Integrated Amino Acids and Transcriptome Analysis Reveals Arginine Transporter SLC7A2 Is a Novel Regulator of Myogenic Differentiation
Abstract
1. Introduction
2. Results
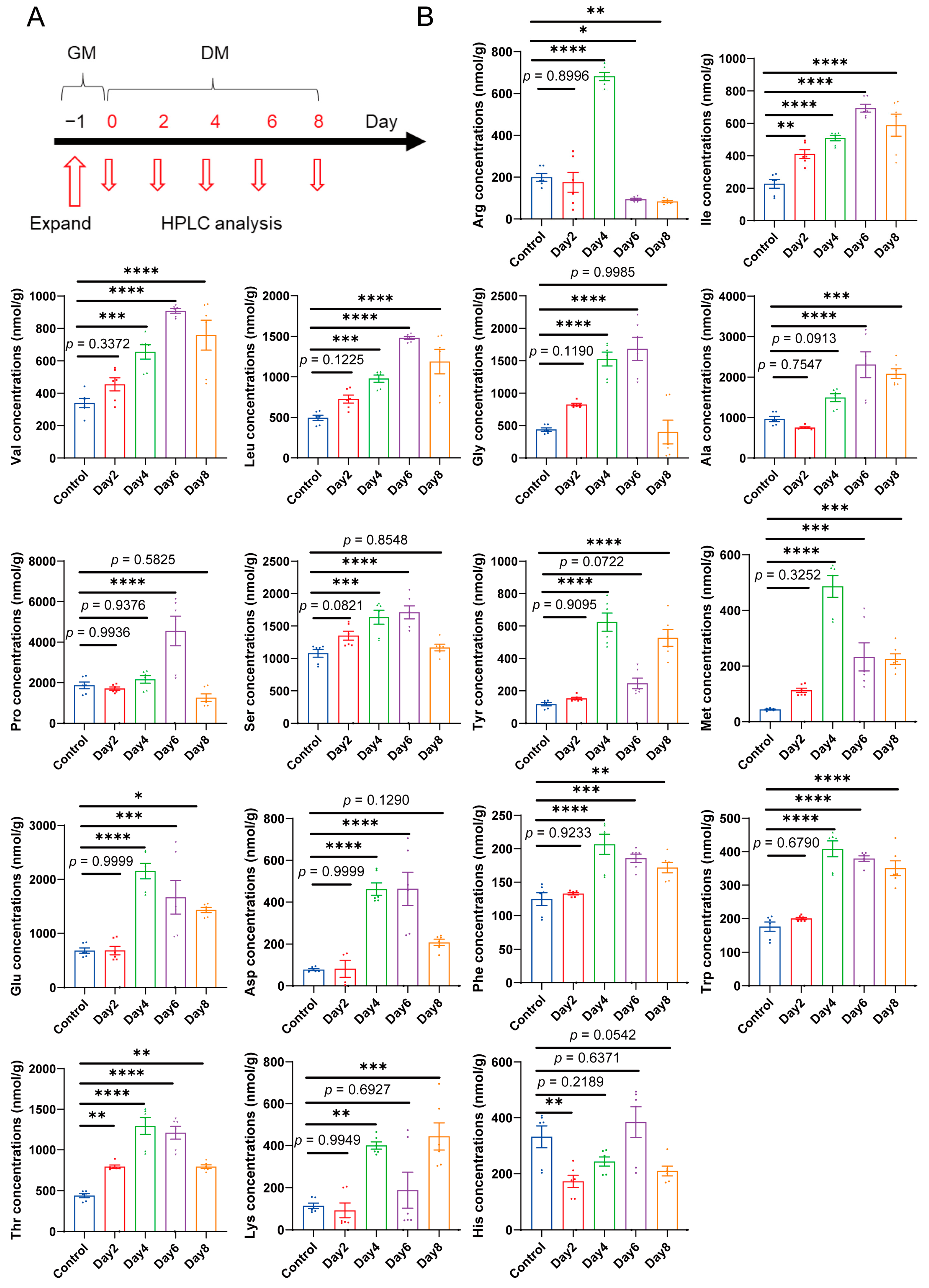
2.1. Amino Acids Levels Exhibit Distinct Changes during C2C12 Myoblast Differentiation
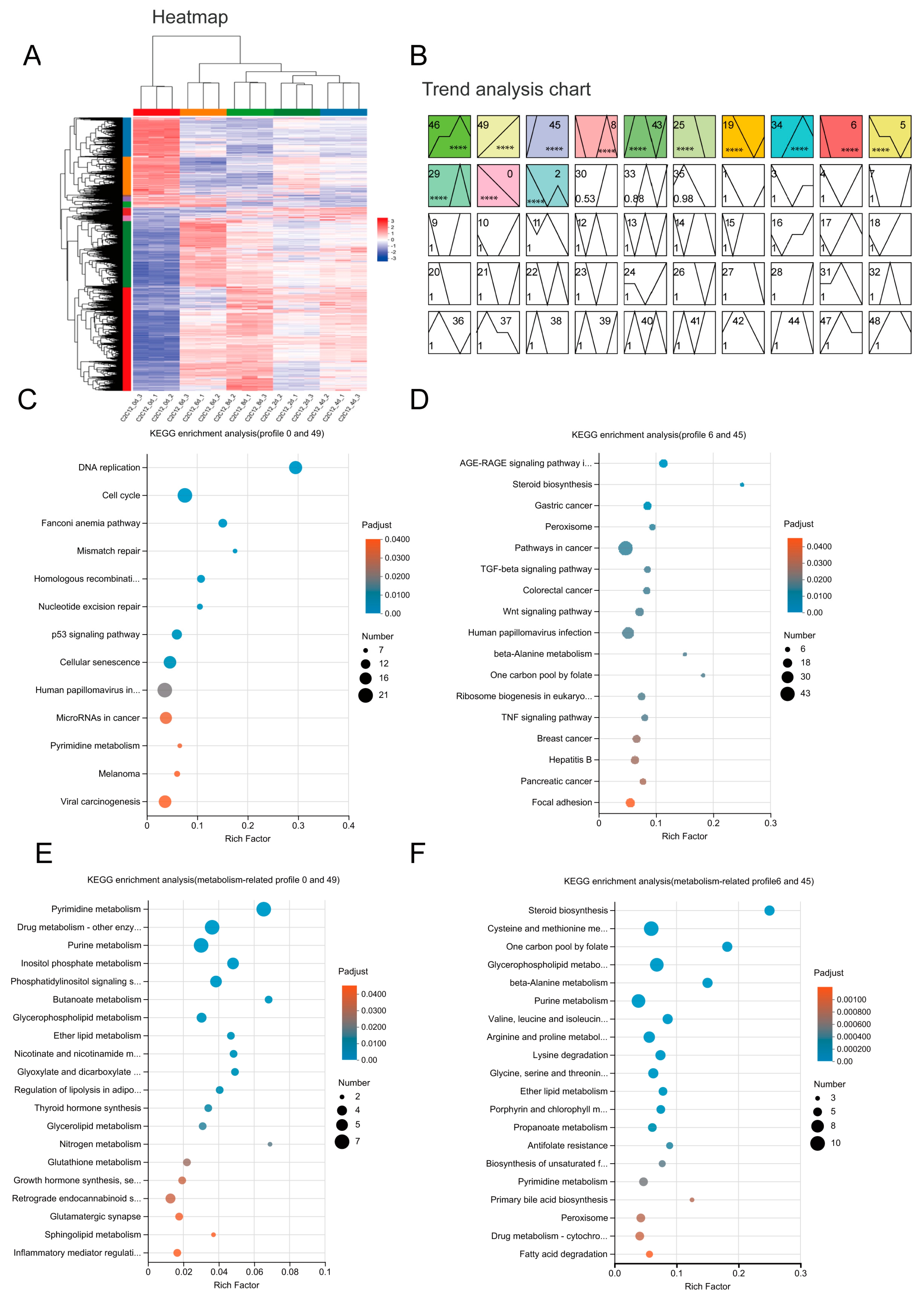
2.2. Amino Acid Metabolism Related Genes Are Enriched during C2C12 Myoblast Differentiation
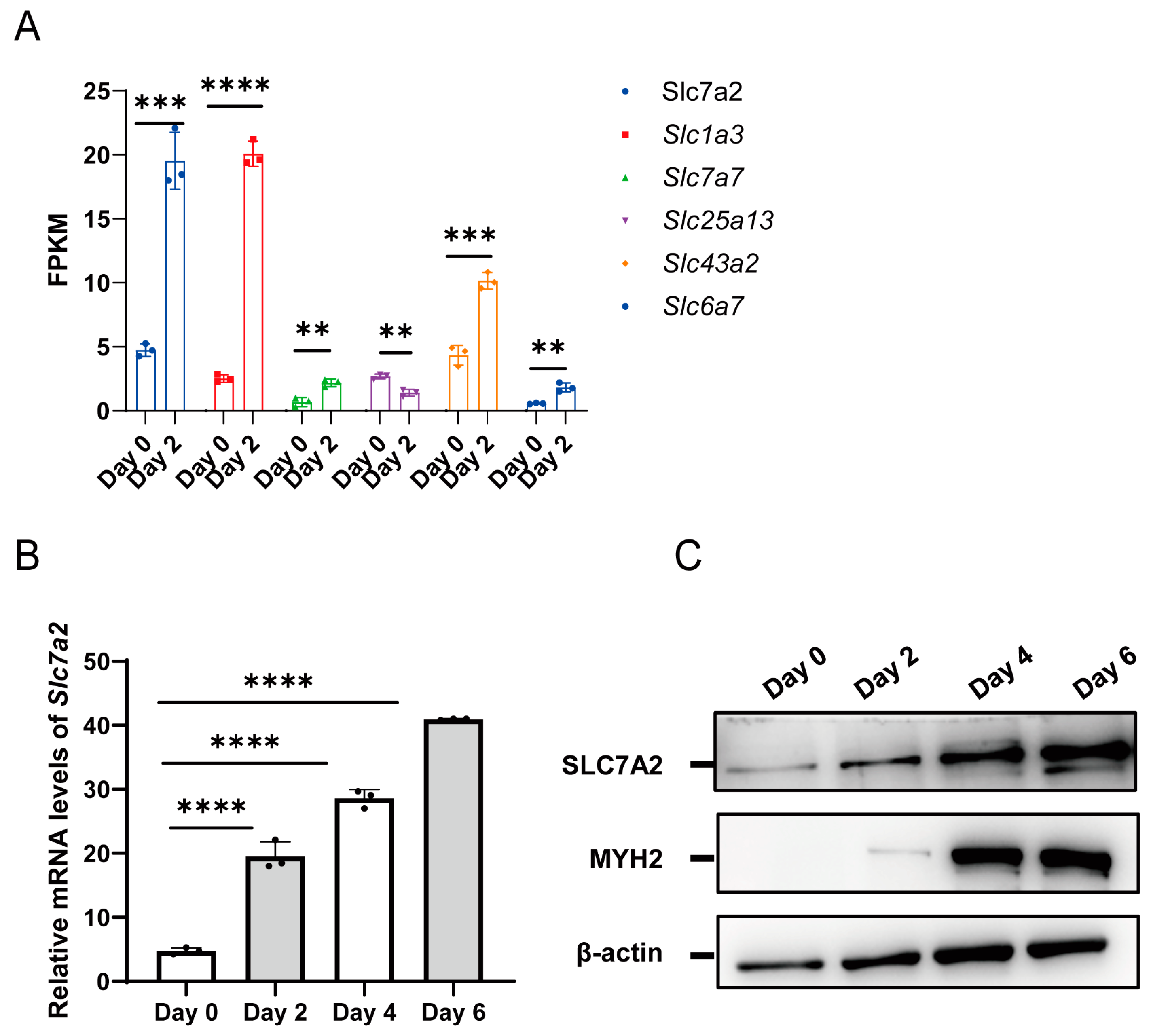
2.3. Arginine Transporter SLC7A2 Is Upregulated during Myogenic Differentiation
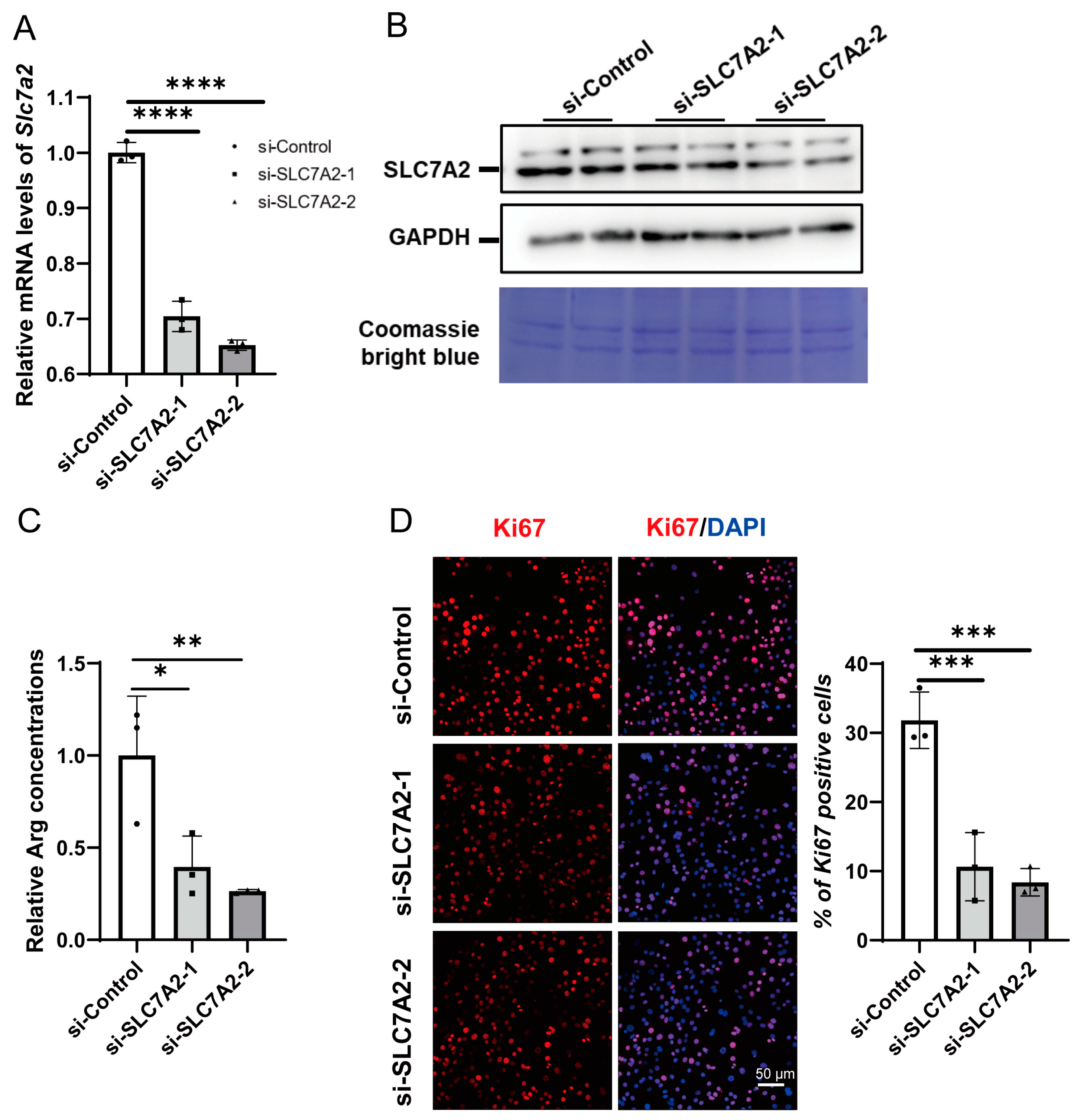
2.4. Knockdown of SLC7A2 Inhibits C2C12 Myoblast Proliferation
2.5. Knockdown of SLC7A2 Inhibits Myoblast Differentiation
2.6. The Expression of SLC7A2 Is Upregulated during Aging
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. RNA Sequencing
4.3. Time-Course Differential Expression Gene Analysis
4.4. RNA Interference
4.5. Quantitative RT-PCR
4.6. Western Blot
4.7. Immunofluorescence Assay
4.8. HPLC Method for Quantification of Intracellular Amino Acid Concentrations in C2C12 Cell
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Correction Statement
References
- Janssen, I.; Heymsfield, S.B.; Wang, Z.M.; Ross, R. Skeletal Muscle Mass and Distribution in 468 Men and Women Aged 18–88 Yr. J. Appl. Physiol. 2000, 89, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Thomas, D.R. Loss of Skeletal Muscle Mass in Aging: Examining the Relationship of Starvation, Sarcopenia and Cachexia. Clin. Nutr. 2007, 26, 389–399. [Google Scholar] [CrossRef] [PubMed]
- Tipton, K.D.; Wolfe, R.R. Exercise, Protein Metabolism, and Muscle Growth. Int. J. Sport Nutr. Exerc. Metab. 2001, 11, 109–132. [Google Scholar] [CrossRef] [PubMed]
- Rahman, N.I.A.; Lam, C.L.; Sulaiman, N.; Abdullah, N.A.H.; Nordin, F.; Ariffin, S.H.Z.; Yazid, M.D. Pax7, a Key for Myogenesis Modulation in Muscular Dystrophies through Multiple Signaling Pathways: A Systematic Review. Int. J. Mol. Sci. 2023, 24, 13051. [Google Scholar] [CrossRef] [PubMed]
- Bentzinger, C.F.; Wang, Y.X.; Rudnicki, M.A. Building Muscle: Molecular Regulation of Myogenesis. Cold Spring Harb. Perspect. Biol. 2012, 4, a008342. [Google Scholar] [CrossRef] [PubMed]
- Kamei, Y.; Hatazawa, Y.; Uchitomi, R.; Yoshimura, R.; Miura, S. Regulation of Skeletal Muscle Function by Amino Acids. Nutrients 2020, 12, 261. [Google Scholar] [CrossRef] [PubMed]
- Attaix, D.; Ventadour, S.; Codran, A.; Béchet, D.; Taillandier, D.; Combaret, L. The Ubiquitin-Proteasome System and Skeletal Muscle Wasting. Essays Biochem. 2005, 41, 173–186. [Google Scholar] [CrossRef] [PubMed]
- Church, D.D.; Hirsch, K.R.; Park, S.; Kim, I.Y.; Gwin, J.A.; Pasiakos, S.M.; Wolfe, R.R.; Ferrando, A.A. Essential Amino Acids and Protein Synthesis: Insights into Maximizing the Muscle and Whole-Body Response to Feeding. Nutrients 2020, 12, 3717. [Google Scholar] [CrossRef]
- Mieulet, V.; Roceri, M.; Espeillac, C.; Sotiropoulos, A.; Ohanna, M.; Oorschot, V.; Klumperman, J.; Sandri, M.; Pende, M. S6 kinase Inactivation Impairs Growth and Translational Target Phosphorylation in Muscle Cells Maintaining Proper Regulation of Protein Turnover. Am. J. Physiol. Cell Physiol. 2007, 293, C712–C722. [Google Scholar] [CrossRef]
- Kimball, S.R.; Jefferson, L.S.; Nguyen, H.V.; Suryawan, A.; Bush, J.A.; Davis, T.A. Feeding Stimulates Protein Synthesis in Muscle and Liver of Neonatal Pigs through an MTOR-Dependent Process. Am. J. Physiol. Endocrinol. Metab. 2000, 279, E1080–E1087. [Google Scholar] [CrossRef]
- Bolster, D.R.; Jefferson, L.S.; Kimball, S.R. Regulation of Protein Synthesis Associated with Skeletal Muscle Hypertrophy by Insulin-, Amino Acid- and Exercise-Induced Signalling. Proc. Nutr. Soc. 2004, 63, 351–356. [Google Scholar] [CrossRef] [PubMed]
- Jia, S.; Li, X.; Zheng, S.; Wu, G. Amino Acids Are Major Energy Substrates for Tissues of Hybrid Striped Bass and Zebrafish. Amino Acids 2017, 49, 2053–2063. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, N.; Maruyama, T.; Yoshikawa, N.; Matsumiya, R.; Ma, Y.; Ito, N.; Tasaka, Y.; Kuribara-Souta, A.; Miyata, K.; Oike, Y.; et al. A Muscle-Liver-Fat Signalling Axis Is Essential for Central Control of Adaptive Adipose Remodelling. Nat. Commun. 2015, 6, 6693. [Google Scholar] [CrossRef] [PubMed]
- Dyar, K.A.; Hubert, M.J.; Mir, A.A.; Ciciliot, S.; Lutter, D.; Greulich, F.; Quagliarini, F.; Kleinert, M.; Fischer, K.; Eichmann, T.O.; et al. Transcriptional Programming of Lipid and Amino Acid Metabolism by the Skeletal Muscle Circadian Clock. PLoS Biol. 2018, 16, e2005886. [Google Scholar] [CrossRef] [PubMed]
- Lv, Z.; Shi, W.; Zhang, Q. Role of Essential Amino Acids in Age-Induced Bone Loss. Int. J. Mol. Sci. 2022, 23, 11281. [Google Scholar] [CrossRef] [PubMed]
- Javed, K.; Fairweather, S.J. Amino Acid Transporters in the Regulation of Insulin Secretion and Signalling. Biochem. Soc. Trans. 2019, 47, 571–590. [Google Scholar] [CrossRef] [PubMed]
- Kandasamy, P.; Gyimesi, G.; Kanai, Y.; Hediger, M.A. Amino Acid Transporters Revisited: New Views in Health and Disease. Trends Biochem. Sci. 2018, 43, 752–789. [Google Scholar] [CrossRef] [PubMed]
- Verrey, F.; Closs, E.I.; Wagner, C.A.; Palacin, M.; Endou, H.; Kanai, Y. Cats and Hats: The Slc7 Family of Amino Acid Transporters. Pflug. Arch. Eur. J. Physiol. 2004, 447, 532–542. [Google Scholar] [CrossRef]
- Jungnickel, K.E.J.; Parker, J.L.; Newstead, S. Structural Basis for Amino Acid Transport by the Cat Family of Slc7 Transporters. Nat. Commun. 2018, 9, 550. [Google Scholar] [CrossRef]
- Bröer, S. Amino Acid Transporters as Modulators of Glucose Homeostasis. Trends Endocrinol. Metab. 2022, 33, 120–135. [Google Scholar] [CrossRef]
- Ren, W.; Liu, G.; Yin, J.; Tan, B.; Wu, G.; Bazer, F.W.; Peng, Y.; Yin, Y. Amino-Acid Transporters in T-Cell Activation and Differentiation. Cell Death Dis. 2017, 8, e2655. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Wang, J.; Garibsingh, R.A.; Hutchinson, K.; Shi, Y.; Eisenberg, G.; Yu, X.; Schlessinger, A.; Grewer, C. Conserved Allosteric Inhibition Mechanism in Slc1 Transporters. Elife 2023, 12, e83464. [Google Scholar] [CrossRef] [PubMed]
- Kanai, Y.; Clémençon, B.; Simonin, A.; Leuenberger, M.; Lochner, M.; Weisstanner, M.; Hediger, M.A. The Slc1 High-Affinity Glutamate and Neutral Amino Acid Transporter Family. Mol. Asp. Med. 2013, 34, 108–120. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.H.; Reith, M.E.; Quick, M.W. Synaptic Uptake and Beyond: The Sodium- and Chloride-Dependent Neurotransmitter Transporter Family Slc6. Pflug. Arch. Eur. J. Physiol. 2004, 447, 519–531. [Google Scholar] [CrossRef] [PubMed]
- Jayaraman, K.; Das, A.K.; Luethi, D.; Szöllősi, D.; Schütz, G.J.; Reith, M.E.A.; Sitte, H.H.; Stockner, T. Slc6 Transporter Oligomerization. J. Neurochem. 2021, 157, 919–929. [Google Scholar] [CrossRef] [PubMed]
- Yan, R.; Zhao, X.; Lei, J.; Zhou, Q. Structure of the Human Lat1-4f2hc Heteromeric Amino Acid Transporter Complex. Nature 2019, 568, 127–130. [Google Scholar] [CrossRef] [PubMed]
- Yaffe, D.; Saxel, O. Serial Passaging and Differentiation of Myogenic Cells Isolated from Dystrophic Mouse Muscle. Nature 1977, 270, 725–727. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Bermudez, J.; Baudrier, L.; La, K.; Zhu, X.G.; Fidelin, J.; Sviderskiy, V.O.; Papagiannakopoulos, T.; Molina, H.; Snuderl, M.; Lewis, C.A.; et al. Aspartate Is a Limiting Metabolite for Cancer Cell Proliferation under Hypoxia and in Tumours. Nat. Cell Biol. 2018, 20, 775–781. [Google Scholar] [CrossRef]
- Cai, Z.; Liu, D.; Yang, Y.; Xie, W.; He, M.; Yu, D.; Wu, Y.; Wang, X.; Xiao, W.; Li, Y. The Role and Therapeutic Potential of Stem Cells in Skeletal Muscle in Sarcopenia. Stem Cell Res. Ther. 2022, 13, 28. [Google Scholar] [CrossRef]
- Chen, Z.; Li, L.; Wu, W.; Liu, Z.; Huang, Y.; Yang, L.; Luo, Q.; Chen, J.; Hou, Y.; Song, G. Exercise Protects Proliferative Muscle Satellite Cells against Exhaustion Via the Igfbp7-Akt-Mtor Axis. Theranostics 2020, 10, 6448–6466. [Google Scholar] [CrossRef]
- Felig, P. Amino Acid Metabolism in Man. Annu. Rev. Biochem. 1975, 44, 933–955. [Google Scholar] [CrossRef] [PubMed]
- Wu, G. Amino Acids: Metabolism, Functions, and Nutrition. Amino Acids 2009, 37, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Nie, C.; He, T.; Zhang, W.; Zhang, G.; Ma, X. Branched Chain Amino Acids: Beyond Nutrition Metabolism. Int. J. Mol. Sci. 2018, 19, 954. [Google Scholar] [CrossRef] [PubMed]
- Montanari, A.; Simoni, I.; Vallisa, D.; Trifirò, A.; Colla, R.; Abbiati, R.; Borghi, L.; Novarini, A. Free Amino Acids in Plasma and Skeletal Muscle of Patients with Liver Cirrhosis. Hepatology 1988, 8, 1034–1039. [Google Scholar] [CrossRef]
- Baños, G.; Daniel, P.M.; Moorhouse, S.R.; Pratt, O.E. The Movement of Amino Acids between Blood and Skeletal Muscle in the Rat. J. Physiol. 1973, 235, 459–475. [Google Scholar] [CrossRef] [PubMed]
- Schiaffino, S.; Blaauw, B.; Dyar, K.A. The Functional Significance of the Skeletal Muscle Clock: Lessons from Bmal1 Knockout Models. Skelet. Muscle 2016, 6, 33. [Google Scholar] [CrossRef] [PubMed]
- Yeramian, A.; Martin, L.; Serrat, N.; Arpa, L.; Soler, C.; Bertran, J.; McLeod, C.; Palacín, M.; Modolell, M.; Lloberas, J.; et al. Arginine Transport Via Cationic Amino Acid Transporter 2 Plays a Critical Regulatory Role in Classical or Alternative Activation of Macrophages. J. Immunol. 2006, 176, 5918–5924. [Google Scholar] [CrossRef] [PubMed]
- Szondy, Z.; Al-Zaeed, N.; Tarban, N.; Fige, É.; Garabuczi, É.; Sarang, Z. Involvement of Phosphatidylserine Receptors in the Skeletal Muscle Regeneration: Therapeutic Implications. J. Cachexia Sarcopenia Muscle 2022, 13, 1961–1973. [Google Scholar] [CrossRef]
- Sancho-Muñoz, A.; Guitart, M.; Rodríguez, D.A.; Gea, J.; Martínez-Llorens, J.; Barreiro, E. Deficient Muscle Regeneration Potential in Sarcopenic Copd Patients: Role of Satellite Cells. J. Cell Physiol. 2021, 236, 3083–3098. [Google Scholar] [CrossRef]
- Pasini, E.; Corsetti, G.; Aquilani, R.; Romano, C.; Picca, A.; Calvani, R.; Dioguardi, F.S. Protein-Amino Acid Metabolism Disarrangements: The Hidden Enemy of Chronic Age-Related Conditions. Nutrients 2018, 10, 391. [Google Scholar] [CrossRef]
- Taylor, S.C.; Berkelman, T.; Yadav, G.; Hammond, M. A Defined Methodology for Reliable Quantification of Western Blot Data. Mol. Biotechnol. 2013, 55, 217–226. [Google Scholar] [CrossRef]


| Gene Name | Primer Sequence (Forward) | Primer Sequence (Reverse) |
|---|---|---|
| Slc7a2 | 5′-GAGTAAGAGGCAGTCACCCG-3′ | 5′-ACTCGCTCTTCAAAGTCGCA-3′ |
| Myogenin | 5′-AATGGATTTGGACGCATTGGT-3′ | 5′-TTTGCACTGGTACGTGTTGAT-3′ |
| MyoD | 5′-GCACTACAGTGGCGACTCAGAT-3′ | 5′-TAGTAGGCGGTGTCGTAGCCAT-3′ |
| Myh1 | 5′-GCGAATCGAGGCTCAGAACAA-3′ | 5′-GTAGTTCCGCCTTCGGTCTTG-3′ |
| Myh2 | 5′-CAGAGGCAAGTAGTGGTGGA-3′ | 5′-CAAATTCTCTCTGAACAGGGCA-3′ |
| Myh4 | 5′-ACACAGAGTCAGGCGAGTTT-3′ | 5′-CAGTGCGTTCTTGGCCTT-3′ |
| Myh7 | 5′-CTTGCTACCCTCAGGTGGCT-3′ | 5′-GAGCCTTGGATTCTCAAACG-3′ |
| Gapdh | 5′-TGTCAAGCTCATTTCCTGGT-3′ | 5′-TAGGGCCTCTCTTGCTCAGT-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, T.; Zhou, J.; Wang, B.; Wang, X.; Xiao, W.; Yang, M.; Liu, Y.; Wang, Q.; Xiang, Y.; Lan, X. Integrated Amino Acids and Transcriptome Analysis Reveals Arginine Transporter SLC7A2 Is a Novel Regulator of Myogenic Differentiation. Int. J. Mol. Sci. 2024, 25, 95. https://doi.org/10.3390/ijms25010095
Huang T, Zhou J, Wang B, Wang X, Xiao W, Yang M, Liu Y, Wang Q, Xiang Y, Lan X. Integrated Amino Acids and Transcriptome Analysis Reveals Arginine Transporter SLC7A2 Is a Novel Regulator of Myogenic Differentiation. International Journal of Molecular Sciences. 2024; 25(1):95. https://doi.org/10.3390/ijms25010095
Chicago/Turabian StyleHuang, Tiane, Jing Zhou, Benhui Wang, Xiang Wang, Wanli Xiao, Mengqi Yang, Yan Liu, Qiquan Wang, Yang Xiang, and Xinqiang Lan. 2024. "Integrated Amino Acids and Transcriptome Analysis Reveals Arginine Transporter SLC7A2 Is a Novel Regulator of Myogenic Differentiation" International Journal of Molecular Sciences 25, no. 1: 95. https://doi.org/10.3390/ijms25010095
APA StyleHuang, T., Zhou, J., Wang, B., Wang, X., Xiao, W., Yang, M., Liu, Y., Wang, Q., Xiang, Y., & Lan, X. (2024). Integrated Amino Acids and Transcriptome Analysis Reveals Arginine Transporter SLC7A2 Is a Novel Regulator of Myogenic Differentiation. International Journal of Molecular Sciences, 25(1), 95. https://doi.org/10.3390/ijms25010095






