. [Google Scholar] [CrossRef] [PubMed]
- Little, R.R.; Hiller, G.B.; Son, E. Differential effect of dilute alkali on 25 varieties of milled white rice. Cereal Chem. 1958, 35, 111–126. Available online: https://www.semanticscholar.org/paper/Differential-effect-of-dilute-alkali-on-25-of-white-Little-Hilder/07558177695bd2e5eb3f623c01b420203af04f16 (accessed on 8 April 2021).
- Cagampang, G.B.; Perez, C.M.; Bo, J.O. A gel consistency test for eating quality of rice. J. Sci. Food Agric. 1973, 24, 1589–1594. [Google Scholar] [CrossRef]
- Dang, X.J.; Yang, Y.; Zhang, Y.Q.; Chen, X.G.; Fan, Z.L.; Liu, Q.M.; Ji, J.; Li, D.L.; Li, Y.H.; Fang, B.J.; et al. OsSYL2AA, an allele identified by gene-based association, increases style length in rice (Oryza sativa L.). Plant J. 2020, 104, 1491–1503. [Google Scholar] [CrossRef]
- Lipka, A.E.; Tian, F.; Wang, Q.S.; Peiffer, J.; Li, M.; Bradbury, P.J.; Gore, M.A.; Buckler, E.S.; Zhang, Z.W. GAPIT: Genome association and prediction integrated tool. Bioinformatics 2012, 28, 2397–2399. [Google Scholar] [CrossRef]
- Turner, S.D. qqman: An R package for visualizing GWAS results using QQ and Manhattan plots. BioRxiv 2014. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.H.; Blay, S.; McNeney, B.; Graham, J. LDheatmap: An R function for graphical display of pairwise linkage disequilibria between single nucleotide polymorphisms. J. Stat. Softw. 2006, 16, 1–10. [Google Scholar] [CrossRef]
- Huang, Z.B.; Ying, J.F.; Peng, L.L.; Sun, S.; Huang, C.W.; Li, C.; Wang, Z.F.; He, Y.Q. A genome-wide association study reveals that the cytochrome b5 involved in seed reserve mobilization during seed germination in rice. Theor. Appl. Genet. 2021, 134, 4067–4076. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, J.; Song, S.; Hu, C.; Jing, C.; Xu, Q.; Li, X.; Zhang, M.; Hai, M.; Shen, J.; Zhang, Y.; et al. QTL Detection and Candidate Gene Identification for Eating and Cooking Quality Traits in Rice (Oryza sativa L.) via a Genome-Wide Association Study. Int. J. Mol. Sci. 2024, 25, 630. https://doi.org/10.3390/ijms25010630
Jiang J, Song S, Hu C, Jing C, Xu Q, Li X, Zhang M, Hai M, Shen J, Zhang Y, et al. QTL Detection and Candidate Gene Identification for Eating and Cooking Quality Traits in Rice (Oryza sativa L.) via a Genome-Wide Association Study. International Journal of Molecular Sciences. 2024; 25(1):630. https://doi.org/10.3390/ijms25010630
Chicago/Turabian StyleJiang, Jianhua, Shaojie Song, Changmin Hu, Chunyu Jing, Qing Xu, Xinru Li, Mengyuan Zhang, Mei Hai, Jiaming Shen, Ying Zhang, and et al. 2024. "QTL Detection and Candidate Gene Identification for Eating and Cooking Quality Traits in Rice (Oryza sativa L.) via a Genome-Wide Association Study" International Journal of Molecular Sciences 25, no. 1: 630. https://doi.org/10.3390/ijms25010630



