Abstract
miR-129 plays a crucial role in regulating various cellular processes, including adipogenesis; however, its downstream molecular mechanisms remain unclear. In this study, we demonstrated that miR-129 promotes yak adipogenesis in vitro via the PI3K/AKT pathway. Overexpression and interference of miR-129 in yak intramuscular preadipocytes (YIMAs) enhanced and inhibited cell differentiation, respectively, with corresponding changes in cell proliferation. Further investigation revealed that miR-129 enhances AKT and p-AKT activity in the AKT pathway without affecting cell apoptosis, and a specific inhibitor (LY294002) was used to confirm that miR-129 regulates YIMAs proliferation and differentiation through the PI3K/AKT pathway. Our findings suggest that miR-129 promotes yak adipogenesis by enhancing PI3K/AKT pathway activity. This study provides the foundation to precisely elucidate the molecular mechanism of miR-129 in YIMAs adipogenesis and develop advanced miRNA-based strategies to improve meat nutrition and obesity-related ailments in beef production.
1. Introduction
Yaks (Bos grunniens), a unique breed of livestock in the Qinghai–Tibetan Plateau region, hold a significant position in China’s beef market. Yaks are the third largest breed of cattle in China, accounting for over 95% of the world’s yak population [1]. Studies indicate that fat content, particularly intramuscular fat (IMF) content, plays a significant role in the tenderness, juiciness, and flavor of meat [2,3]. However, the low IMF content of yak meat has become a major bottleneck, restricting high-quality meat development in the yak industry. Recent studies report that non-coding RNAs (ncRNAs) regulate key aspects of lipid metabolism [4,5,6,7].
MicroRNAs (miRNAs) are a subclass of ncRNAs consisting of approximately 20 nucleotides that play crucial roles in regulating adipocyte proliferation, differentiation, and apoptosis [8]. In particular, miR-129-5p has been identified as a potential biomarker because it is a crucial tumor suppressor [9,10]. Several studies have highlighted the significant regulatory potential of miR-129 in lipid metabolism and adipogenesis and its potential as a biomarker and therapeutic target for obesity [11,12,13,14]. In our previous studies on adipogenesis in YIMAs, we noted that miR-129 was significantly down-regulated after overexpression of the adipogenesis-promoting factor lncFAM200B [4]. This suggests that miR-129 may have a key regulatory role in yak adipogenesis, but further experimental validation is required.
The PI3K/AKT pathway is crucial for insulin stress, metabolic regulation, and cell apoptosis [15]. Several studies have demonstrated that miR-129 regulates cell growth and development by participating in the PI3K/AKT pathway, either at the post-transcriptional or post-translational level. For example, miR-129-5p overexpression inhibits the development of retinoblastoma by directly targeting PAX6 via the PI3K/AKT signaling pathway [16]. Similarly, the miR-129-5p/SLC2A3 axis regulates glucose metabolism and gastric cancer growth and is influenced by the PI3K/AKT pathway [17]. Additionally, Xu et al. showed that miR-129 inhibits prostate cancer cell proliferation and metastasis by targeting ETS1 via the PI3K/AKT/mTOR pathway [18]. Therefore, we investigated the potential interactions between miR-129 and the PI3K/AKT pathway during the proliferation and differentiation of YIMAs.
In this study, we performed overexpression and interference techniques to manipulate miR-129 levels and investigated its regulatory effects on lipid deposition and the PI3K/AKT signaling pathway in yaks to further explore the relationship between miR-129 and the activity of the PI3K/AKT pathway during YIMAs proliferation and differentiation. The current work provides valuable insights and a theoretical basis for understanding the molecular mechanism of miR-129 in adipogenesis and exploring its biological functions.
2. Results
2.1. miR-129 Promotes Differentiation of YIMAs
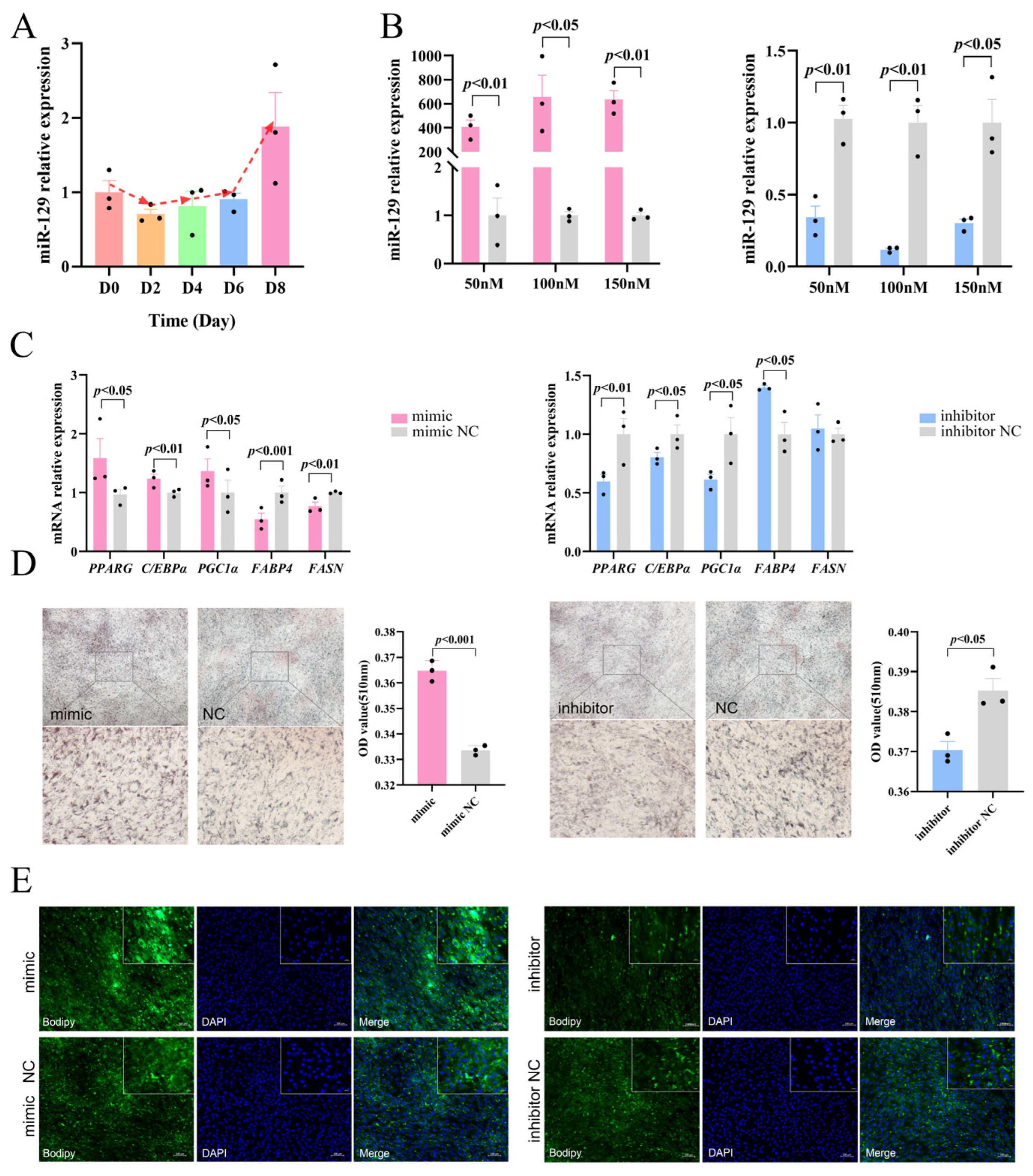
To preliminarily investigate the regulatory role of miR-129 in YIMAs adipogenesis, we measured the miR-129 expression levels during YIMAs differentiation. The results showed that the expression of miR-129 gradually increased, indicating that miR-129 has a potential regulatory role in YIMA differentiation (Figure 1A). Subsequently, we manipulated the miR-129 expression in YIMAs using transfection mimics and inhibitors. An optimal concentration of 100 nM was selected for subsequent tests. (Figure 1B). We further analyzed the effects of miR-129 overexpression and knockdown on YIMAs differentiation and lipid deposition in vitro. The reverse transcriptase–-quantitative polymerase chain reaction (RT-qPCR) results showed that overexpression of miR-129 increased the expression of differentiation marker genes (PPARG and C/EBPα), while interference with miR-129 decreased the mRNA expression of these marker genes. PGC1α, a coactivator of PPARG, showed a similar trend (Figure 1C). Additionally, we observed that the overexpression of miR-129 suppressed the expression of FABP4 and FASN, which are key targets of PPARG during fatty acid synthesis. In contrast, miR-129 interference upregulated the mRNA expression of FABP4. Furthermore, Oil Red O and Bodipy staining experiments revealed that overexpression of miR-129 significantly enhanced lipid deposition in YIMAs during differentiation, whereas interference with miR-129 inhibited lipid deposition (Figure 1D,E). These results suggest that miR-129 plays a positive role by regulating the expression of lipid-related genes during YIMAs lipid deposition.
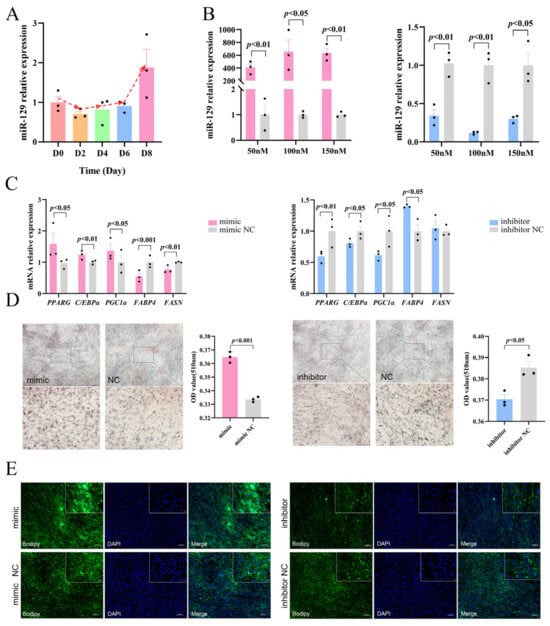
Figure 1.
Effects of miR-129 on YIMAs differentiation and lipid deposition. (A) Expression of miR-129 in different stages of YIMAs differentiation. (B) Concentration screening of miR-129 mimic and inhibitor after miR-129 overexpression and interference. (C) mRNA expression of differentiation genes. (D) Oil Red O staining. Scale bar: 200 μm, 50 μm. (E) Bodipy staining. Scale bar: 100 μm, 20 μm.
2.2. miR-129 Inhibits Proliferation of YIMAs
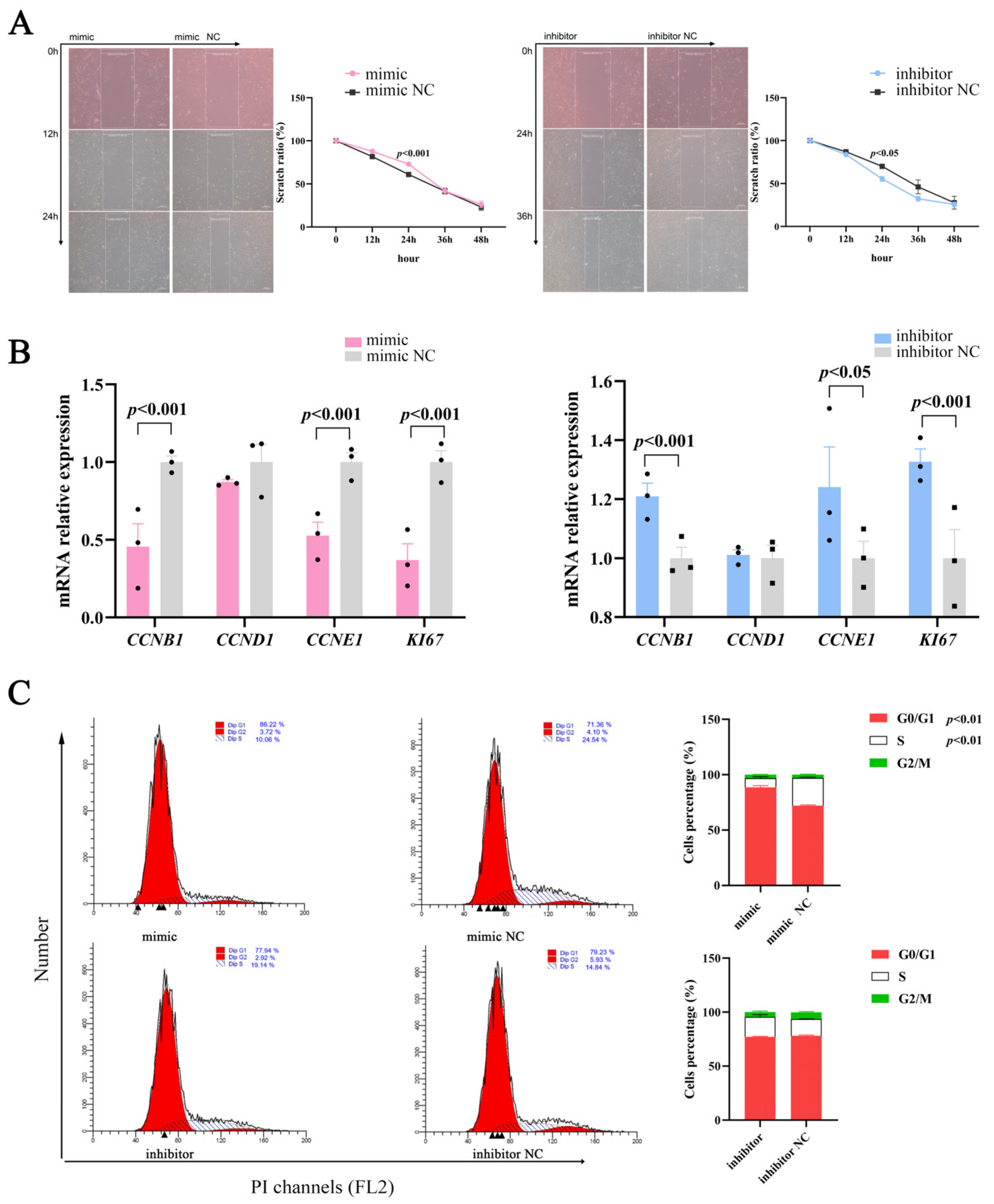
Cell proliferation and differentiation are highly coordinated processes [19]; therefore, we studied the effects of miR-129 on proliferation during YIMAs differentiation. The scratch results showed that proliferation was significantly altered in the overexpression or inhibition groups compared to their respective controls (Figure 2A). Moreover, miR-129 overexpression significantly suppressed the expression of cell-proliferation-associated marker genes, whereas miR-129 inhibition had the opposite effect (Figure 2B). Overexpression of miR-129 resulted in cell cycle arrest in the G0/G1 phase and decreased the number of cells in the S phase. Conversely, interference with miR-129 had the opposite effect on the number of cells in the G0/G1 and S phases (Figure 2C). These results indicated that miR-129 may impede YIMAs proliferation by inhibiting the transition of cells from the G0/G1 phase to the S phase.
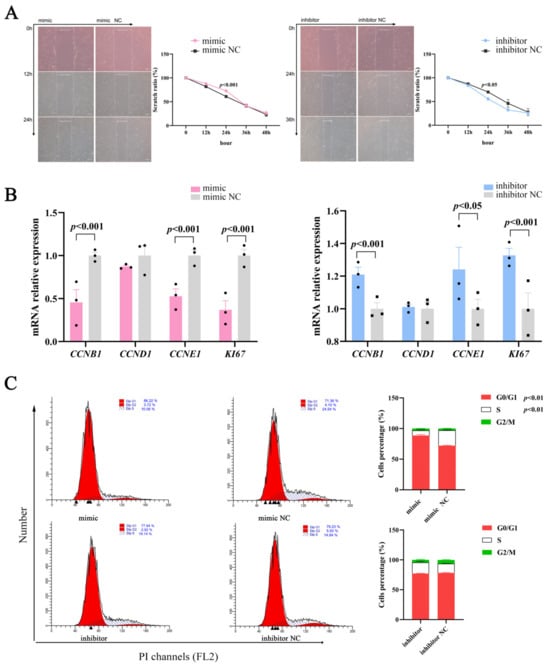
Figure 2.
Effect of miR-129 on YIMAs proliferation after miR-129 overexpression and interference. (A) Scratch test. Scale bar: 200 μm. (B) mRNA expression of proliferative genes. (C) Flow cytometry. ▲: The peak with the highest DNA content in G1 or G2.
2.3. miR-129 Positively Regulates PI3K/AKT Pathway Activity in YIMAs
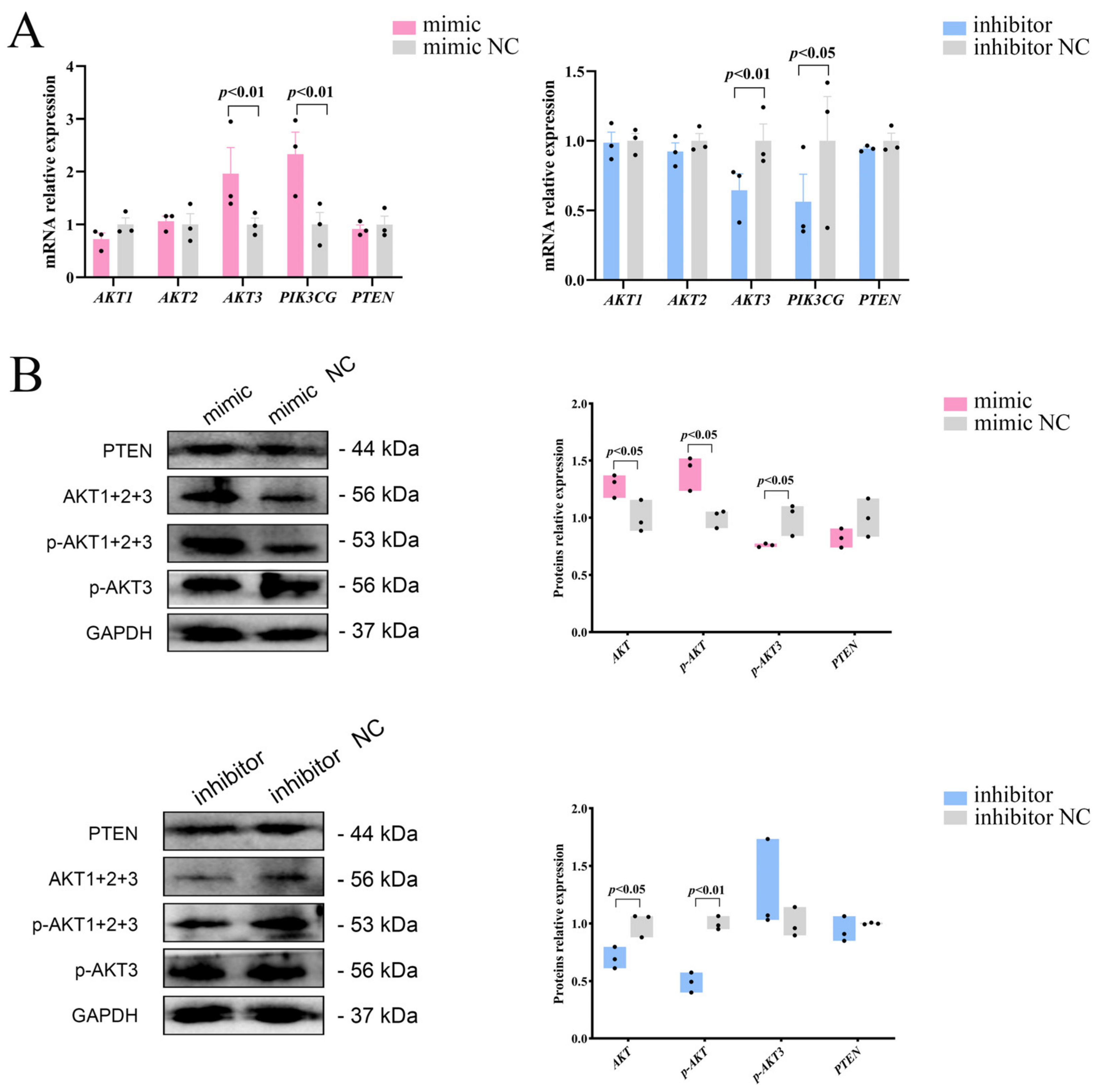
To investigate whether miR-129 influences the proliferation and differentiation of YIMAs via the PI3K/AKT pathway, we initially analyzed the mRNA expression of three isoforms of AKT (AKT1, AKT2, and AKT3) after overexpression or interference with miR-129. We observed significant upregulation and downregulation of AKT3 mRNA expression upon overexpression and interference, respectively. As a nonclassical activator of the PI3K/AKT pathway, PIK3CG showed the same expression pattern as AKT3 (Figure 3A). Western blot results showed that miR-129 overexpression increased the expression of AKT and p-AKT proteins but decreased the expression of p-AKT3 protein. In contrast, miR-129 inhibition significantly reduced the expression of AKT and p-AKT proteins, with no significant change in the expression of p-AKT3 (Figure 3B). This suggests that miR-129 has different effects on the mRNA and protein levels of AKT isoforms in the PI3K/AKT pathway.
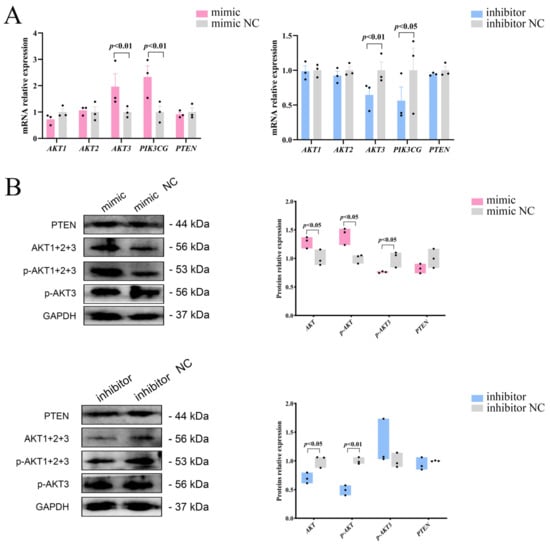
Figure 3.
Effects of miR-129 on PI3K/AKT pathway after miR-129 overexpression and interference. (A) mRNA expression of AKT1, AKT2, AKT3, PTEN, and PIK3CG. (B) Protein expression of AKT, p-AKT, p-AKT3, and PTEN.
2.4. miR-129 Has No Significant Effect on Apoptosis of YIMAs
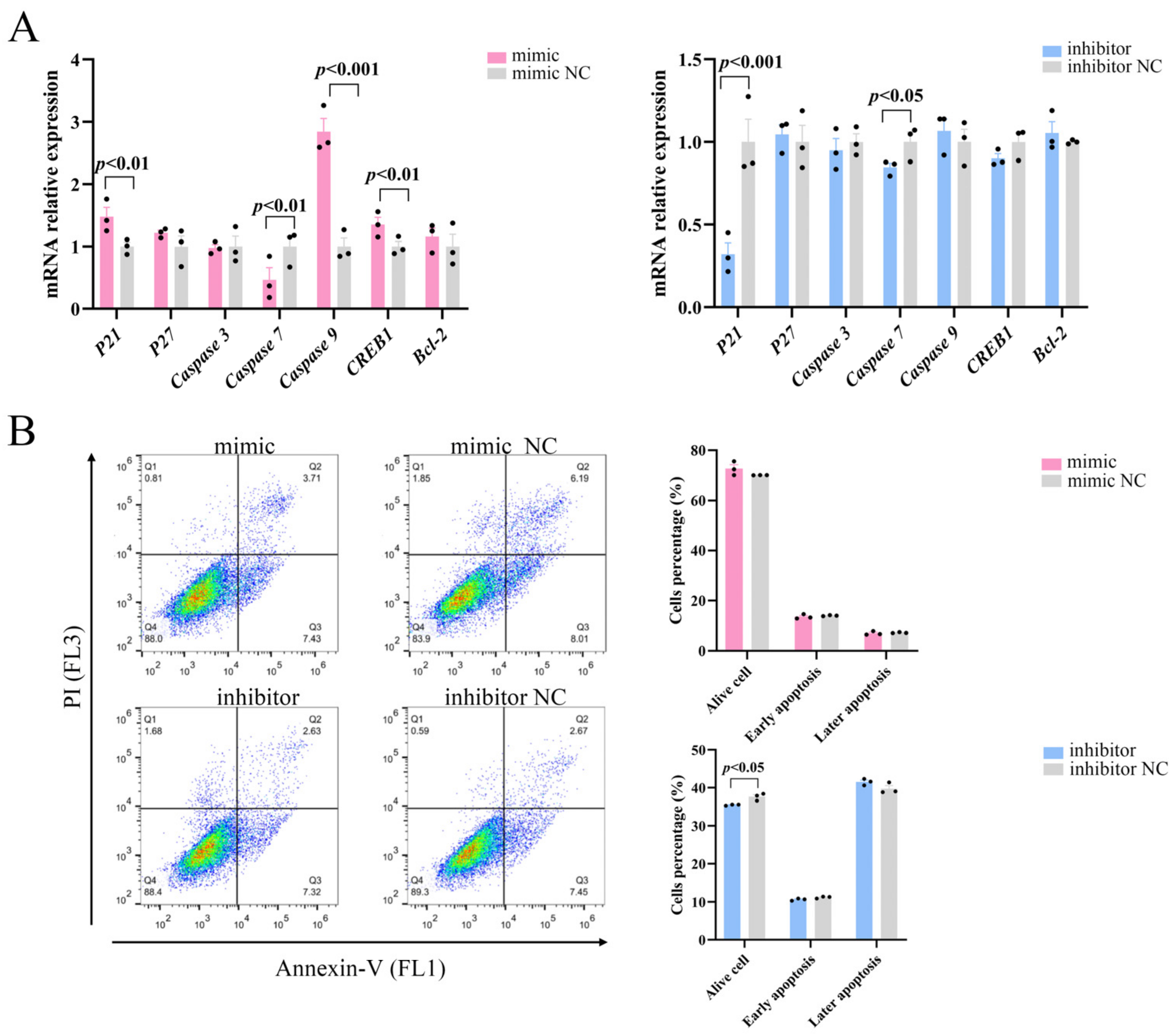
The role of the PI3K/AKT pathway in controlling apoptosis and survival is well-established [20]. We examined whether miR-129 also affects apoptosis in YIMAs via the PI3K/AKT pathway and regulates cell proliferation by examining the expression of essential apoptosis regulators downstream of PI3K/AKT. The results demonstrated that overexpression of miR-129 significantly increased the mRNA expression levels of the apoptosis regulators P21, Caspase9, and CREB1 while reducing the expression of Caspase7. Simultaneously, inhibition of miR-129 led to a decrease in the P21 and Caspase7 mRNA expression levels (Figure 4A). Flow cytometric analysis showed that miR-129 overexpression did not significantly affect early or late apoptosis. However, miR-129 interference resulted in a reduction in the number of viable cells but did not have a significant impact on the ratio of apoptotic cells (Figure 4B). This difference could be attributed to the interference of cell debris during the experiment. Overall, these findings suggested that miR-129 does not significantly impact apoptosis.
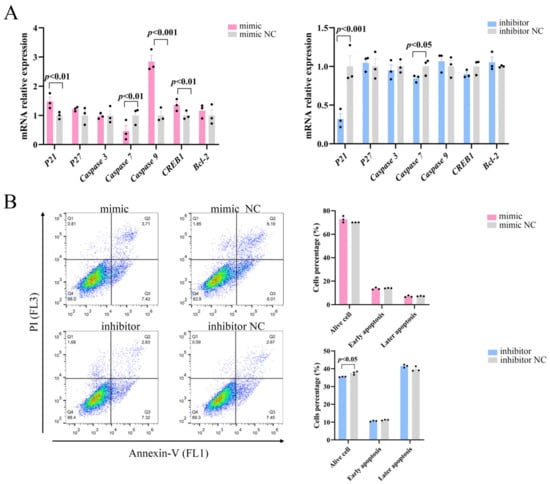
Figure 4.
Effect of miR-129 on YIMAs apoptosis after miR-129 overexpression and interference. (A) mRNA expression of apoptotic regulatory factors. (B) Flow cytometry.
2.5. miR-129 Promotes Differentiation and Inhibits the Proliferation of YIMAs through the PI3K/AKT Pathway
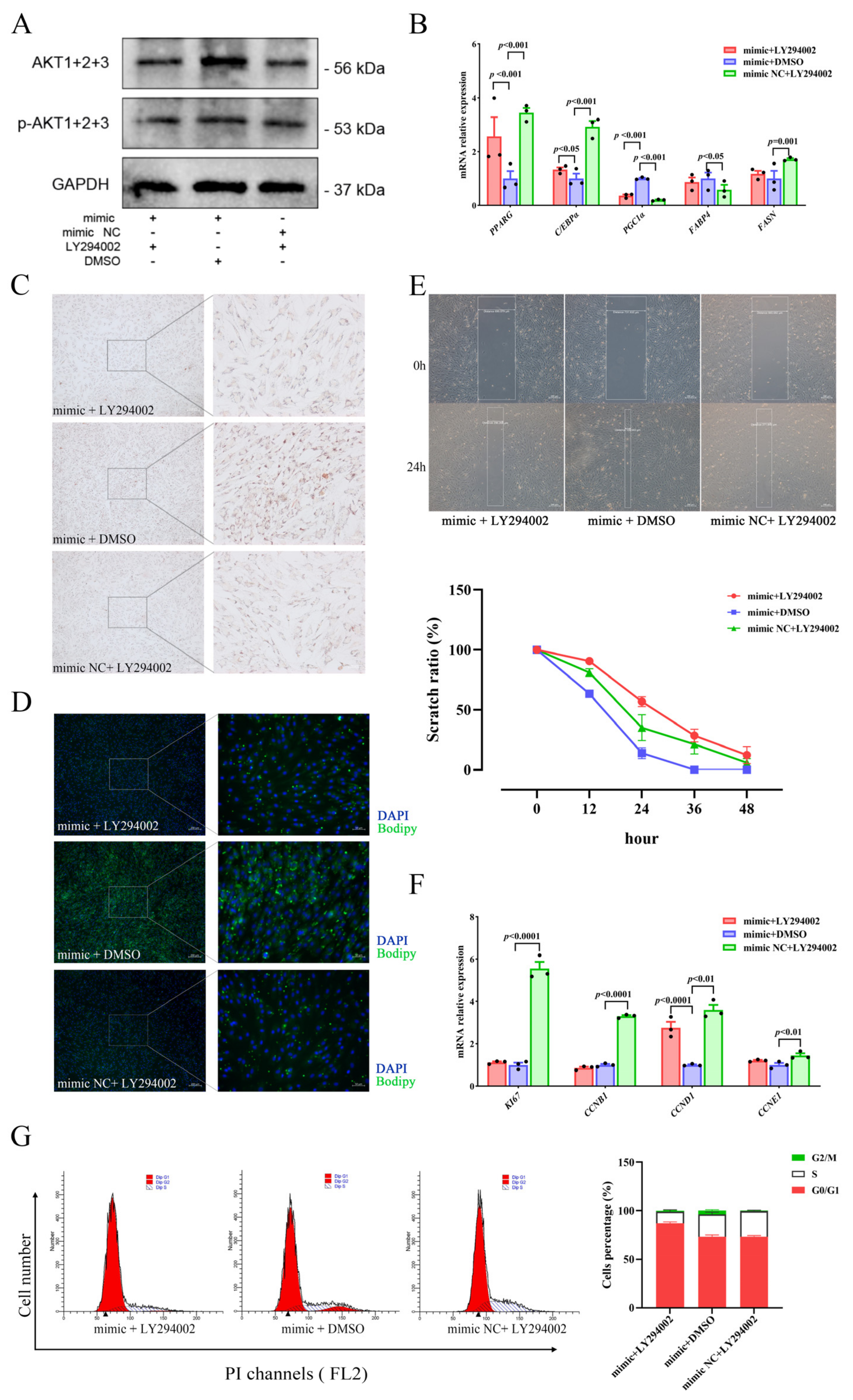
To investigate the relationship regarding regulation between miR-129 and PI3K/AKT pathway activity in YIMAs proliferation and differentiation, we inhibited the PI3K/AKT pathway activity after overexpressing miR-129. As expected, the results showed that overexpression of miR-129, followed by injection of LY294002 (a PI3K/AKT pathway inhibitor), significantly suppressed the expression of AKT and p-AKT proteins (Figure 5A). Meanwhile, RT-qPCR results revealed that inhibiting the PI3K/AKT pathway after miR-129 overexpression significantly upregulated PGC1α expression (Figure 5B). Additionally, Oil Red O and Bodipy staining results indicated that inhibiting the PI3K/AKT pathway after miR-129 overexpression significantly impeded the enhancing effect of miR-129 on cell differentiation (Figure 5C,D). Scratch assay and flow cytometry results indicated an intensification of the inhibitory impact of miR-129 on cell proliferation (Figure 5E–G). Overall, our findings suggest that miR-129 regulates cell differentiation and proliferation by modulating the activity of the PI3K/AKT pathway, and PGC1α may be involved.
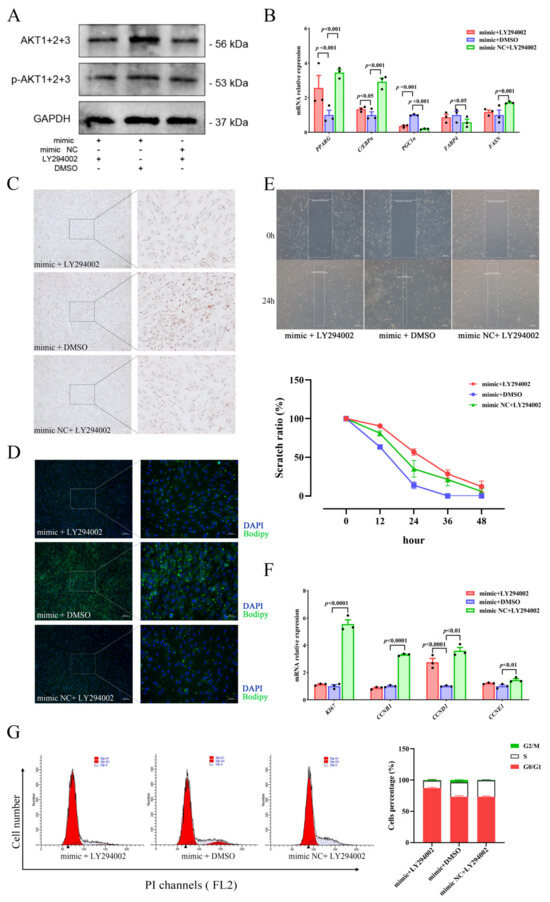
Figure 5.
miR-129 regulates YIMAs proliferation and differentiation through the PI3K/AKT pathway after co-transfection of miR-129 mimic or NC and LY294002 or DMSO. (A) Protein expression of AKT and p-AKT. (B) mRNA expression of differentiation genes. (C) Oil Red O staining. Scale bar: 200 μm, 50 μm. (D) Bodipy staining. Scale bar: 200 μm, 50 μm. (E) Scratch assay. Scale bar: 200 μm. (F) mRNA expression of proliferation genes. (G) Flow cytometry. ▲: The peak with the highest DNA content in G1 or G2.
3. Discussion
Lipid deposition, particularly in intramuscular fat, is a crucial factor influencing beef quality and is considered one of the most economically significant traits in beef production [21]. This process is primarily controlled by preadipocyte turnover, which involves cell proliferation, differentiation, and apoptosis [22]. NcRNAs, particularly miRNAs, have been found to play crucial roles in regulating adipocyte proliferation, differentiation, and apoptosis [23,24,25,26]. miR-129 has now been identified as a potential biomarker of tumor cell growth and development. Numerous studies have demonstrated that miR-129 indirectly regulates adipose tissue development and metabolism [27,28,29,30,31]. In our previous study, we observed that the expression of miR-129 significantly decreased after the overexpression of lncFAM200B, which promotes adipogenesis, and that the expression of miR-129 exhibited an upward trend during the differentiation of YIMAs. These findings suggest that miR-129 plays a role in intramuscular lipid deposition in yaks. We conducted miR-129 overexpression and knockdown experiments using YIMAs. Our results revealed that miR-129 promoted the differentiation and inhibited the proliferation of YIMAs, as observed through Oil Red O, Bodipy, and flow cytometry experiments. However, miR-129 did not affect apoptosis.
The PI3K/AKT signaling pathway, known for its role in insulin signaling, is closely associated with adipogenesis [32]. Although previous studies on yaks have primarily focused on fibroblast proliferation [33], intestinal inflammation [34], and reproduction [35], there is limited research on fat metabolism. Numerous studies have indicated that miR-129 is involved in regulating the PI3K/AKT pathway [16,17,18]. Based on these findings, we proposed that miR-129 affects lipid deposition in yaks via the PI3K/AKT signaling pathway. As expected, our experiments showed that miR-129 positively regulates the activity of the PI3K/AKT pathway by promoting AKT phosphorylation, thereby promoting YIMAs cell differentiation and inhibiting proliferation. miRNAs function by inhibiting translation or causing the degradation of messenger RNA (mRNA) [36]. The PTEN/PI3K/AKT pathway plays a crucial role in cell proliferation, apoptosis, and tumor growth [37]. In a study involving rats with chronic heart failure, miR-129-5p was found to inhibit PTEN ubiquitination and enhance PTEN expression by targeting Smurf1 [38]. The restoration of cardiac function led us to investigate whether miR-129 regulates yak lipid deposition by influencing the PI3K/AKT pathway via PTEN. However, we discovered that the impact of miR-129 on intramuscular fat lipid deposition was not influenced by PTEN (Figure 3A,B) and that this inconsistency may be species-specific.
To investigate the relationship between miR-129 and the PI3K/AKT pathway in regulating the proliferation and differentiation of YIMAs, we identified 290 putative target genes of miR-129 by analyzing the overlapping intersections of three databases (miRDB, TargetScan, and miRWalk). However, we did not find any typical targets related to the PI3K/AKT pathway, indicating that miR-129 regulates this pathway indirectly. Instead, we identified the classical pathways that regulate adipocyte differentiation and lipid metabolism. These pathways included the Wnt, calcium, Hippo, and cAMP signaling pathways (Table S1). Notably, the cAMP signaling pathway is considered to be upstream of the PI3K/AKT pathway. We observed that CAMK4, a serine/threonine kinase family member, is enriched in the cAMP signaling pathway and regulates the expression of various genes [39,40]. Increased lipid deposition in tissues is closely associated with insulin resistance, and CAMK4 enhances insulin sensitivity and indirectly modulates lipid metabolism [41,42,43,44]. Furthermore, studies have shown that miR-129 can target the CAMK4 and that PI3K/AKT acts as a downstream mediator of CAMK4 in saffronin signaling [45,46]. Therefore, we hypothesized that miR-129 affects lipid deposition through the cAMP-PI3K/AKT signaling pathway involving CAMK4. However, the exact mechanism underlying this process requires further experimental validation.
In this study, we observed that miR-129 affects PPARG, its coactivators (PGC1α), and downstream target genes (FABP4), indicating that miR-129 may regulate the expression of downstream target genes through PPARG and subsequently regulate adipose deposition in yaks. However, inhibition of the PI3K/AKT pathway led to increased PPARG expression when miR-129 was overexpressed, suggesting that miR-129 independently regulates intramuscular lipid deposition in yaks through the PI3K/AKT pathway without involving PPARG. The upregulation of PPARG may result from the disrupted lipid balance caused by the inhibition of the PI3K/AKT pathway. It is worth noting that PGC1α, a downstream target of the PI3K/AKT signaling pathway, is positively influenced by the phosphorylated PI3K/AKT signaling pathway, which enhances its activity [47]. Conversely, when the PI3K/AKT pathway was inhibited, the expression of PGC1α decreased; this conclusion is consistent with the results of our analysis. Based on these findings, we propose that miR-129 may regulate lipid deposition in yaks by influencing the PI3K/AKT pathway and subsequently modulating the expression of PGC1α. However, further investigations are required to elucidate this mechanism fully.
In conclusion, our study showed that miR-129 promotes the differentiation of YIMAs and inhibits their proliferation by activating the PI3K/AKT pathway. This study provides a theoretical foundation for further exploration of the regulatory mechanisms of miRNAs in the adipogenesis of YIMAs.
4. Materials and Methods
4.1. Cell Culture and Transfection
Based on a previous study [48], yak preadipocytes were isolated from the longissimus dorsi muscle tissue. The cells were then cultured in a complete culture medium consisting of Dulbecco’s modified Eagle medium (DMEM/F12) (HyClone, Logan, UT, USA), 10% fetal bovine serum (Gibco, Waltham, MA, USA), and 1% penicillin–streptomycin (Boster, Wuhan, China) under sterile conditions at 37 °C and 5% CO2 until they reached a cell density of 80–90%. Transfection was performed using the Lipofectamine 3000 reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions. The miR-129 mimic, inhibitor, and corresponding NC were synthesized by Tsingke Biotech (Tsingke, Beijing, China). The miR-129 sequence was available in the NCBI for Biotechnology Information BioProject database (accession number PRJNA831897). The treated YIMAs were incubated in either a complete culture medium or induced differentiation medium (complete culture medium containing 100 µM oleic acid) (Sigma Aldrich, St. Louis, MO, USA) for 48 h, after which mRNA and protein expression were detected. The primer sequences are provided in Table S2.
4.2. RT-qPCR
Total RNA was extracted using TRIzol (Takara, Shiga, Japan), and complementary DNA (cDNA) was prepared using the PrimeScript RT Reagent Kit (Takara). Stem-loop reverse transcription (RT) primers were designed based on previous studies on miRNA and U6 cDNA synthesis [49]. RT-qPCR was conducted using the SYBR Premix Ex Taq kit (Takara) following the manufacturer’s instructions. Gene expression levels and miRNA levels were normalized using the 2−∆∆Ct method with GAPDH and U6 as internal references, respectively. The primer sequences are shown in Table S3.
4.3. Oil Red O and Bodipy Stain
In 6-well plates, cells were seeded and transfected with the miR-129 mimic or inhibitor (with their respective controls). Additionally, the miR-129 mimic was co-transfected with either the PI3K/AKT pathway inhibitor, LY294002 (Beyotime, Shanghai, China), or dimethyl sulfoxide (Boster) to investigate the relationship between miR-129 and the PI3K/AKT pathway in regulating the proliferative differentiation of YIMAs. Differentiated and mature YIMAs were washed thrice with PBS and fixed with 4% paraformaldehyde (Biosharp, Hefei, China) at room temperature for 1 h. After removing the fixative with PBS, the cells were stained with either Oil Red O (Sigma Aldrich) or Bodipy stain (Invitrogen) at room temperature for 30 min each and micrographed (Carl Zeiss, Oberkochen, Germany). Next, 200 µL of isopropanol was added to each well to extract the Oil Red O staining solution, and the optical density (OD) value was measured at 510 nm.
4.4. Scratch Test
A scratch assay was used to examine the impact of each treatment on cell proliferation, and the transfection pattern was consistent with the Oil Red O and BODIPY staining. Uniform scratching along the central axis of the well plate was performed using a sterile pipette tip after transfection. The scratch width was measured every 12 h to calculate the scratch ratio.
4.5. Flow Cytometry
Flow cytometry was conducted using Sysmex Cube 8.0 (Sysmex, Kobe, Japan). Transfection was performed in accordance with Oil Red O and Bodipy staining. For apoptosis analysis, cells were collected after digestion with 0.25% trypsin (Boster), washed with pre-cooled PBS, centrifuged at 1500 rpm for 5 min, and stained with the Annexin V-FITC/PI Apoptosis Detection Kit (Vazyme, Nanjing, China) at room temperature. For cell cycle analysis, the washed cells were fixed with 70% anhydrous ethanol at 4 °C overnight. After washing with pre-cooled PBS solution, 50 ng/mL PI reagent (Solarbio, Beijing, China) was added and incubated at room temperature with light protection for 30 min before assay. Flow cytometry data were de-adhered and graphically fitted using ModFit 32 software, whereas flow apoptosis data were analyzed using FlowJo (v10) software.
4.6. Western Blot
Total cellular proteins were extracted using cell lysis buffer containing protease inhibitors (Boster) and phosphatase inhibitors (Biosharp) for western blotting and IP (Beyotime). The protein content of each sample group was determined using the BCA protein quantification kit (Thermo Fisher Scientific, Waltham, MA, USA). Subsequently, 20 μg of protein from each group of samples was separated on 8% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a polyvinylidene fluoride (PVDF) membrane. Then, 5% skim milk powder was used to seal at room temperature for 1.5 h. After sealing, the PVDF membrane was treated with primary antibodies (Anti-phospho-PIK3CG (Bioss, Beijing, China, 1:2000), Anti-PTEN (Bioss, 1:2000), Anti-AKT1+2+3 (Bioss, 1:2000), Anti-phospho-AKT1+2+3 (Bioss, 1:2000), Anti-phospho-AKT3 (Bioss, 1:2000), GAPDH Antibody (Affinity, Liyang, China, 1:2000), and secondary antibodies (Goat Anti-Rabbit IgG H&L (HRP), Abcam, Cambridge, England, 1:5000). Photographic observations were made using an ultrasensitive ECL chemiluminescent substrate (Biosharp). Western blot results were analyzed using ImageJ v1.54d software (National Institute of Health, Bethesda, MD, USA).
4.7. Statistical Analysis
Three independent biological samples were used for each experiment. Student’s t-test or one-way analysis of variance (ANOVA) was performed using SPSS version 25.0 (IBM Corp, Armonk, NY, USA). Duncan’s test was employed to determine significance, and values are presented as mean ± standard error of the mean (SEM). The data were visualized using GraphPad Prism v8.0 (GraphPad Software, La Jolla, CA, USA).
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms25010632/s1.
Author Contributions
Conceptualization, H.W. and W.P.; Methodology, C.Q. and H.R.; Formal analysis, J.Z. and H.R.; Resources, H.W.; Writing—original draft preparation, C.Q.; Writing—review and editing, H.W. and C.Q. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (No. 32372849); the National Key Research and Development Program of China (2022YFD1601600); the Qinghai Science and Technology Program, China (No. 2022-NK-110); the Fundamental Research Funds for the Central Universities, Southwest Minzu University (ZYN2023101); and the Program of National Beef Cattle and Yak Industrial Technology System (No. CARS-37). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Institutional Review Board Statement
All animal experimental procedures were conducted in compliance with the guidelines of the Regional Ethics Committee for Animal Experimentation and the care regulations approved by the Animal Protection and Utilisation Committee of the Southwest Minzu University for Nationalities (Permit No. SMU 202106010).
Informed Consent Statement
All authors have approved of the contents of this manuscript and provided consent for publication.
Data Availability Statement
All data generated or analyzed during this study are included in this published article. The data that support the findings of this study are available from the corresponding author upon request.
Acknowledgments
All the authors thank the Key Laboratory of Qinghai–Tibetan Plateau Animal Genetic Resource Reservation and Utilization (Sichuan Province and Ministry of Education) of Southwest Minzu University for providing the experimental platform and conditions.
Conflicts of Interest
Authors declare no conflict of interest.
References
- Chang, X.; Zhang, J.; Liu, Z.; Luo, Z.; Chen, L.; Wang, J.; Geng, F. Integrated proteomic, phosphoproteomic, and N-glycoproteomic analyses of the longissimus thoracis of yaks. Curr. Res. Food Sci. 2022, 5, 1494–1507. [Google Scholar] [CrossRef] [PubMed]
- Yan, E.; Wang, Y.; He, L.; Guo, J.; Zhang, X.; Yin, J. Effects of Dietary L-malic Acid Supplementation on Meat Quality, Antioxidant Capacity and Muscle Fiber Characteristics of Finishing Pigs. Foods 2022, 11, 3335. [Google Scholar] [CrossRef]
- Wang, H.; Zhong, J.; Zhang, C.; Chai, Z.; Cao, H.; Wang, J.; Zhu, J.; Wang, J.; Ji, Q. The whole-transcriptome landscape of muscle and adipose tissues reveals the ceRNA regulation network related to intramuscular fat deposition in yak. BMC Genom. 2020, 21, 347. [Google Scholar] [CrossRef] [PubMed]
- Ran, H.; Yang, Y.; Luo, M.; Liu, X.; Yue, B.; Chai, Z.; Zhong, J.; Wang, H. Molecular Regulation of Yak Preadipocyte Differentiation and Proliferation by LncFAM200B and ceRNA Regulatory Network Analysis. Cells 2022, 11, 2366. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Jiang, E.; Kang, Z.; Bi, Y.; Liu, H.; Xu, H.; Wang, Z.; Lei, C.; Chen, H.; Lan, X. CircRNA Profiling Reveals an Abundant circBDP1 that Regulates Bovine Fat Development by Sponging miR-181b/miR-204 Targeting Sirt1/TRARG1. J. Agric. Food Chem. 2022, 70, 14312–14328. [Google Scholar] [CrossRef]
- Long, F.; Wang, X.; Wan, Y.; Zhang, Z.; Zhang, W.; Zan, L.; Cheng, G. Bta-miR-493 Inhibits Bovine Preadipocytes Differentiation by Targeting BMPR1A via the TGFbeta/BMP and p38MAPK Signaling Pathways. J. Agric. Food Chem. 2022, 70, 14641–14653. [Google Scholar] [CrossRef]
- Feng, X.; Zhao, J.; Li, F.; Aloufi, B.H.; Alshammari, A.M.; Ma, Y. Weighted Gene Co-expression Network Analysis Revealed That CircMARK3 Is a Potential CircRNA Affects Fat Deposition in Buffalo. Front. Vet. Sci. 2022, 9, 946447. [Google Scholar] [CrossRef]
- Ma, F.; Zhan, Y.; Bartolome-Cabrero, R.; Ying, W.; Asano, M.; Huang, Z.; Xiao, C.; Gonzalez-Martin, A. Analysis of a miR-148a Targetome in B Cell Central Tolerance. Front. Immunol. 2022, 13, 861655. [Google Scholar] [CrossRef]
- Meng, Q.; Wu, W.; Pei, T.; Xue, J.; Xiao, P.; Sun, L.; Li, L.; Liang, D. miRNA-129/FBW7/NF-kappaB, a Novel Regulatory Pathway in Inflammatory Bowel Disease. Mol. Ther.-Nucl. Acids 2020, 19, 731–740. [Google Scholar] [CrossRef]
- Liao, C.; Long, Z.; Zhang, X.; Cheng, J.; Qi, F.; Wu, S.; Huang, T. LncARSR sponges miR-129-5p to promote proliferation and metastasis of bladder cancer cells through increasing SOX4 expression. Int. J. Biol. Sci. 2020, 16, 1–11. [Google Scholar] [CrossRef]
- Liang, Y.; Wang, Y.; Ma, L.; Zhong, Z.; Yang, X.; Tao, X.; Chen, X.; He, Z.; Yang, Y.; Zeng, K.; et al. Comparison of microRNAs in adipose and muscle tissue from seven indigenous Chinese breeds and Yorkshire pigs. Anim. Genet. 2019, 50, 439–448. [Google Scholar] [CrossRef] [PubMed]
- Pahlavani, M.; Wijayatunga, N.N.; Kalupahana, N.S.; Ramalingam, L.; Gunaratne, P.H.; Coarfa, C.; Rajapakshe, K.; Kottapalli, P.; Moustaid-Moussa, N. Transcriptomic and microRNA analyses of gene networks regulated by eicosapentaenoic acid in brown adipose tissue of diet-induced obese mice. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2018, 1863, 1523–1531. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Feng, Y.; Zhang, H.; Niu, Q.; Liang, K.; Bian, C.; Li, H. Clinical Value and Role of miR-129-5p in Non-Alcoholic Fatty Liver Disease. Horm. Metab. Res. 2021, 53, 692–698. [Google Scholar] [CrossRef] [PubMed]
- Cerda, A.; Bortolin, R.H.; Manriquez, V.; Salazar, L.; Zambrano, T.; Fajardo, C.M.; Hirata, M.H.; Hirata, R. Effect of statins on lipid metabolism-related microRNA expression in HepG2 cells. Pharmacol. Rep. 2021, 73, 868–880. [Google Scholar] [CrossRef] [PubMed]
- Aierken, A.; Li, B.; Liu, P.; Cheng, X.; Kou, Z.; Tan, N.; Zhang, M.; Yu, S.; Shen, Q.; Du, X.; et al. Melatonin treatment improves human umbilical cord mesenchymal stem cell therapy in a mouse model of type II diabetes mellitus via the PI3K/AKT signaling pathway. Stem Cell Res. Ther. 2022, 13, 164. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liang, G.; Wang, H.; Liu, Z. MicroRNA-129-5p suppresses proliferation, migration and invasion of retinoblastoma cells through PI3K/AKT signaling pathway by targeting PAX6. Pathol. Res. Pract. 2019, 215, 152641. [Google Scholar] [CrossRef]
- Chen, D.; Wang, H.; Chen, J.; Li, Z.; Li, S.; Hu, Z.; Huang, S.; Zhao, Y.; He, X. MicroRNA-129-5p Regulates Glycolysis and Cell Proliferation by Targeting the Glucose Transporter SLC2A3 in Gastric Cancer Cells. Front. Pharmacol. 2018, 9, 502. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Ge, J.; Zhang, Z.; Zhou, W. MiR-129 inhibits cell proliferation and metastasis by targeting ETS1 via PI3K/AKT/mTOR pathway in prostate cancer. Biomed. Pharmacother. 2017, 96, 634–641. [Google Scholar] [CrossRef]
- Ruan, C.; Li, X.; Hu, J.; Zhang, Y.; Zhao, X. MITF and PU.1 inhibit adipogenesis of ovine primary preadipocytes by restraining C/EBPbeta. Cell. Mol. Biol. Lett. 2017, 22, 2. [Google Scholar] [CrossRef]
- Wang, L.; Wang, Z.; Yang, Z.; Yang, K.; Yang, H. Study of the Active Components and Molecular Mechanism of Tripterygium wilfordii in the Treatment of Diabetic Nephropathy. Front. Mol. Biosci. 2021, 8, 664416. [Google Scholar] [CrossRef]
- Khan, R.; Raza, S.; Junjvlieke, Z.; Xiaoyu, W.; Garcia, M.; Elnour, I.E.; Hongbao, W.; Linsen, Z. Function and Transcriptional Regulation of Bovine TORC2 Gene in Adipocytes: Roles of C/EBP, XBP1, INSM1 and ZNF263. Int. J. Mol. Sci. 2019, 20, 4338. [Google Scholar] [CrossRef]
- Zhang, Q.; Cai, R.; Tang, G.; Zhang, W.; Pang, W. MiR-146a-5p targeting SMAD4 and TRAF6 inhibits adipogenensis through TGF-beta and AKT/mTORC1 signal pathways in porcine intramuscular preadipocytes. J. Anim. Sci. Biotechnol. 2021, 12, 12. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Zhang, P.; Gan, M.; Zhao, X.; Xu, Y.; Li, Q.; Jiang, Y.; Tang, G.; Li, M.; Wang, J.; et al. MicroRNA-204-5p regulates 3T3-L1 preadipocyte proliferation, apoptosis and differentiation. Gene 2018, 668, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Xu, Y.; Zhang, P.; Zhao, X.; Gan, M.; Li, Q.; Ma, J.; Tang, G.; Jiang, Y.; Wang, J.; et al. MicroRNA-125a-5p Affects Adipocytes Proliferation, Differentiation and Fatty Acid Composition of Porcine Intramuscular Fat. Int. J. Mol. Sci. 2018, 19, 501. [Google Scholar] [CrossRef]
- Khan, R.; Raza, S.; Junjvlieke, Z.; Wang, X.; Wang, H.; Cheng, G.; Mei, C.; Elsaeid, E.I.; Zan, L. Bta-miR-149-5p inhibits proliferation and differentiation of bovine adipocytes through targeting CRTCs at both transcriptional and posttranscriptional levels. J. Cell. Physiol. 2020, 235, 5796–5810. [Google Scholar] [CrossRef] [PubMed]
- Lv, S.; Ma, M.; Sun, Y.; Wang, X.; Qimuge, N.; Qin, J.; Pang, W. MicroRNA-129-5p inhibits 3T3-L1 preadipocyte proliferation by targeting G3BP1. Anim. Cells Syst. 2017, 21, 269–277. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Yi, Y.; Xia, G.; Zhao, Y.; Yu, Y.; Li, L.; Hua, C.; He, B.; Yang, B.; Yu, C.; et al. Nrf2-miR-129-3p-mTOR Axis Controls an miRNA Regulatory Network Involved in HDACi-Induced Autophagy. Mol. Ther. 2019, 27, 1039–1050. [Google Scholar] [CrossRef]
- Fu, R.; Yang, P.; Sajid, A.; Li, Z. Avenanthramide A Induces Cellular Senescence via miR-129-3p/Pirh2/p53 Signaling Pathway To Suppress Colon Cancer Growth. J. Agric. Food Chem. 2019, 67, 4808–4816. [Google Scholar] [CrossRef]
- Wang, X.; Li, J.; Xu, X.; Zheng, J.; Li, Q. miR-129 inhibits tumor growth and potentiates chemosensitivity of neuroblastoma by targeting MYO10. Biomed. Pharmacother. 2018, 103, 1312–1318. [Google Scholar] [CrossRef]
- Fu, X.; Jin, L.; Han, L.; Yuan, Y.; Mu, Q.; Wang, H.; Yang, J.; Ning, G.; Zhou, D.; Zhang, Z. miR-129-5p Inhibits Adipogenesis through Autophagy and May Be a Potential Biomarker for Obesity. Int. J. Endocrinol. 2019, 2019, 5069578. [Google Scholar] [CrossRef]
- Mi, J.Q. LncRNA-420/MiR-129-5p Competitively Target DLK1 to Regulate the Differentiation of Bovine Preadipocytes. Ph.D. Thesis, Jilin University, Changchun, China, 2021. [Google Scholar]
- Cheng, F.; Han, L.; Xiao, Y.; Pan, C.; Li, Y.; Ge, X.; Zhang, Y.; Yan, S.; Wang, M. d-chiro-Inositol Ameliorates High Fat Diet-Induced Hepatic Steatosis and Insulin Resistance via PKCepsilon-PI3K/AKT Pathway. J. Agric. Food Chem. 2019, 67, 5957–5967. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, Q.; Zhang, Y.; Zhao, X. Yak OXGR1 promotes fibroblast proliferation via the PI3K/AKT pathways. J. Cell. Biochem. 2019, 120, 6729–6740. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.N.; Hu, H.; Wen, P.C.; Lian, S.; Xie, X.L.; Song, H.L.; Yang, Z.N.; Ren, F.Z. Yak milk-derived exosomes alleviate lipopolysaccharide-induced intestinal inflammation by inhibiting PI3K/AKT/C3 pathway activation. J. Dairy Sci. 2021, 104, 8411–8424. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Pan, Y.; Wang, M.; Xu, R.; Han, X.; Ma, R.; Zhao, L.; Zhang, T.; Wang, Y.; Zhao, T.; et al. Follicular fluid exosomes regulate OVGP1 secretion in yak oviduct epithelial cells via autophagy in vitro. J. Cell. Physiol. 2023, 238, 1020–1035. [Google Scholar] [CrossRef] [PubMed]
- Shukla, G.C.; Singh, J.; Barik, S. MicroRNAs: Processing, Maturation, Target Recognition and Regulatory Functions. Mol. Cell Pharmacol. 2011, 3, 83–92. [Google Scholar] [PubMed]
- Selvakumar, S.C.; Preethi, K.A.; Sekar, D. MicroRNAs as important players in regulating cancer through PTEN/PI3K/AKT signalling pathways. Biochim. Biophys. Acta-Rev. Cancer 2023, 1878, 188904. [Google Scholar] [CrossRef]
- Qi, Y.; Tang, Y.; Yin, L.; Ding, K.; Zhao, C.; Yan, W.; Yao, Y. miR-129-5p restores cardiac function in rats with chronic heart failure by targeting the E3 ubiquitin ligase Smurf1 and promoting PTEN expression. Bioengineered 2022, 13, 2371–2386. [Google Scholar] [CrossRef]
- Yong, L.; Yu, Y.; Li, B.; Ge, H.; Zhen, Q.; Mao, Y.; Yu, Y.; Cao, L.; Zhang, R.; Li, Z.; et al. Calcium/calmodulin-dependent protein kinase IV promotes imiquimod-induced psoriatic inflammation via macrophages and keratinocytes in mice. Nat. Commun. 2022, 13, 4255. [Google Scholar] [CrossRef] [PubMed]
- Racioppi, L.; Means, A.R. Calcium/calmodulin-dependent kinase IV in immune and inflammatory responses: Novel routes for an ancient traveller. Trends Immunol. 2008, 29, 600–607. [Google Scholar] [CrossRef]
- Samuel, V.T.; Petersen, K.F.; Shulman, G.I. Lipid-induced insulin resistance: Unravelling the mechanism. Lancet 2010, 375, 2267–2277. [Google Scholar] [CrossRef]
- Morino, K.; Petersen, K.F.; Sono, S.; Choi, C.S.; Samuel, V.T.; Lin, A.; Gallo, A.; Zhao, H.; Kashiwagi, A.; Goldberg, I.J.; et al. Regulation of mitochondrial biogenesis by lipoprotein lipase in muscle of insulin-resistant offspring of parents with type 2 diabetes. Diabetes 2012, 61, 877–887. [Google Scholar] [CrossRef]
- Samuel, V.T.; Shulman, G.I. Mechanisms for insulin resistance: Common threads and missing links. Cell 2012, 148, 852–871. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.Y.; Gattu, A.K.; Camporez, J.P.; Kanda, S.; Guigni, B.; Kahn, M.; Zhang, D.; Galbo, T.; Birkenfeld, A.L.; Jornayvaz, F.R.; et al. Muscle-specific activation of Ca(2+)/calmodulin-dependent protein kinase IV increases whole-body insulin action in mice. Diabetologia 2014, 57, 1232–1241. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Lu, J.; Zeng, G.; Pang, J.; Zheng, X.; Feng, J.; Zhang, J. MiR-129-5p inhibits liver cancer growth by targeting calcium calmodulin-dependent protein kinase IV (CAMK4). Cell Death Dis. 2019, 10, 789. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Park, G.Y.; Bang, S.Y.; Park, S.Y.; Bae, S.K.; Kim, Y. Crocin suppresses LPS-stimulated expression of inducible nitric oxide synthase by upregulation of heme oxygenase-1 via calcium/calmodulin-dependent protein kinase 4. Mediat. Inflamm. 2014, 2014, 728709. [Google Scholar] [CrossRef]
- Wang, L.; Yang, M.; Jin, H. PI3K/AKT phosphorylation activates ERRalpha by upregulating PGC-1alpha and PGC-1beta in gallbladder cancer. Mol. Med. Rep. 2021, 24, 613. [Google Scholar] [CrossRef]
- Khan, R.; Raza, S.; Junjvlieke, Z.; Wang, H.; Cheng, G.; Smith, S.B.; Jiang, Z.; Li, A.; Zan, L. RNA-seq reveal role of bovine TORC2 in the regulation of adipogenesis. Arch. Biochem. Biophys. 2020, 680, 108236. [Google Scholar] [CrossRef]
- Zhou, C.; Han, L.; Fu, C.; Wen, J.; Cheng, X.; Nakashima, J.; Ma, J.; Tang, Y.; Tan, Y.; Tadege, M.; et al. The trans-acting short interfering RNA3 pathway and no apical meristem antagonistically regulate leaf margin development and lateral organ separation, as revealed by analysis of an argonaute7/lobed leaflet1 mutant in Medicago truncatula. Plant Cell 2013, 25, 4845–4862. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).