Copper(II) Complexes with 1-(Isoquinolin-3-yl)heteroalkyl-2-ones: Synthesis, Structure and Evaluation of Anticancer, Antimicrobial and Antioxidant Potential
Abstract
:1. Introduction
2. Results and Discussion
2.1. Chemistry
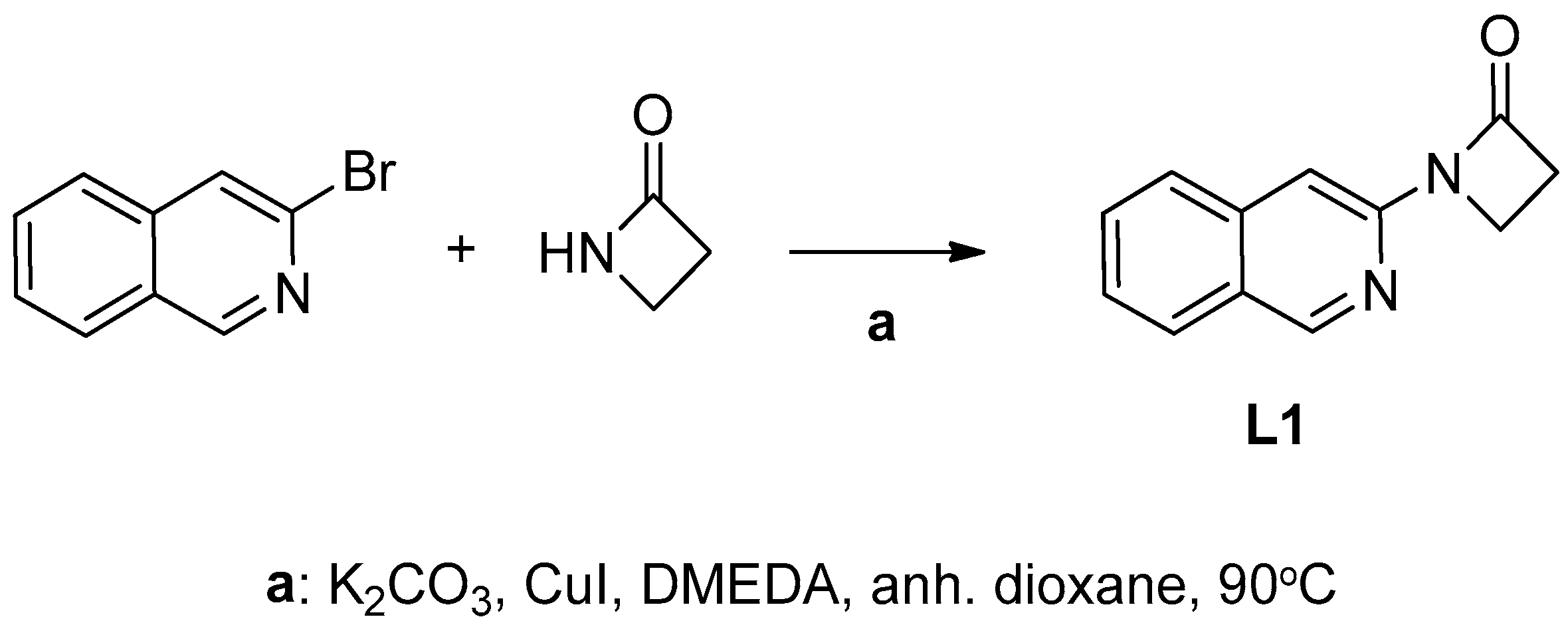
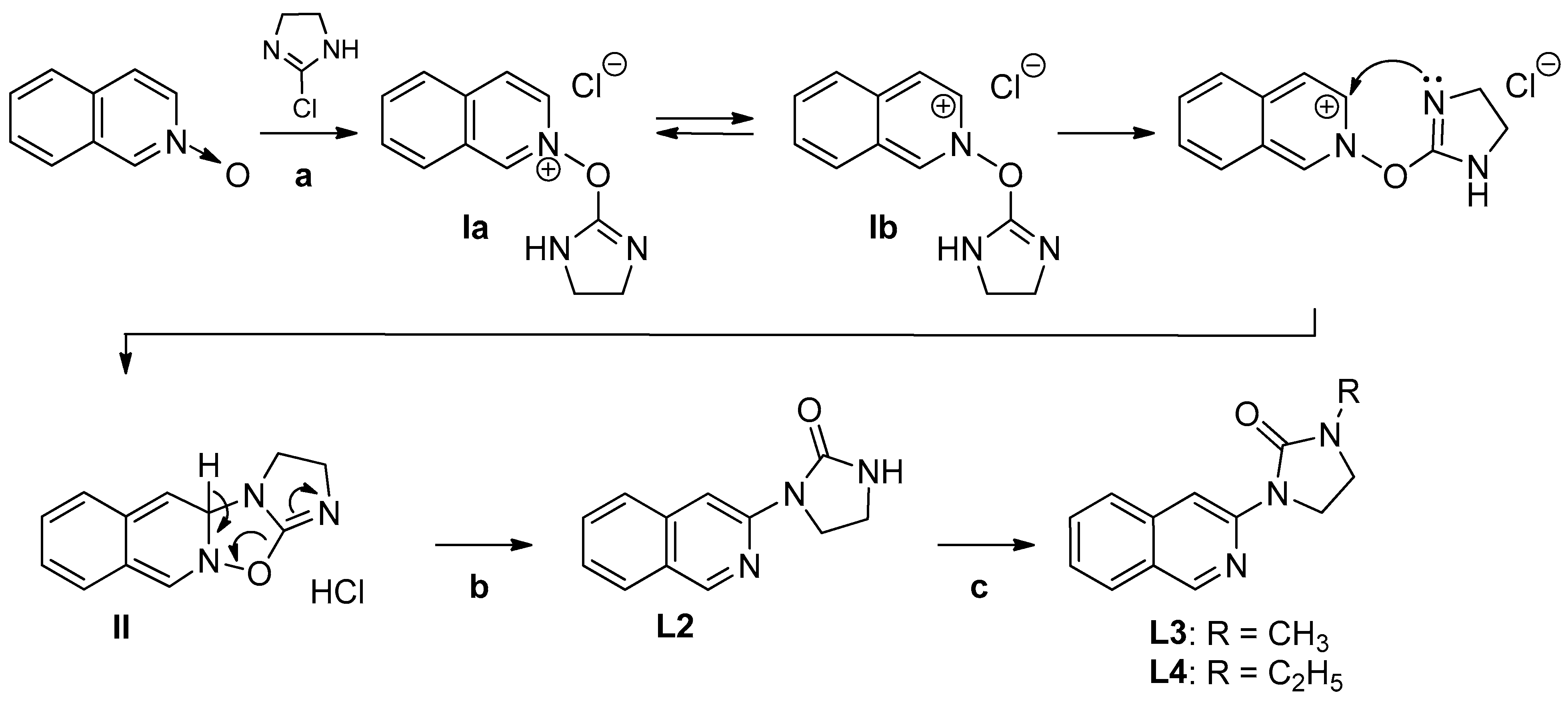
2.1.1. Synthesis of 1-(Isoquinolin-3-yl)heteroalkyl-2-one Ligands L1–4
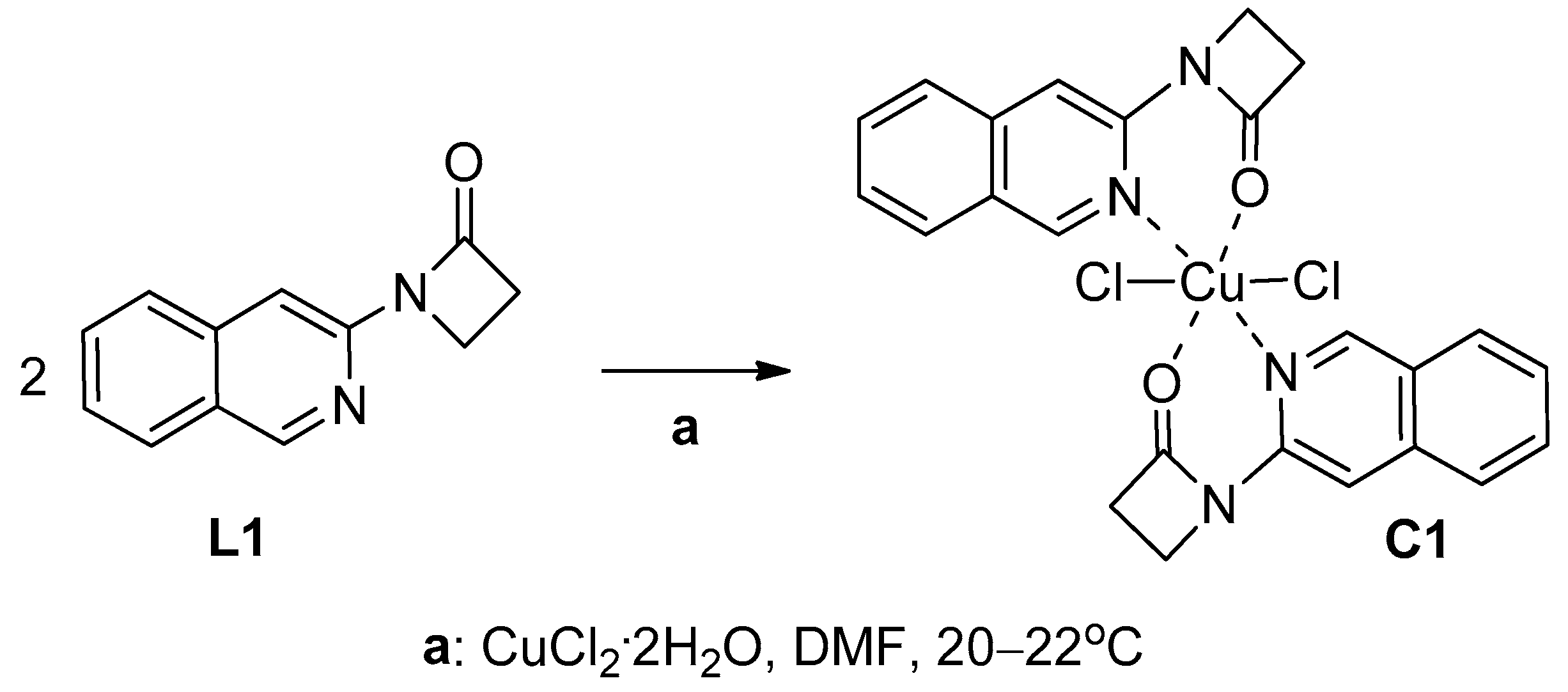
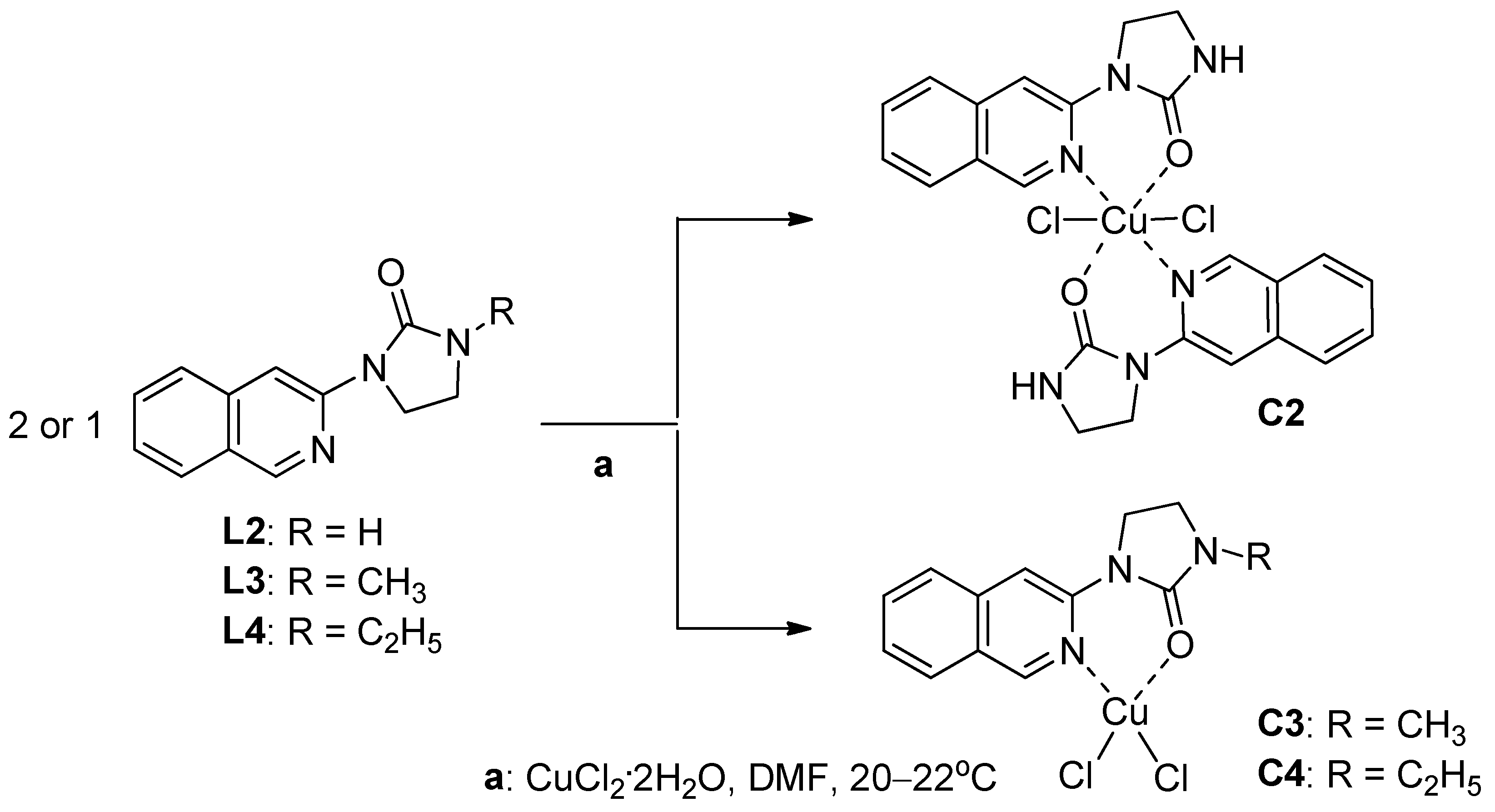
2.1.2. Synthesis of Copper(II) Complexes of 1-(Isoquinolin-3-yl)heteroalkyl-2-ones C1–4
2.2. Structural Analysis of Copper(II) Complexes C1–4
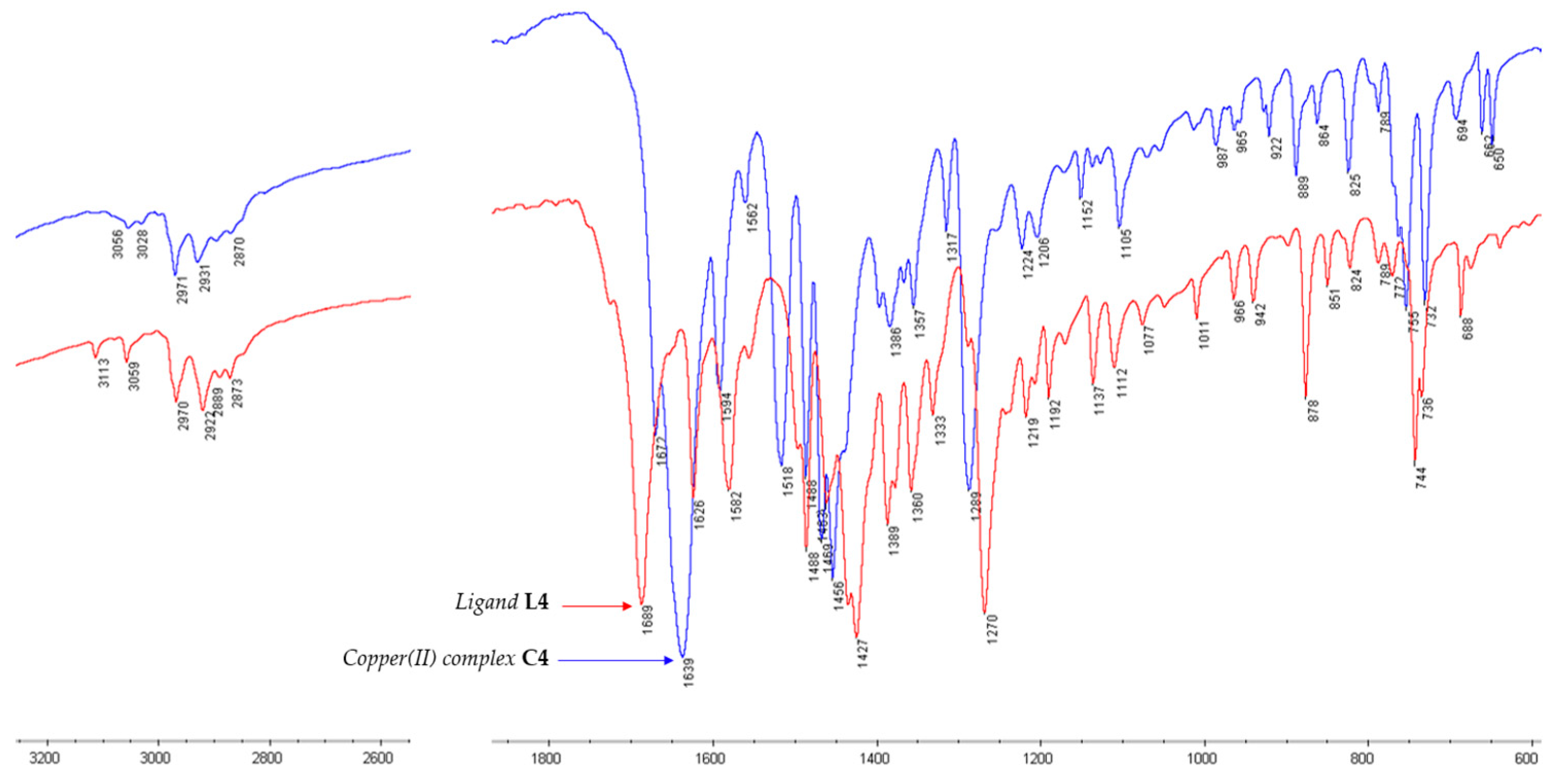
2.2.1. Infrared Spectra
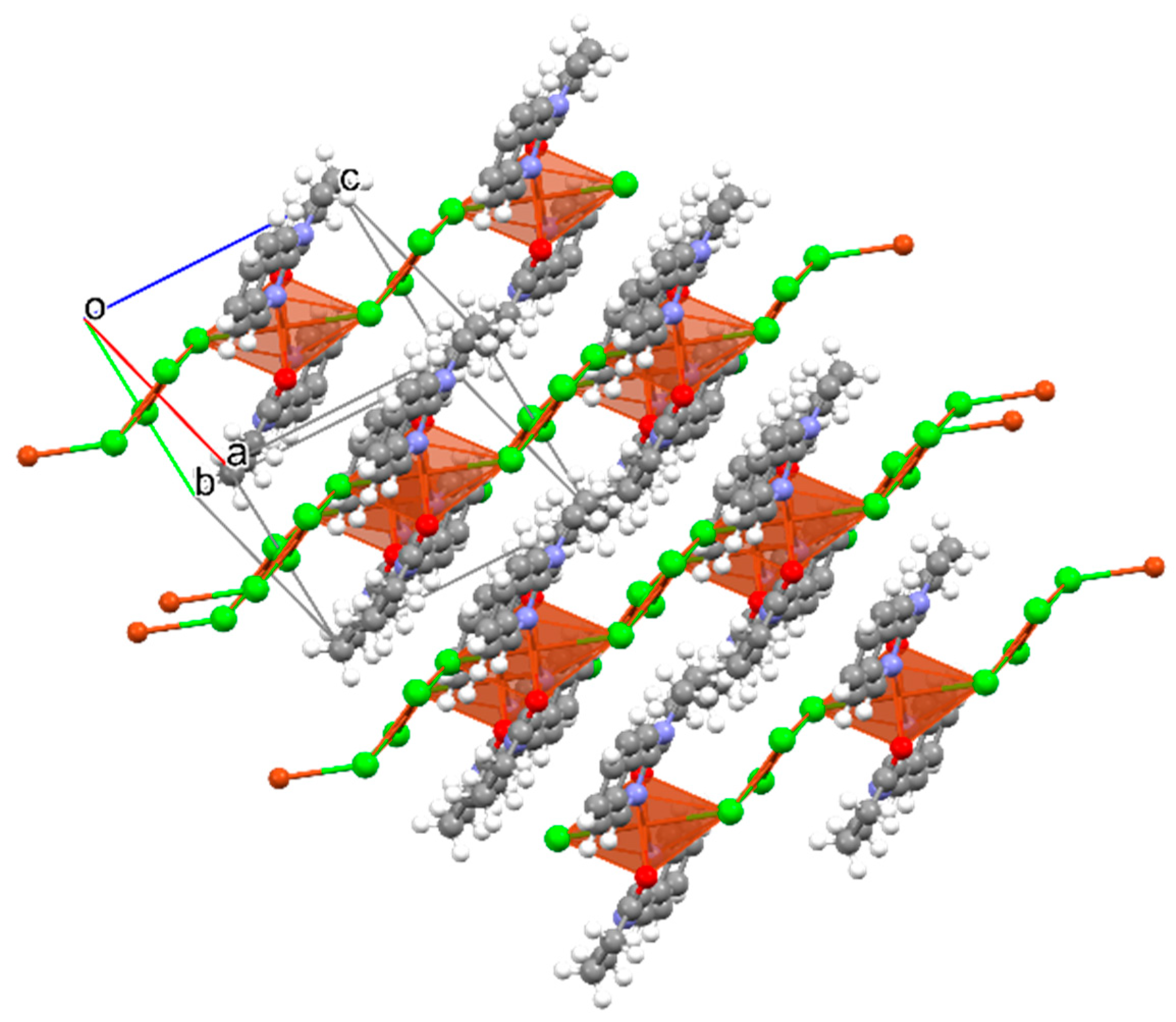
2.2.2. X-ray Crystallographic Studies
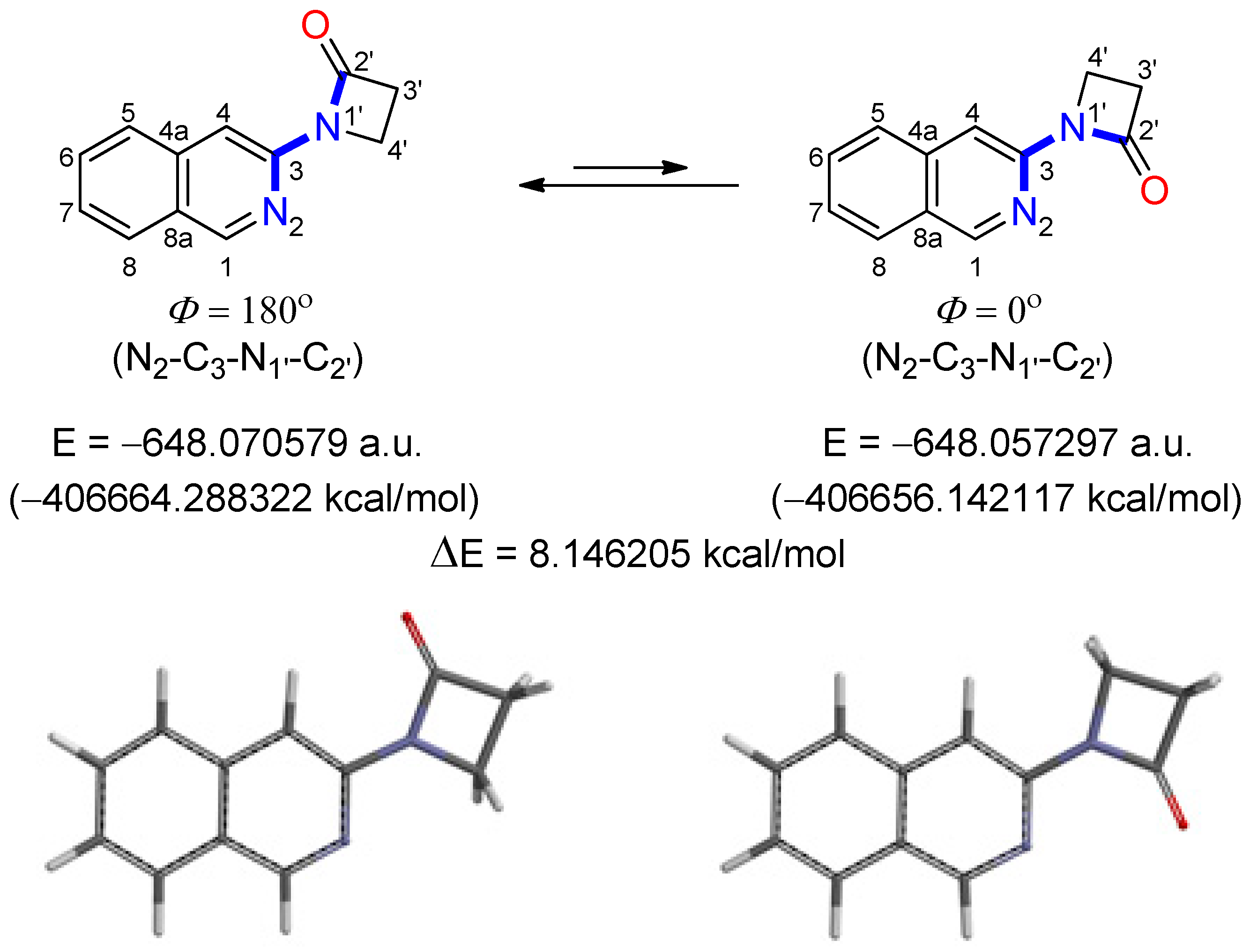
2.2.3. Molecular Modeling Studies of Ligands L1 and L2
2.3. Stability Studies of Copper(II) Complexes C1–4 in Aqueous Buffer
2.4. Biological Evaluation
2.4.1. In Vitro Cytotoxic Activity
2.4.2. Cell Cycle Analysis
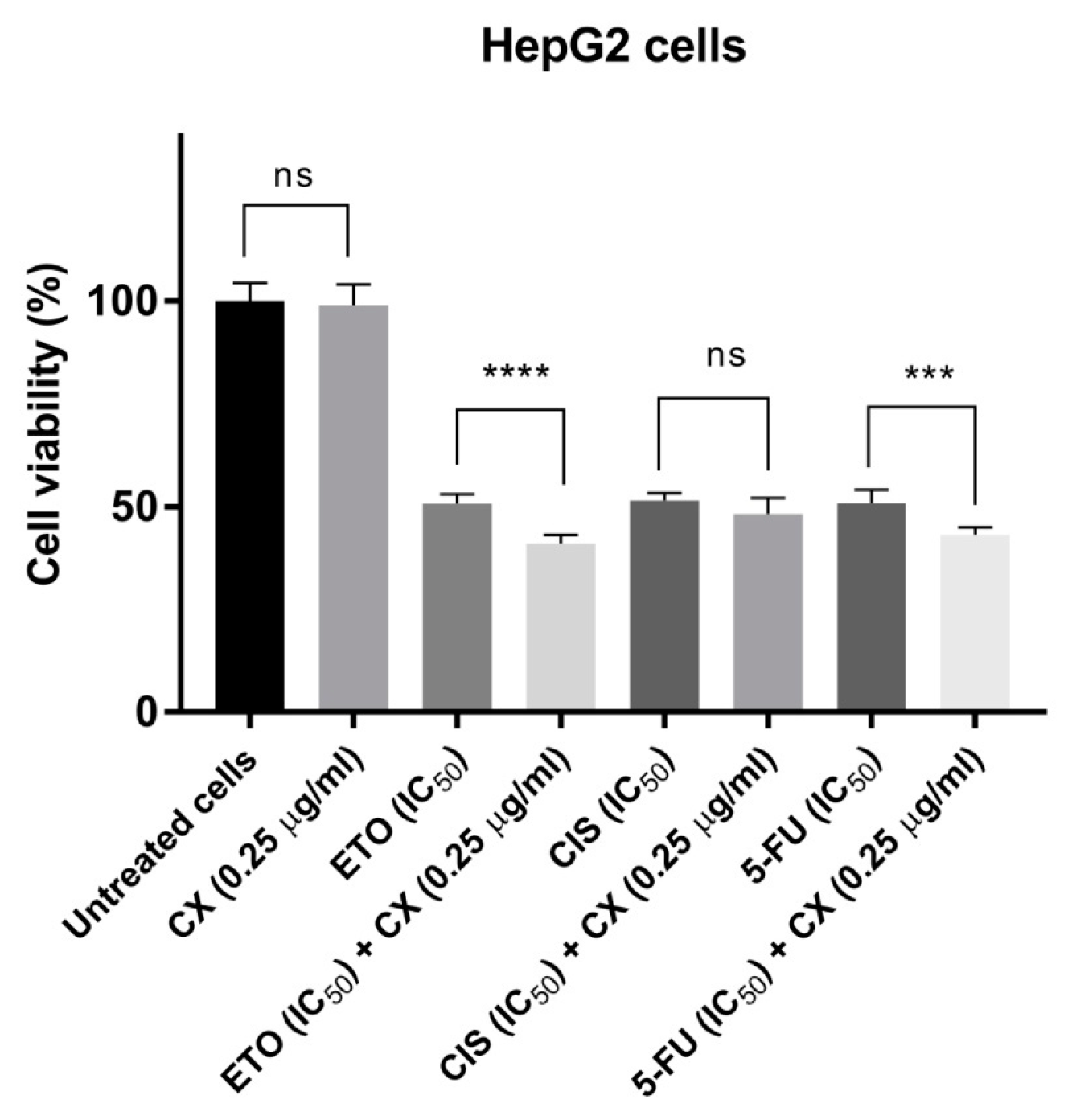
2.4.3. Interaction of Copper(II) Complex C2 with Clinically Used Anticancer Agents
2.4.4. Antimicrobial Activity
2.5. Determination of Free Radical Scavenging Capacity
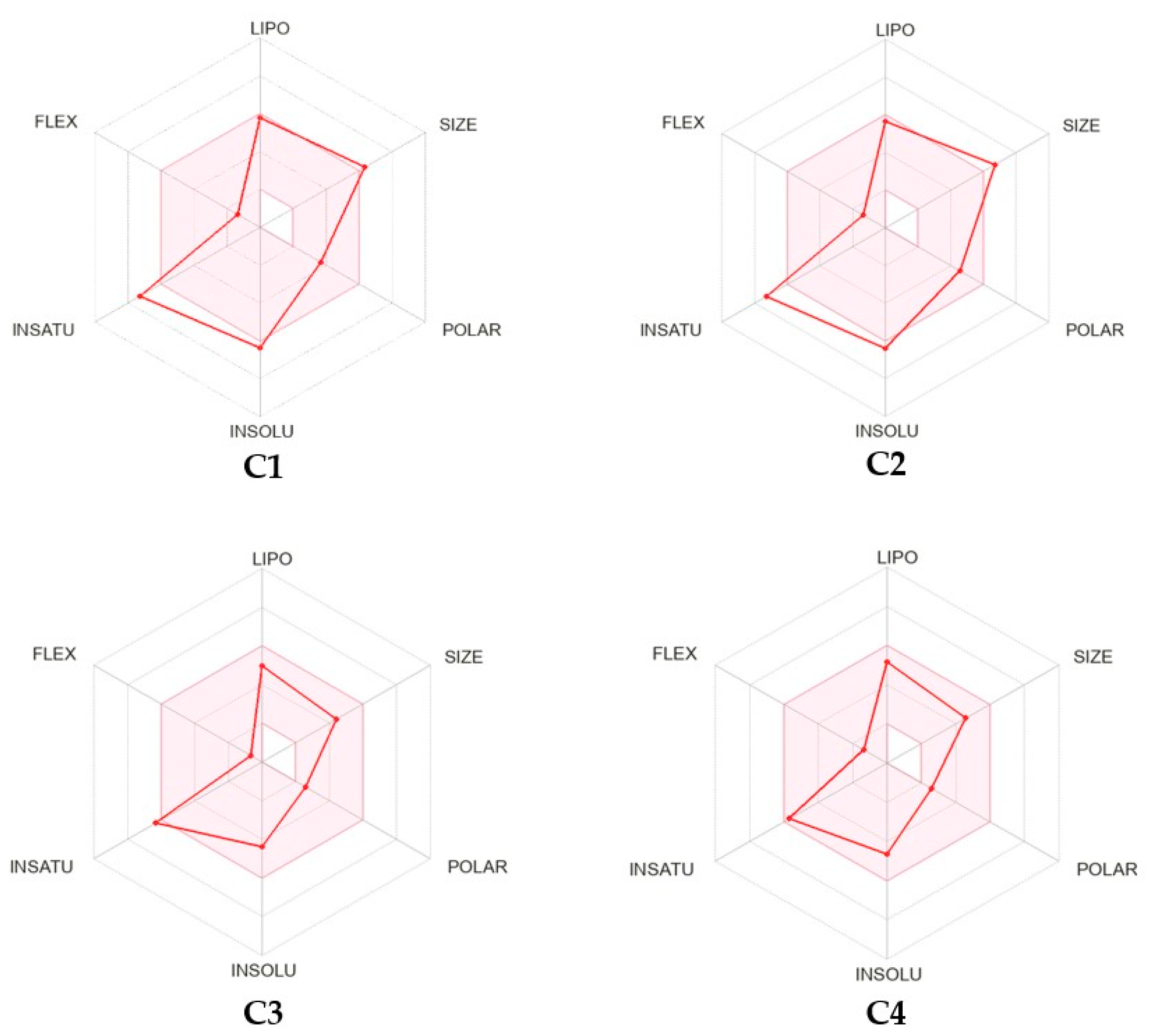
2.6. In Silico Physicochemical, Pharmacokinetic and Drug-Likeness Predictions
3. Materials and Methods
3.1. Chemistry
3.1.1. General Information
3.1.2. Synthesis of 1-(Isoquinolin-3-yl)imidazolidin-2-one (L2)
3.1.3. Synthesis of 1-Ethyl-3-(isoquinolin-3-yl)imidazolidin-2-one (L4)
3.1.4. Synthesis of Copper(II) Complexes of 1-(Isoquinolin-3-yl)heteroalkyl-2-ones C1–4 (General Method)
Dichloro{bis[1-(Isoquinolin-3-yl)azetidin-2-one]}copper(II) (C1)
Dichloro{bis[1-(Isoquinolin-3-yl)imidazolidin-2-one]}copper(II) (C2)
Dichloro[1-(Isoquinolin-3-yl)-3-methylimidazolidin-2-one]copper(II) (C3)
Dichloro[1-ethyl-3-(isoquinolin-3-yl)imidazolidin-2-one]copper(II) (C4)
3.1.5. Single Crystal X-ray Diffraction Studies
3.2. Aqueous Stability Studies
3.3. Biological Studies
3.3.1. Examination of the Cytotoxic Activity with Assessment by MTT Assay
3.3.2. Cell Cycle Analysis
3.3.3. Analysis of Interaction of Copper(II) Complex C2 (CX) with Clinically Used Anticancer Agents
3.3.4. In Vitro Antimicrobial Activity
3.4. Antioxidant
3.4.1. Materials
3.4.2. DPPH Assay
3.4.3. ABTS Assay
3.5. ADME/Drug-Likeness Calculation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gaynor, D.; Griffith, D.M. The prevalence of metal-based drugs as therapeutic or diagnostic agents: Beyond platinum. Dalton Trans. 2012, 41, 13239. [Google Scholar] [CrossRef] [PubMed]
- Tesauro, D. Metal complexes in diagnosis and therapy. Int. J. Mol. Sci. 2022, 23, 4377. [Google Scholar] [CrossRef] [PubMed]
- Johnstone, T.C.; Suntharalingam, K. Lippard. The next generation of platinum drugs: Targeted Pt(II) agents, nanoparticle delivery, and Pt(IV) prodrugs. Chem. Rev. 2016, 116, 3436–3486. [Google Scholar] [CrossRef] [PubMed]
- Zhong, T.; Yu, J.; Pan, Y.; Zhang, N.; Qi, Y.; Huang, Y. Recent advances of platinum-based anticancer complexes in combinational multimodal therapy. Adv. Healthcare Mater. 2023, 12, 2300253. [Google Scholar] [CrossRef] [PubMed]
- Oun, R.; Moussa, Y.E.; Wheate, N.J. The side effects of platinum-based chemotherapy drugs: A review for chemists. Dalton Trans. 2018, 47, 6645–6653. [Google Scholar] [CrossRef] [PubMed]
- Lugones, Y.; Loren, P.; Salazar, L.A. Cisplatin resistance: Genetic and epigenetic factors involved. Biomolecules 2022, 12, 1365. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Kang, Y.; Chen, L.; Wang, H.; Liu, J.; Zeng, S.; Yu, L. The drug-resistance mechanisms of five platinum-based antitumor agents. Front. Pharmacol. 2020, 11, 343. [Google Scholar] [CrossRef]
- Waters, J.E.; Stevens-Cullinane, L.; Siebenmann, L.; Hess, J. Recent advances in the development of metal complexes as antibacterial agents with metal-specific modes of action. Curr. Opin. Microbiol. 2023, 75, 102347. [Google Scholar] [CrossRef]
- Sharma, B.; Shukla, S.; Rattan, R.; Fatima, M.; Goel, M.; Bhat, M.; Dutta, S.; Ranjan, R.K.; Sharma, M. Antimicrobial agents based on metal complexes: Present situation and future prospects. Int. J. Biomater. 2022, 2022, 6819080. [Google Scholar] [CrossRef]
- Lucaciu, R.L.; Hangan, A.C.; Sevastre, B.; Oprean, L.S. Metallo-drugs in cancer therapy: Past, present and future. Molecules 2022, 27, 6485. [Google Scholar] [CrossRef]
- Ndagi, U.; Mhlongo, N.; Soliman, M.E. Metal complexes in cancer therapy—An update from drug design perspective. Drug Des. Devel. Ther. 2017, 11, 599–616. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Dai, F.; Mao, Y.; Zhang, K.; Qin, Y.; Zheng, J. Metallodrugs in the battle against non-small cell lung cancer: Unlocking the potential for improved therapeutic outcomes. Front. Pharmacol. 2023, 14, 1242488. [Google Scholar] [CrossRef] [PubMed]
- Muhammad, N.; Hanif, M.; Yang, P. Beyond cisplatin: New frontiers in metallodrugs for hard-to-treat triple negative breast cancer. Coord. Chem. Rev. 2024, 499, 215507. [Google Scholar] [CrossRef]
- Gaál, A.; Orgován, G.; Mihucz, V.G.; Pape, I.; Ingerle, D.; Streli, C.; Szoboszlai, N.J. Metal transport capabilities of anticancer copper chelators. Trace Elem. Med. Biol. 2018, 47, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Mahendiran, D.; Amuthakala, S.; Bhuvanesh, N.S.P.; Kumar, R.S.; Rahiman, A.K. Copper complexes as prospective anticancer agents: In vitro and in vivo evaluation, selective targeting of cancer cells by DNA damage and S phase arrest. RSC Adv. 2018, 8, 16973–16990. [Google Scholar] [CrossRef] [PubMed]
- Lelièvre, P.; Sancey, L.; Coll, J.-L.; Deniaud, A.; Busser, B. The multifaceted roles of copper in cancer: A trace metal element with dysregulated metabolism, but also a target or a bullet for therapy. Cancers 2020, 12, 3594. [Google Scholar] [CrossRef] [PubMed]
- Krasnovskaya, O.; Naumov, A.; Dmitry Guk, D.; Gorelkin, P.; Erofeev, A.; Beloglazkina, E.; Majouga, A. Copper coordination compounds as biologically active agents. Int. J. Mol. Sci. 2020, 21, 3965. [Google Scholar] [CrossRef]
- Ji, P.; Wang, P.; Chen, H.; Xu, Y.; Ge, J.; Tian, Z.; Yan, Z. Potential of copper and copper compounds for anticancer applications. Pharmaceuticals 2023, 16, 234. [Google Scholar] [CrossRef]
- Jiang, Y.; Huo, Z.; Qi, X.; Zuo, T.; Wu, Z. Copper-induced tumor cell death mechanisms and antitumor theragnostic applications of copper complexes. Nanomedicine 2022, 17, 303–324. [Google Scholar] [CrossRef]
- Carcelli, M.; Tegoni, M.; Bartoli, J.; Marzano, C.; Pelosi, G.; Salvalaio, M.; Rogolino, D.; Gandin, V. In vitro and in vivo anticancer activity of tridentate thiosemicarbazone copper complexes: Unravelling an unexplored pharmacological target. Eur. J. Med. Chem. 2020, 194, 112266. [Google Scholar] [CrossRef]
- Khan, R.A.; Usman, M.; Dhivya, R.; Balaji, P.; Alsalme, A.; AlLohedan, H.; Arjmand, F.; AlFarhan, K.; Akbarsha, M.A.; Marchetti, F.; et al. Heteroleptic copper(I) complexes of “scorpionate” bis-pyrazolyl carboxylate ligand with auxiliary phosphine as potential anticancer agents: An insight into cytotoxic mode. Sci. Rep. 2017, 7, 45229–45246. [Google Scholar] [CrossRef] [PubMed]
- Molinaro, C.; Martoriati, A.; Pelinski, L.; Cailliau, K. Cooper complexes as anticancer agents targeting topoisomerases I and II. Cancers 2020, 12, 2863. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; Zheng, Y.; Li, B.; Ai, Y.; Chen, M.; Zheng, X. Pyridoxal hydrochloride thiosemicarbazones with copper ions inhibit cell division via Topo-I and Topo-IIα. J. Inorg. Biochem. 2022, 232, 111816. [Google Scholar] [CrossRef] [PubMed]
- Qin, Q.-P.; Meng, T.; Tan, M.-X.; Liu, Y.-C.; Luo, X.-J.; Zou, B.-Q.; Liang, H. Synthesis, crystal structure and biological evaluation of a new dasatinib copper(II) complex as telomerase inhibitor. Eur. J. Med. Chem. 2018, 143, 1597–1603. [Google Scholar] [CrossRef] [PubMed]
- Xin, C.; Xiao, Z.; Jinghong, C.; Qianqi, Y.; Li, Y. Hinokitiol copper complex inhibits proteasomal deubiquitination and induces paraptosis-like cell death in human cancer cells. Eur. J. Pharmacol. 2017, 815, 147–155. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, H.; Yan, M.; Wang, H.; Zhang, C. Novel copper complexes as potential proteasome inhibitors for cancer treatment. Mol. Med. Rep. 2017, 15, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Konarikova, K.; Frivaldska, J.; Gbelcova, H.; Sveda, M.; Ruml, T.; Janubova, M.; Zitnanova, I. Schiff base Cu(II) complexes as inhibitors of proteasome in human cancer cells. Bratisl. Lek Listy 2019, 120, 646–649. [Google Scholar] [CrossRef] [PubMed]
- Anu, D.; Naveen, P.; Vijaya, P.B.; Frampton, C.S.; Kaveri, M.V. An unexpected mixed valence tetranuclear copper (I/II) complex: Synthesis, structural characterization, DNA/protein binding, antioxidant and anticancer properties. Polyhedron 2019, 167, 137–150. [Google Scholar] [CrossRef]
- Bollua, V.S.; Bathinib, T.; Baruia, A.K.; Roya, A.; Ragic, N.C.; Maloth, S.; Sripadic, P.; Sreedharb, B.; Nagababub, P.; Patra, C.R. Design of DNA-intercalators based copper(II) complexes, investigation of their potential anti-cancer activity and sub-chronic toxicity. Mater. Sci. Eng. C 2019, 105, 110079. [Google Scholar] [CrossRef]
- Movahedi, E.; Razmazma, H.; Rezvani, A.; Nowroozi, A.; Ebrahimi, A.; Eigner, V.; Dusek, M.; Arjmand, F. A novel Cu(II)-based DNA-intercalating agent: Structural and biological insights using biophysical and in silico techniques. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2023, 293, 122438. [Google Scholar] [CrossRef]
- Lesiów, M.K.; Bieńko, A.; Sobańska, K.; Kowalik-Jankowska, T.; Rolka, K.; Łęgowska, A.; Ptaszyńska, N. Cu(II) complexes with peptides from FomA protein containing –His-Xaa-Yaa-Zaa-His and –His-His-motifs. ROS generation and DNA degradation. J. Inorg. Biochem. 2020, 212, 111250. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, M.R.; Lavecchia, M.J.; Parajón-Costa, B.S.; González-Baró, A.C.; González-Baró, M.R.; Cattáneo, E.R. DNA cleavage mechanism by metal complexes of Cu(II), Zn(II) and VO(IV) with a schiff-base ligand. Biochemie 2021, 186, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Shao, J.; Li, M.; Guo, Z.; Jin, C.; Zhang, F.; Ou, C.; Xie, Y.; Tan, S.; Wang, Z.; Zheng, S.; et al. TPP-related mitochondrial targeting copper(II) complex induces p53-dependent apoptosis in hepatoma cells through ROS-mediated activation of Drp1. Cell Commun. Signal. 2019, 17, 149. [Google Scholar] [CrossRef] [PubMed]
- Done, G.; Ari, F.; Akgun, O.; Akgun, H.; Cevatemre, B.; Gençkal, H.M. The mechanism for anticancer and apoptosis-inducing properties of Cu(II) complex with quercetin and 1,10-phenanthroline. ChemistrySelect 2022, 7, e202203242. [Google Scholar] [CrossRef]
- Martinez-Bulit, P.; Garza-Ortíz, A.; Mijangos, E.; Barrón-Sosa, L.; Sánchez-Bartéz, F.; Gracia-Mora, I.; Flores-Parra, A.; Contreras, R.; Reedijk, J.; Barba-Behrens, N. 2,6-Bis(2,6-diethylphenyliminomethyl)pyridine coordination compounds with cobalt(II), nickel(II), copper(II), and zinc(II): Synthesis, spectroscopic characterization, X-ray study and in vitro cytotoxicity. J. Inorg. Biochem. 2015, 142, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Sîrbu, A.; Palamarciuc, O.; Babak, M.V.; Lim, J.M.; Ohui, K.; Enyedy, E.A.; Shova, S.; Darvasiová, D.; Rapta, P.; Ang, W.H.; et al. Copper(II) thiosemicarbazone complexes induce marked ROS accumulation and promote nrf2-mediated antioxidant response in highly resistant breast cancer cells. Dalton Trans. 2017, 46, 3833–3847. [Google Scholar] [CrossRef] [PubMed]
- Peña, Q.; Lorenzo, J.; Sciortino, G.; Rodríguez-Calado, S.; Maréchal, J.-D.; Bayón, P.; Simaan, A.J.; Iranzo, O.; Capdevila, M.; Palacios, O. Studying the reactivity of “old” Cu(II) complexes for “novel” anticancer purpose. J. Inorg. Biochem. 2019, 195, 51–60. [Google Scholar] [CrossRef]
- Zehra, S.; Tabassum, S.; Arjmand, F. Biochemical pathways of copper complexes: Progress over the past 5 years. Drug Discov. Today 2021, 26, 1086–1096. [Google Scholar] [CrossRef]
- Meraz-Torres, F.; Plöger, S.; Garbe, C.; Niessner, H.; Sinnberg, T. Disulfiram as a therapeutic agent for metastatic malignant melanoma-old myth or new logos? Cancers 2020, 12, 3538. [Google Scholar] [CrossRef]
- Xu, B.; Shi, P.; Fombon, I.S.; Zhang, Y.; Huang, F.; Wang, W.; Zhou, S. Disulfiram/copper complex activated JNK/c-jun pathway and sensitized cytotoxicity of doxorubicin in doxorubicin resistant leukemia HL60 cells. Blood Cells Mol. Dis. 2011, 47, 264–269. [Google Scholar] [CrossRef]
- Climova, A.; Pivovarova, E.; Szczesio, M.; Gobis, K.; Ziembicka, D.; Korga-Plewko, A.; Kubik, J.; Iwan, M.; Antos-Bielska, M.; Krzyżowska, M.; Czylkowska, A. Anticancer and antimicrobial activity of new copper (II) complexes. J Inorg Biochem. 2023, 240, 112108. [Google Scholar] [CrossRef] [PubMed]
- Gul, N.S.; Khan, T.M.; Chen, M.; Huang, K.B.; Hou, C.; Choudhary, M.I.; Liang, H.; Chen, Z.F. New copper complexes inducing bimodal death through apoptosis and autophagy in A549 cancer cells. J. Inorg. Biochem. 2020, 213, 111260. [Google Scholar] [CrossRef] [PubMed]
- Heffeter, P.; Jungwirth, U.; Jakupec, M.; Hartinger, C.; Galanski, M.; Elbling, L.; Micksche, M.; Keppler, B. Resistance against novel anticancer metal compounds: Differences and similarities. Drug Resist. Updates 2008, 11, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.-Y.; Chang, Y.-L.; Liu, S.-T.; Chen, G.-S.; Lee, S.-P.; Huang, S.-M. Differential cytotoxicity mechanisms of copper complexed with disulfiram in oral cancer cells. Int. J. Mol. Sci. 2021, 22, 3711. [Google Scholar] [CrossRef] [PubMed]
- da Silva, D.A.; De Luca, A.; Squitti, R.; Rongioletti, M.; Rossi, L.; Machado, C.M.L.; Cerchiaro, G. Copper in tumors and the use of copper-based compounds in cancer treatment. J. Inorg. Biochem. 2022, 226, 111634. [Google Scholar] [CrossRef]
- Hussain, A.; AlAjmi, M.F.; Rehman, M.T.; Amir, S.; Husain, F.M.; Alsalme, A.; Siddiqui, M.A.; AlKhedhairy, A.A.; Khan, R.A. Copper(II) complexes as potential anticancer and nonsteroidal anti-inflammatory agents: In vitro and in vivo studies. Sci. Rep. 2019, 9, 5237. [Google Scholar] [CrossRef]
- Malis, G.; Geromichalou, E.; Geromichalos, G.D.; Hatzidimitriou, A.G.; Psomas, G. Copper(II) complexes with non-steroidal anti-inflammatory drugs: Structural characterization, in vitro and in silico biological profile. J. Inorg. Biochem. 2021, 224, 111563. [Google Scholar] [CrossRef]
- Bellia, F.; Lanza, V.; Naletova, I.; Tomasello, B.; Ciaffaglione, V.; Greco, V.; Sciuto, S.; Amico, P.; Inturri, R.; Vaccaro, S.; et al. Copper(II) complexes with carnosine conjugates of hyaluronic acids at different dipeptide loading percentages behave as multiple SOD mimics and stimulate Nrf2 translocation and antioxidant response in in vitro inflammatory model. Antioxidants 2023, 12, 1632. [Google Scholar] [CrossRef]
- Gordon, N.A.; McGuire, K.L.; Wallentine, S.K.; Mohl, G.A.; Lynch, J.D.; Harrison, R.G.; Busath, D.D. Divalent copper complexes as influenza A M2 inhibitors. Antivir. Res. 2017, 147, 100–106. [Google Scholar] [CrossRef]
- Kowalczyk, M.; Golonko, A.; Świsłocka, R.; Kalinowska, M.; Parcheta, M.; Swiergiel, A.; Lewandowski, W. Drug design strategies for the treatment of viral disease. Plant phenolic compounds and their derivatives. Front. Pharmacol. 2021, 12, 709104. [Google Scholar] [CrossRef]
- Salah, I.; Parkin, I.P.; Allan, E. Copper as an antimicrobial agent: Recent advances. RSC Adv. 2021, 11, 18179. [Google Scholar] [CrossRef] [PubMed]
- Pereira, A.L.; Vasconcelos, M.A.; Andrade, A.L.; Martins, I.M.; Holanda, A.K.M.; Gondim, A.C.S.; Penha, D.P.S.; Bruno, K.L.; Silva, F.O.N.; Teixeira, E.H. Antimicrobial and antibiofilm activity of copper-based metallic compounds against bacteria related with healthcare-associated infections. Curr. Microbiol. 2023, 80, 133. [Google Scholar] [CrossRef] [PubMed]
- Iranshahy, M.; Quinnb, R.J.; Iranshahi, M. Biologically active isoquinoline alkaloids with drug-like properties from the genus Corydalis. RSC Adv. 2014, 4, 15900–15913. [Google Scholar] [CrossRef]
- Yuan, H.L.; Zhao, Y.L.; Qin, X.J.; Liu, Y.P.; Yang, X.W.; Luo, X.D. Diverse isoquinolines with anti-inflammatory and analgesic bioactivities from Hypecoum erectum. J. Ethnopharmacol. 2021, 270, 113811. [Google Scholar] [CrossRef] [PubMed]
- Alagumuthu, M.; Sathiyanarayanan, K.I.; Arumugam, S. Molecular docking, design, synthesis, in vitro antioxidant and anti-inflammatory evaluations of new isoquinoline derivatives. Int. J. Pharm. Pharm. Sci. 2015, 7, 200–208. [Google Scholar]
- Zajdel, P.; Marciniec, K.; Maślankiewicz, A.; Grychowska, K.; Satała, G.; Duszyńska, B.; Lenda, T.; Siwek, A.; Nowak, G.; Partyka, A.; et al. Antidepressant and antipsychotic activity of new quinoline- and isoquinoline-sulfonamide analogs of aripiprazole targeting serotonin 5-HT1A/5-HT2A/5-HT7 and dopamine D2/D3 receptors. Eur. J. Med. Chem. 2013, 60, 42–50. [Google Scholar] [CrossRef] [PubMed]
- Luo, C.; Ampomah-Wireko, M.; Wang, H.; Wu, C.; Wang, Q.; Zhang, H.; Cao, Y. Isoquinolines: Important cores in many marketed and clinical drugs. Anticancer Agents Med. Chem. 2021, 21, 811–824. [Google Scholar] [CrossRef] [PubMed]
- Faheem; Kumar, B.K.; Chandra Sekhar, K.V.G.; Chander, S.; Kunjiappan, S.; Murugesan, S. Medicinal chemistry perspectives of 1,2,3,4-tetrahydroisoquinoline analogs—Biological activities and SAR studies. RSC Adv. 2021, 11, 12254. [Google Scholar] [CrossRef]
- Theeramunkong, S.; Thiengsusuk, A.; Vajragupta, O.; Muhamad, P. Synthesis, characterization and antimalarial activity of isoquinoline derivatives. Med. Chem. Res. 2021, 30, 109–119. [Google Scholar] [CrossRef]
- Galán, A.; Moreno, L.; Párraga, J.; Serrano, Á.; Sanz, M.J.; Cortes, D.; Cabedo, N. Novel isoquinoline derivatives as antimicrobial agents. Bioorg. Med. Chem. 2013, 21, 3221–3230. [Google Scholar] [CrossRef]
- Karanja, C.W.; Naganna, N.; Abutaleb, N.S.; Dayal, N.; Onyedibe, K.I.; Aryal, U.; Seleem, M.N.; Sintim, H.O. Isoquinoline antimicrobial agent: Activity against intracellular bacteria and effect on global bacterial proteome. Molecules 2022, 27, 5085. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Soni, K.; Sangani, C.; Yao, Y. An overview of privileged scaffold: Quinolines and isoquinolines in medicinal chemistry as anticancer agents. Curr. Top. Med. Chem. 2020, 20, 2599–2633. [Google Scholar] [CrossRef] [PubMed]
- Yadav, P.; Kumar, A.; Althagafi, I.; Nemaysh, V.; Rai, R.; Pratap, R. The recent development of tetrahydro-quinoline/isoquinoline based compounds as anticancer agents. Curr. Top. Med. Chem. 2021, 21, 1587–1622. [Google Scholar] [CrossRef] [PubMed]
- Balewski, Ł.; Sączewski, F.; Gdaniec, M.; Kornicka, A.; Cicha, K.; Jalińska, A. Synthesis and fluorescent properties of novel isoquinoline derivatives. Molecules 2019, 24, 4070. [Google Scholar] [CrossRef] [PubMed]
- Sączewski, F.; Dziemidowicz-Borys, E.; Bednarski, P.J.; Grünert, R.; Gdaniec, M.; Tabin, P. Synthesis, crystal structure and biological activities of copper(II) complexes with chelating bidentate 2-substituted benzimidazole ligands. J. Inorg. Biochem. 2006, 100, 1389–1398. [Google Scholar] [CrossRef] [PubMed]
- Sączewski, F.; Dziemidowicz-Borys, E.; Bednarski, P.J.; Gdaniec, M. Synthesis, crystal structure, cytotoxic and superoxide dismutase activities of copper(II) complexes of N-(4,5-dihydroimidazol-2-yl)azoles. Arch. Pharm. 2007, 340, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Balewski, Ł.; Sączewski, F.; Bednarski, P.J.; Gdaniec, M.; Borys, E.; Makowska, A. Structural diversity of copper(II) complexes with N-(2-pyridyl)imidazolidin-2-ones(thiones) and their in vitro antitumor activity. Molecules 2014, 19, 17026–17051. [Google Scholar] [CrossRef]
- Makowska, A.; Sączewski, F.; Bednarski, P.J.; Gdaniec, M.; Balewski, Ł.; Warmbier, M.; Kornicka, A. Synthesis, structure and cytotoxic properties of copper(II) complexes of 2-iminocoumarins bearing a 1,3,5-triazine or benzoxazole/benzothiazole moiety. Molecules 2022, 27, 7155. [Google Scholar] [CrossRef]
- Sączewski, F.; Bułakowska, A.; Gdaniec, M. 2-Chloro-4,5-dihydroimidazole, Part X. Revisiting route to N-heteroarylimidazolidin-2-ones. J. Heterocycl. Chem. 2002, 39, 911–915. [Google Scholar] [CrossRef]
- Balewski, Ł.; Sączewski, F.; Gdaniec, M.; Bednarski, P.J.; Jara, I. Synthesis of N-(2-pyridyl)imidazolidin-2-ones and 1-(2-pyridyl)-2,3,7,8-tetrahydro-1H-imidazo[2,1-b][1,3,5]-triazepin-5(6H)-ones with potential biological activity. Heterocycl. Commun. 2013, 19, 331–341. [Google Scholar] [CrossRef]
- Becke, A.D. Density-functional thermochemistry. I. The effect of the exchange-only gradient correction. J. Chem. Phys. 1992, 96, 2155–2160. [Google Scholar] [CrossRef]
- Becke, A.D. Density-functional thermochemistry. II. The effect of the Perdew-Wang generalized-gradient correlation correction. J. Chem. Phys. 1992, 97, 9173–9177. [Google Scholar] [CrossRef]
- Becke, A.D. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 1993, 98, 5648–5652. [Google Scholar] [CrossRef]
- Ali, A.A.S.; Khan, D.; Naqvi, A.; Al-blewi, F.F.; Rezki, N.; Aouad, M.R.; Hagar, M. Design, synthesis, molecular modeling, anticancer studies, and density functional theory calculations of 4-(1,2,4-triazol-3-ylsulfanylmethyl)-1,2,3-triazole derivatives. ACS Omega 2021, 6, 301–316. [Google Scholar] [CrossRef]
- Zhang, Y.L.; Deng, C.X.; Zhou, W.F.; Zhou, L.Y.; Cao, Q.Q.; Shen, W.Y.; Liang, H.; Chen, Z.F. Synthesis and in vitro antitumor activity evaluation of copper(II) complexes with 5-pyridin-2-yl-[1,3]dioxolo[4,5-g]isoquinoline derivatives. J. Inorg. Biochem. 2019, 201, 110820. [Google Scholar] [CrossRef] [PubMed]
- Le, T.T.; Wu, M.; Lee, J.H.; Bhatt, N.; Berger, J.M.; Wang, M.D. Etoposide promotes DNA loop trapping and barrier formation by topoisomerase II. Nat. Chem. Biol. 2023, 19, 641–650. [Google Scholar] [CrossRef] [PubMed]
- Dasari, S.; Tchounwou, P.B. Cisplatin in cancer therapy: Molecular mechanisms of action. Eur. J. Pharmacol. 2014, 740, 364–378. [Google Scholar] [CrossRef]
- Strobel, H.; Baisch, T.; Fitzel, R.; Schilberg, K.; Siegelin, M.D.; Karpel-Massler, G.; Debatin, K.M.; Westhoff, M.A. Temozolomide and other alkylating agents in glioblastoma therapy. Biomedicines 2019, 7, 69. [Google Scholar] [CrossRef]
- Longley, D.; Harkin, D.; Johnston, P. 5-Fluorouracil: Mechanisms of action and clinical strategies. Nat. Rev. Cancer 2003, 3, 330–338. [Google Scholar] [CrossRef]
- Khan, T.M.; Gul, N.S.; Lu, X.; Wei, J.H.; Liu, Y.C.; Sun, H.; Liang, H.; Orvig, C.; Chen, Z.F. In vitro and in vivo anti-tumor activity of two gold(III) complexes with isoquinoline derivatives as ligands. Eur. J. Med. Chem. 2019, 163, 333–343. [Google Scholar] [CrossRef]
- Huang, K.B.; Mo, H.Y.; Chen, Z.F.; Wei, J.H.; Liu, Y.C.; Liang, H. Isoquinoline derivatives Zn(II)/Ni(II) complexes: Crystal structures, cytotoxicity, and their action mechanism. Eur. J. Med. Chem. 2015, 100, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.Y.; Xi, Q.Y.; Huang, K.B.; Tang, X.M.; Chen, Z.F.; Liu, Y.C.; Liang, H. Crystal structure, cytotoxicity and action mechanism of Zn(II)/Mn(II) complexes with isoquinoline ligands. J. Inorg. Biochem. 2017, 169, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.B.; Chen, Z.F.; Liu, Y.C.; Wang, M.; Wei, J.H.; Xie, X.L.; Zhang, J.L.; Hu, K.; Liang, H. Copper(II/I) complexes of 5-pyridin-2-yl-[1,3]dioxolo[4,5-g]isoquinoline: Synthesis, crystal structure, antitumor activity and DNA interaction. Eur. J. Med. Chem. 2013, 70, 640–648. [Google Scholar] [CrossRef] [PubMed]
- Gomaa, E.Z. Human gut microbiota/microbiome in health and diseases: A review. Antonie Van Leeuwenhoek 2020, 113, 2019–2040. [Google Scholar] [CrossRef] [PubMed]
- George, S.; Abrahamse, H. Redox potential of antioxidants in cancer progression and prevention. Antioxidants 2020, 9, 1156. [Google Scholar] [CrossRef] [PubMed]
- Fox, J.T.; Sakamuru, S.; Huang, R.L.; Teneva, N.; Simmons, S.O.; Xia, M.H.; Tice, R.R.; Austin, C.P.; Myung, K. High-throughput genotoxicity assay identifies antioxidants as inducers of DNA damage response and cell death. Proc. Natl. Acad. Sci. USA 2012, 109, 5423–5428. [Google Scholar] [CrossRef] [PubMed]
- Veiga, N.; Alvarez, N.; Castellano, E.E.; Ellena, J.; Facchin, G.; Torre, M.H. Comparative study of antioxidant and pro-oxidant properties of homoleptic and heteroleptic copper complexes with amino acids, dipeptides and 1,10-phenanthroline: The quest for antitumor compounds. Molecules 2021, 26, 6520. [Google Scholar] [CrossRef]
- Pinheiro, A.C.; Nunes, I.J.; Ferreira, W.V.; Tomasini, P.P.; Trindade, C.; Martins, C.C.; Wilhelm, E.A.; Oliboni, R.D.S.; Netz, P.A.; Stieler, R.; et al. Antioxidant and anticancer potential of the new Cu(II) complexes bearing imine-phenolate ligands with pendant amine N-donor groups. Pharmaceutics 2023, 15, 376. [Google Scholar] [CrossRef]
- Yusuf, T.L.; Waziri, I.; Olofinsan, K.A.; Akintemi, E.O.; Hosten, E.C.; Muller, A.J. Evaluating the in vitro antidiabetic, antibacterial and antioxidant properties of copper(II) Schiff base complexes: An experimental and computational studies. J. Mol. Liq. 2023, 389, 122845. [Google Scholar] [CrossRef]
- Gurgul, I.; Hricovíniovà, J.; Mazuryk, O.; Hricovíniovà, Z.; Brindell, M. Enhancement of the cytotoxicity of quinazolinone Schiff base derivatives with copper coordination. Inorganics 2023, 11, 391. [Google Scholar] [CrossRef]
- Hazra, M.; Dolai, T.; Pandey, A.; Dey, S.K.; Patra, A. Synthesis and characterisation of copper(II) complexes with tridentate NNO functionalized ligand: Density Function Theory study, DNA binding mechanism, optical properties, and biological application. Bioinorg. Chem. Appl. 2014, 2014, 104046. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [PubMed]
- Shityakov, S. Analysing molecular polar surface descriptors to predict blood-brain barrier permeation. Int. J. Comput. Biol. Drug Des. 2013, 6, 146–156. [Google Scholar] [CrossRef] [PubMed]
- Trani, A.; Bellasio, E. Synthesis of 2-chloro-2-imidazoline and its reactivity with aromatic amines, phenols, and thiophenols. J. Heterocycl. Chem. 1974, 11, 257–262. [Google Scholar] [CrossRef]
- Sheldrick, G.M. SHELXT—Integrated space-group and crystal-structure determination. Acta Crystallogr. Sect. Found. Adv. 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Sheldrick, G.M. A short history of SHELX. Acta Crystallogr. A 2008, 64, 112–122. [Google Scholar] [CrossRef]
- Hübschle, C.B.; Sheldrick, G.M.; Dittrich, B. ShelXle: A Qt graphical user interface for SHELXL. J. Appl. Cryst. 2011, 44, 1281–1284. [Google Scholar] [CrossRef]
- Macrae, C.F.; Sovago, I.; Cottrell, S.J.; Galek, P.T.A.; McCabe, P.; Pidcock, E.; Platings, M.; Shields, G.P.; Stevens, J.S.; Towler, M.; et al. Mercury 4.0: From visualization to analysis, design and prediction. J. Appl. Cryst. 2020, 53, 226–235. [Google Scholar] [CrossRef]
- Dolomanov, O.V.; Bourhis, L.J.; Gildea, R.J.; Howard, J.A.K.; Puschmann, H. OLEX2: A complete structure solution, refinement and analysis program. J. Appl. Crystallogr. 2009, 42, 339–341. [Google Scholar] [CrossRef]
- Westrip, S.P. publCIF: Software for editing, validating and formatting crystallographic information files. J. Apply. Cryst. 2010, 43, 920–925. [Google Scholar] [CrossRef]
- IC50 Tool Kit: A Free Web Server. Available online: http://www.ic50.tk (accessed on 20 September 2023).
- Motyka, S.; Kusznierewicz, B.; Ekiert, H.; Korona-Głowniak, I.; Szopa, A. Comparative analysis of metabolic variations, antioxidant profiles and antimicrobial activity of Salvia hispanica (Chia) seed, sprout, leaf, flower, root and herb extracts. Molecules 2023, 28, 2728. [Google Scholar] [CrossRef] [PubMed]
- SwissADME: A Free Web Tool to Compute Physicochemical Descriptors as Well as to Predict ADME Parameters, Pharmacokinetic Properties, Druglike Nature and Medicinal Chemistry Friendliness of One or Multiple Small Molecules to Support Drug Discovery. Available online: http://www.swissadme.ch (accessed on 10 November 2023).








| Compound | (C1_1_auto) |
|---|---|
| Chemical formula | C24H20Cl2CuN4O2 |
| Mr | 530.89 |
| Crystal system, space group | Triclinic, |
| Temperature (K) | 100 |
| a, b, c (Å) | 7.8288 (2), 8.2173 (2), 9.5081 (2) |
| α, β, γ (°) | 106.092 (2), 111.334 (3), 97.897 (2) |
| V (Å3) | 527.68 (2) |
| Z | 1 |
| Radiation type | Mo Kα |
| μ (mm−1) | 1.32 |
| Crystal size (mm) | 0.22 × 0.2 × 0.09 |
| Diffractometer | XtaLAB Synergy, Dualflex, Pilatus 300 K |
| Tmin, Tmax | 0.586, 1.000 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 15,833, 3057, 2752 |
| Rint | 0.040 |
| (sin θ/λ)max (Å−1) | 0.748 |
| R[F2 > 2σ(F2)], wR(F2), S | 0.029, 0.072, 1.09 |
| No. of reflections | 3057 |
| No. of parameters | 151 |
| H-atom treatment | H-atom parameters constrained |
| Δmax, Δmin (e Å−3) | 0.91, −0.35 |
| D—H···A | D—H (Å) | H⋯A (Å) | D···A (Å) | D—H⋯A (º) |
|---|---|---|---|---|
| C9—H9···Cl1 i | 0.95 | 2.86 | 3.7205 (15) | 152 |
| C2—H2···O1 ii | 0.95 | 2.21 | 2.9512 (19) | 135 |
| Ligands/Complexes | IC50 ± SD (μg/mL) | ||||
|---|---|---|---|---|---|
| Cell Line | |||||
| A375 | HepG2 | LS180 | T98G | CCD-1059-Sk | |
| L1 | >200 | >200 | >200 | >200 | >200 |
| C1 | 37.97 ± 3.01 | 14.89 ± 0.56 | 11.46 ± 0.69 | 8.25 ± 0.36 | 18.83 ± 0.91 |
| L2 | >200 | >200 | >200 | >200 | >200 |
| C2 | 28.91 ± 1.79 | 5.04 ± 0.42 | 5.88 ± 0.40 | 6.97 ± 0.52 | 21.86 ± 1.03 |
| L3 | >200 | >200 | >200 | >200 | >200 |
| C3 | 22.78 ± 1.14 | 12.00 ± 0.36 | 7.09 ± 0.53 | 10.55 ± 0.74 | 25.17 ± 0.85 |
| L4 | >200 | >200 | >200 | >200 | >200 |
| C4 | 37.80 ± 3.11 | 6.72. ± 0.12 | 5.92 ± 0.54 | 9.27 ± 0.11 | 18.25 ± 1.37 |
| Etoposide * | 10.20 ± 0.83 | 43.21 ± 2.75 | >100 | >100 | 83.53 ± 3.19 |
| Microorganism/Compounds | L2 | C2 | L3 | C3 | L4 | C4 |
|---|---|---|---|---|---|---|
| MIC (mg/L) * | ||||||
| Yeasts | ||||||
| C. albicans ATCC 102231 | 500 | 1000 | 250 | 1000 | 1000 | 500 |
| C. parapsilosis ATCC 22019 | 1000 | 1000 | 125 | 1000 | >1000 | >1000 |
| C. glabrata ATCC 90030 | >1000 | >1000 | 250 | 1000 | 1000 | >1000 |
| Ligands/ Complexes | DDPH | ABTS |
|---|---|---|
| L1 | NR * | 183.21 ± 2.45 |
| C1 | 37.45 ± 0.66 | 112.67 ± 1.8 |
| L2 | NR * | 82.08 ± 2.77 |
| C2 | 401.52 ± 2.48 | 107.14 ± 1.42 |
| L3 | NR * | 96.67 ± 2.84 |
| C3 | 380.65 ± 2.74 | 106.19 ± 2.55 |
| L4 | NR * | 108.59 ± 1.51 |
| C4 | 26.46 ± 1.04 | 72.5 ± 0.97 |
| Ascorbic acid | 11.65 ± 0.54 | 20.15 ± 0.33 |
| Physicochemical Properties | Lipophilicity | Water Solubility | Pharmacokinetics | Drug Likeness | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| mol. wt. (g/mol) | ROTB (n) | HBA (n) | HBD (n) | TPSA | CLogP o/w | Solubility Class | GI Absorption | BBB Permeant | Lipinski Filter | BS | |
| Rule | <500 | <10 | <10 | <5 | - | <5 | - | - | - | - | - |
| C1 | 530.89 | 2 | 4 | 0 | 66.40 | 3.04 | Soluble(p) | High | Yes | Yes(1) | 0.55 |
| C2 | 560.92 | 2 | 4 | 2 | 90.46 | 2.48 | Soluble(p) | High | No | Yes(1) | 0.55 |
| C3 | 361.71 | 1 | 2 | 0 | 36.44 | 1.87 | Soluble(m) | High | Yes | Yes(0) | 0.55 |
| C4 | 375.74 | 2 | 2 | 0 | 36.44 | 2.14 | Soluble(m) | High | Yes | Yes(0) | 0.55 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Balewski, Ł.; Plech, T.; Korona-Głowniak, I.; Hering, A.; Szczesio, M.; Olczak, A.; Bednarski, P.J.; Kokoszka, J.; Kornicka, A. Copper(II) Complexes with 1-(Isoquinolin-3-yl)heteroalkyl-2-ones: Synthesis, Structure and Evaluation of Anticancer, Antimicrobial and Antioxidant Potential. Int. J. Mol. Sci. 2024, 25, 8. https://doi.org/10.3390/ijms25010008
Balewski Ł, Plech T, Korona-Głowniak I, Hering A, Szczesio M, Olczak A, Bednarski PJ, Kokoszka J, Kornicka A. Copper(II) Complexes with 1-(Isoquinolin-3-yl)heteroalkyl-2-ones: Synthesis, Structure and Evaluation of Anticancer, Antimicrobial and Antioxidant Potential. International Journal of Molecular Sciences. 2024; 25(1):8. https://doi.org/10.3390/ijms25010008
Chicago/Turabian StyleBalewski, Łukasz, Tomasz Plech, Izabela Korona-Głowniak, Anna Hering, Małgorzata Szczesio, Andrzej Olczak, Patrick J. Bednarski, Jakub Kokoszka, and Anita Kornicka. 2024. "Copper(II) Complexes with 1-(Isoquinolin-3-yl)heteroalkyl-2-ones: Synthesis, Structure and Evaluation of Anticancer, Antimicrobial and Antioxidant Potential" International Journal of Molecular Sciences 25, no. 1: 8. https://doi.org/10.3390/ijms25010008
APA StyleBalewski, Ł., Plech, T., Korona-Głowniak, I., Hering, A., Szczesio, M., Olczak, A., Bednarski, P. J., Kokoszka, J., & Kornicka, A. (2024). Copper(II) Complexes with 1-(Isoquinolin-3-yl)heteroalkyl-2-ones: Synthesis, Structure and Evaluation of Anticancer, Antimicrobial and Antioxidant Potential. International Journal of Molecular Sciences, 25(1), 8. https://doi.org/10.3390/ijms25010008












