Highlighting the Major Role of Cyclin C in Cyclin-Dependent Kinase 8 Activity through Molecular Dynamics Simulations
Abstract
:1. Introduction
2. Results and Discussion
2.1. Effect of CycC Exclusion on Structure and Dynamics of CDK8
2.2. Understanding the Molecular Basis of the Interaction between CDK8 and CycC
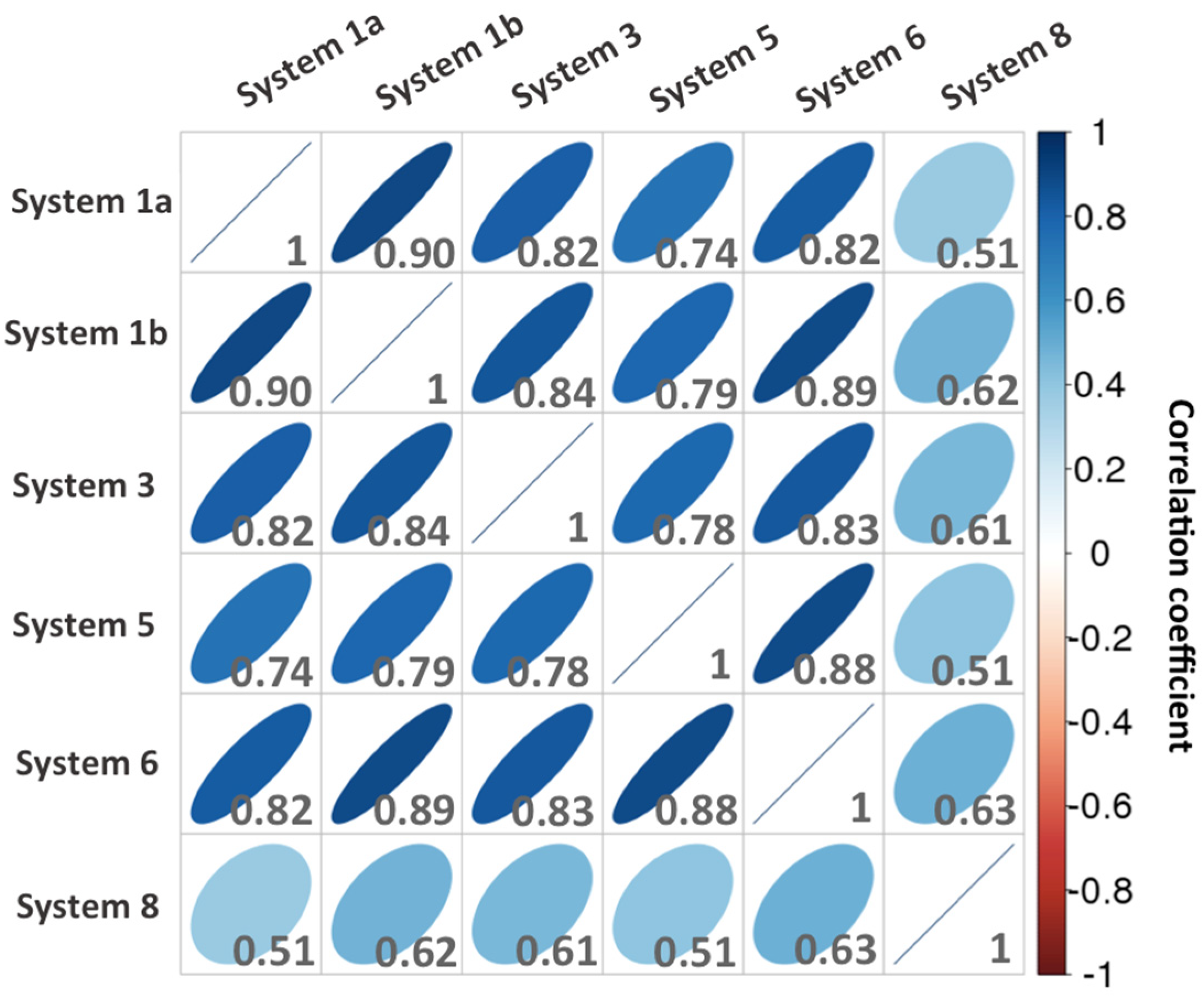
2.2.1. CDK8–CycC Binding Free Energy
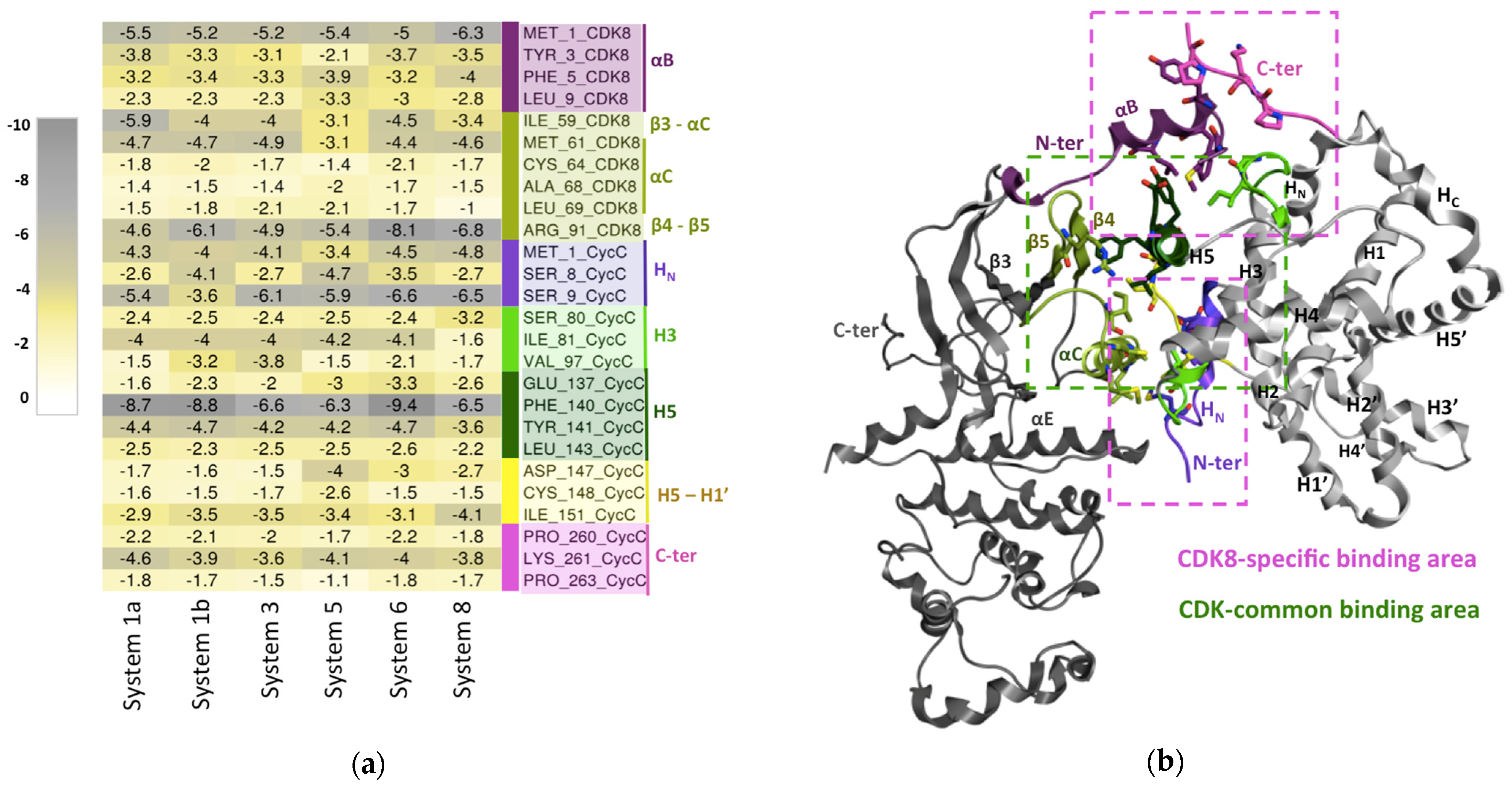
2.2.2. CDK8–CycC Binding Free Energy: Decomposition per Residue
Hotspots Common to All Studied CDK8–CycC Complexes
Hotspots Common to the CDK Family
Hotspots Specific to CDK8–CycC
- Involving the N-terminus segment of CycC
- Involving the CDK8-specific N-terminus helix (αB-helix)
Difference in Binding Surface between the Different Complexes
DMG-out CDK8–CycC Complexes
Difference in Binding Surface between DMG-in and DMG-out
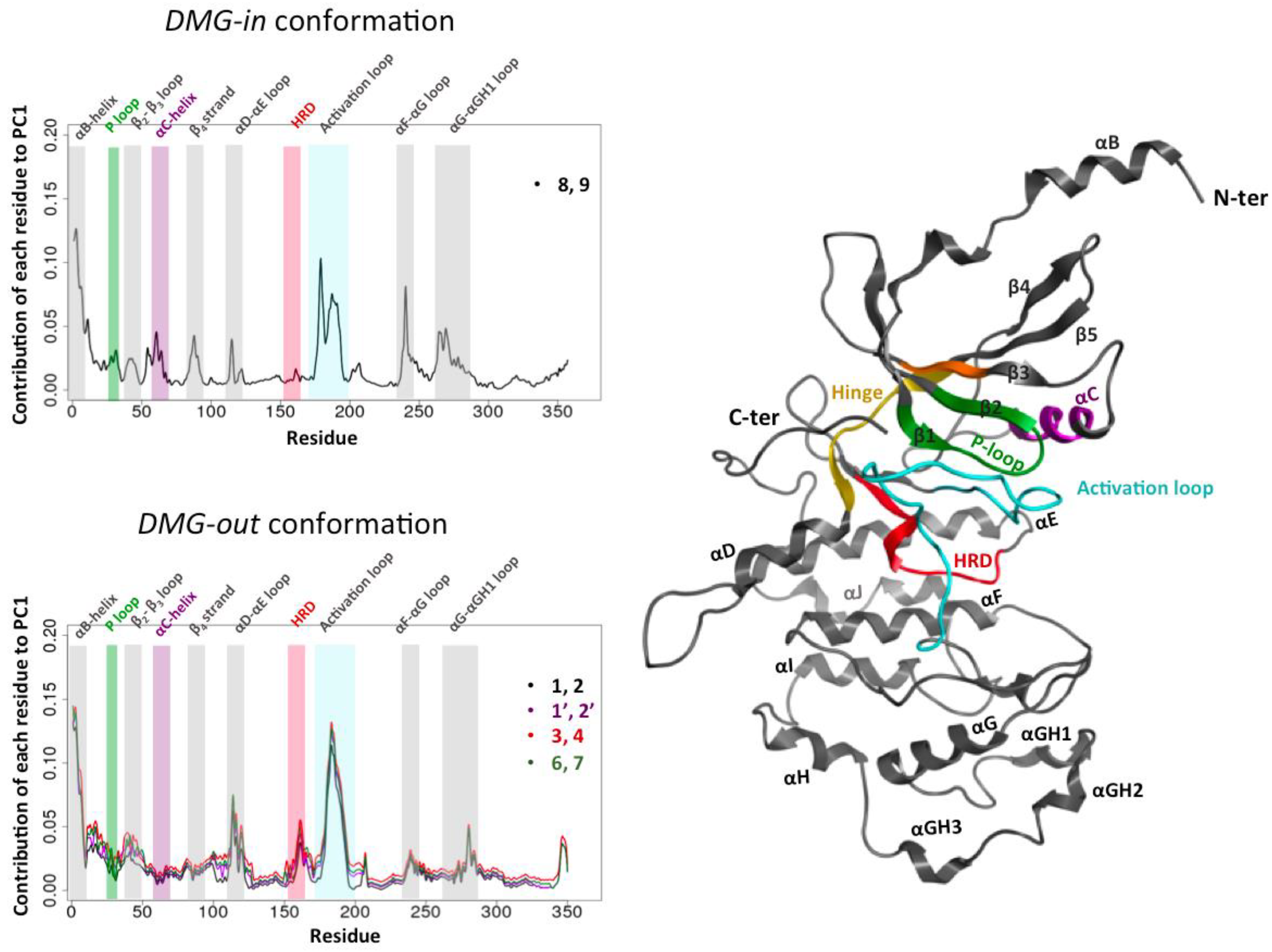
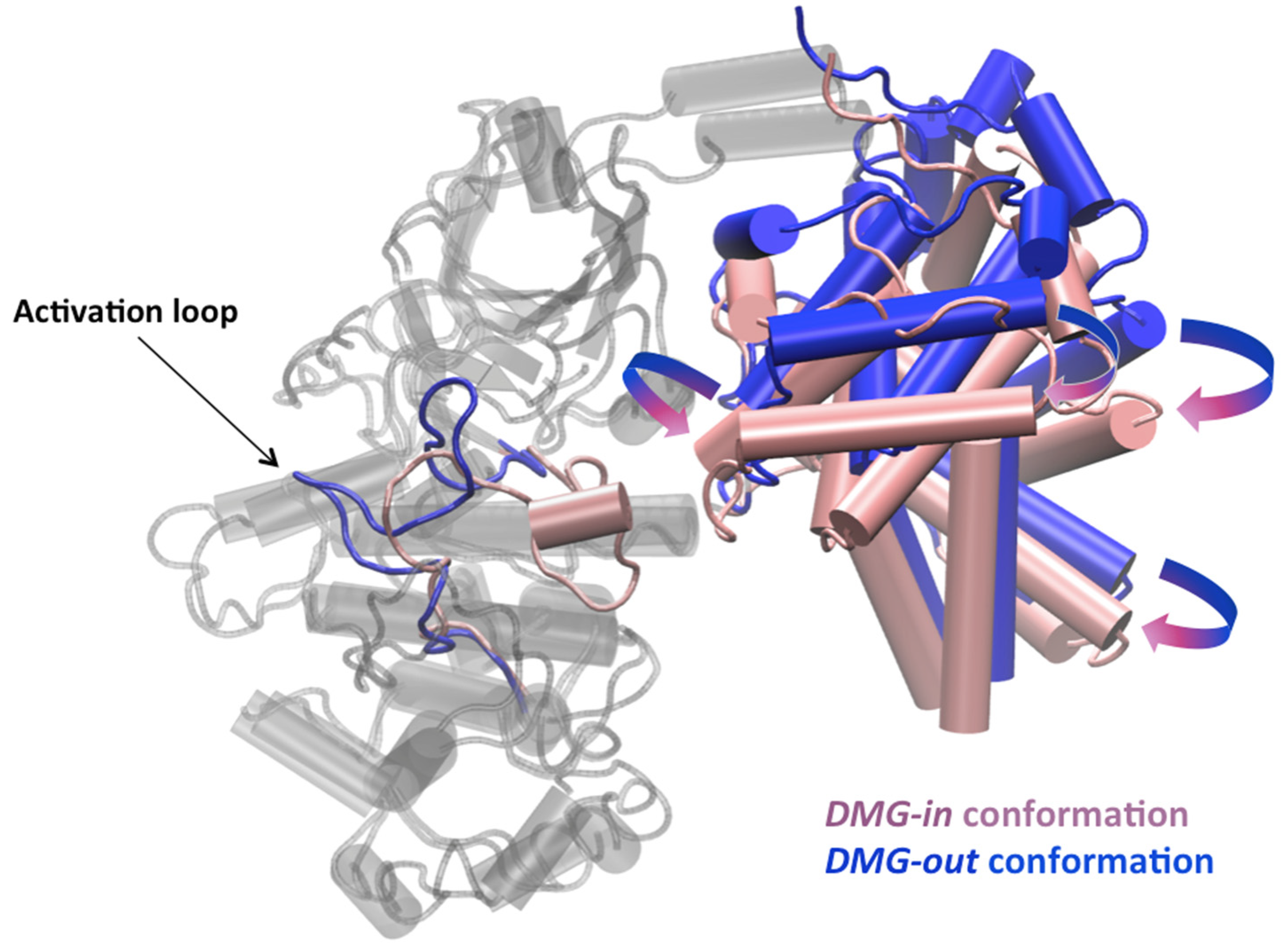
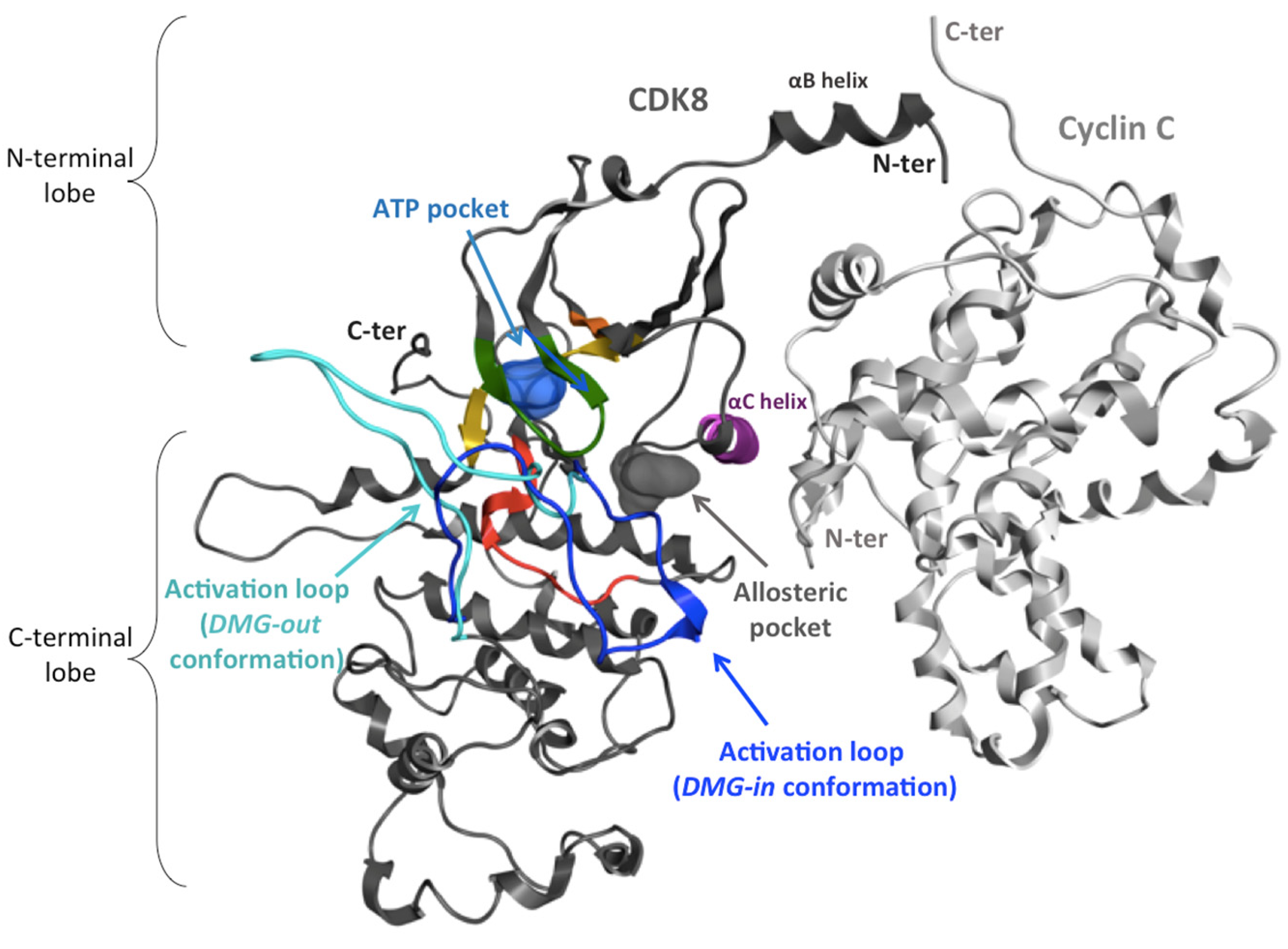
2.3. Activation Mechanism of CDK8
3. Materials and Methods
3.1. Material Description
3.2. Model Building
3.3. System Preparation
3.4. Conventional MD Simulation (cMD)
3.5. Targeted Molecular Dynamics (TMD)
3.6. RMSD, RMSF
3.7. PCA
3.8. MM-GBSA
3.9. Other Analysis Tools
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Malumbres, M. Cyclin-Dependent Kinases. Genome Biol. 2014, 15, 122. [Google Scholar] [CrossRef]
- Zhang, M.; Zhang, L.; Hei, R.; Li, X.; Cai, H.; Wu, X.; Zheng, Q.; Cai, C. CDK Inhibitors in Cancer Therapy, an Overview of Recent Development. Am. J. Cancer Res. 2021, 11, 1913–1935. [Google Scholar]
- Canavese, M.; Santo, L.; Raje, N. Cyclin Dependent Kinases in Cancer: Potential for Therapeutic Intervention. Cancer Biol. Ther. 2012, 13, 451–457. [Google Scholar] [CrossRef]
- Philip, S.; Kumarasiri, M.; Teo, T.; Yu, M.; Wang, S. Cyclin-Dependent Kinase 8: A New Hope in Targeted Cancer Therapy? J. Med. Chem. 2018, 61, 5073–5092. [Google Scholar] [CrossRef] [PubMed]
- Rzymski, T.; Mikula, M.; Wiklik, K.; Brzózka, K. CDK8 Kinase—An Emerging Target in Targeted Cancer Therapy. Biochim. Biophys. Acta (BBA)—Proteins Proteom. 2015, 1854, 1617–1629. [Google Scholar] [CrossRef]
- Allen, B.L.; Taatjes, D.J. The Mediator Complex: A Central Integrator of Transcription. Nat. Rev. Mol. Cell Biol. 2015, 16, 155–166. [Google Scholar] [CrossRef] [PubMed]
- Harper, T.M.; Taatjes, D.J. The Complex Structure and Function of Mediator. J. Biol. Chem. 2018, 293, 13778–13785. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, J.; Ding, Z.; Ji, J.; Sun, Q.; Cai, G. Structural Flexibility and Functional Interaction of Mediator Cdk8 Module. Protein Cell 2013, 4, 911–920. [Google Scholar] [CrossRef]
- Friedson, B.; Cooper, K.F. Cdk8 Kinase Module: A Mediator of Life and Death Decisions in Times of Stress. Microorganisms 2021, 9, 2152. [Google Scholar] [CrossRef]
- Akoulitchev, S.; Chuikov, S.; Reinberg, D. TFIIH Is Negatively Regulated by Cdk8-Containing Mediator Complexes. Nature 2000, 407, 102–106. [Google Scholar] [CrossRef]
- Furumoto, T.; Tanaka, A.; Ito, M.; Malik, S.; Hirose, Y.; Hanaoka, F.; Ohkuma, Y. A Kinase Subunit of the Human Mediator Complex, CDK8, Positively Regulates Transcriptional Activation. Genes Cells 2007, 12, 119–132. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Kung, C.; Fishburn, J.; Ansari, A.Z.; Shokat, K.M.; Hahn, S. Two Cyclin-Dependent Kinases Promote RNA Polymerase II Transcription and Formation of the Scaffold Complex. Mol. Cell. Biol. 2004, 24, 1721–1735. [Google Scholar] [CrossRef] [PubMed]
- Firestein, R.; Bass, A.J.; Kim, S.Y.; Dunn, I.F.; Silver, S.J.; Guney, I.; Freed, E.; Ligon, A.H.; Vena, N.; Ogino, S.; et al. CDK8 Is a Colorectal Cancer Oncogene That Regulates Beta-Catenin Activity. Nature 2008, 455, 547–551. [Google Scholar] [CrossRef] [PubMed]
- McDermott, M.S.J.; Chumanevich, A.A.; Lim, C.-U.; Liang, J.; Chen, M.; Altilia, S.; Oliver, D.; Rae, J.M.; Shtutman, M.; Kiaris, H.; et al. Inhibition of CDK8 Mediator Kinase Suppresses Estrogen Dependent Transcription and the Growth of Estrogen Receptor Positive Breast Cancer. Oncotarget 2017, 8, 12558–12575. [Google Scholar] [CrossRef]
- Schneider, E.V.; Böttcher, J.; Blaesse, M.; Neumann, L.; Huber, R.; Maskos, K. The Structure of CDK8/CycC Implicates Specificity in the CDK/Cyclin Family and Reveals Interaction with a Deep Pocket Binder. J. Mol. Biol. 2011, 412, 251–266. [Google Scholar] [CrossRef] [PubMed]
- Taylor, S.S.; Kornev, A.P. Protein Kinases: Evolution of Dynamic Regulatory Proteins. Trends Biochem. Sci. 2011, 36, 65–77. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Amire-Brahimi, B.; Xie, X.-J.; Huang, L.; Ji, J.-Y. All-Atomic Molecular Dynamic Studies of Human CDK8: Insight into the A-Loop, Point Mutations and Binding with Its Partner CycC. Comput. Biol. Chem. 2014, 51, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Cholko, T.; Chen, W.; Tang, Z.; Chang, C.A. A Molecular Dynamics Investigation of CDK8/CycC and Ligand Binding: Conformational Flexibility and Implication in Drug Discovery. J. Comput. Aided Mol. Des. 2018, 32, 671–685. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Tang, H.-C.; Tsai, K.-L. Unveiling the Noncanonical Activation Mechanism of CDKs: Insights from Recent Structural Studies. Front. Mol. Biosci. 2023, 10, 1290631. [Google Scholar] [CrossRef]
- Ježek, J.; Smethurst, D.G.J.; Stieg, D.C.; Kiss, Z.A.C.; Hanley, S.E.; Ganesan, V.; Chang, K.-T.; Cooper, K.F.; Strich, R. Cyclin C: The Story of a Non-Cycling Cyclin. Biology 2019, 8, 3. [Google Scholar] [CrossRef]
- Pavletich, N.P. Mechanisms of Cyclin-Dependent Kinase Regulation: Structures of Cdks, Their Cyclin Activators, and Cip and INK4 Inhibitors. J. Mol. Biol. 1999, 287, 821–828. [Google Scholar] [CrossRef] [PubMed]
- Hoeppner, S.; Baumli, S.; Cramer, P. Structure of the Mediator Subunit Cyclin C and Its Implications for CDK8 Function. J. Mol. Biol. 2005, 350, 833–842. [Google Scholar] [CrossRef] [PubMed]
- Oppermann, F.S.; Gnad, F.; Olsen, J.V.; Hornberger, R.; Greff, Z.; Kéri, G.; Mann, M.; Daub, H. Large-Scale Proteomics Analysis of the Human Kinome. Mol Cell Proteom. 2009, 8, 1751–1764. [Google Scholar] [CrossRef] [PubMed]
- Schneider, E.V.; Bottcher, J.; Huber, R.; Maskos, K.; Neumann, L. Structure-Kinetic Relationship Study of CDK8/CycC Specific Compounds. Proc. Natl. Acad. Sci. USA 2013, 110, 8081–8086. [Google Scholar] [CrossRef]
- Bergeron, P.; Koehler, M.F.T.; Blackwood, E.M.; Bowman, K.; Clark, K.; Firestein, R.; Kiefer, J.R.; Maskos, K.; McCleland, M.L.; Orren, L.; et al. Design and Development of a Series of Potent and Selective Type II Inhibitors of CDK8. ACS Med. Chem. Lett. 2016, 7, 595–600. [Google Scholar] [CrossRef]
- Roskoski, R. Classification of Small Molecule Pro-Tein Kinase Inhibitors Based upon the Structures of Their Drug-Enzyme Complexes. Pharmacol. Res. 2016, 103, 26–48. [Google Scholar] [CrossRef] [PubMed]
- Alexander, L.T.; Möbitz, H.; Drueckes, P.; Savitsky, P.; Fedorov, O.; Elkins, J.M.; Deane, C.M.; Cowan-Jacob, S.W.; Knapp, S. Type II Inhibitors Targeting CDK2. ACS Chem. Biol. 2015, 10, 2116–2125. [Google Scholar] [CrossRef]
- Barette, C.; Jariel-Encontre, I.; Piechaczyk, M.; Piette, J. Human Cyclin C Protein Is Stabilized by Its Associated Kinase Cdk8, Independently of Its Catalytic Activity. Oncogene 2001, 20, 551–562. [Google Scholar] [CrossRef]
- Coleman, K.G.; Wautlet, B.S.; Morrissey, D.; Mulheron, J.; Sedman, S.A.; Brinkley, P.; Price, S.; Webster, K.R. Identification of CDK4 Sequences Involved in Cyclin D1 and P16 Binding. J. Biol. Chem. 1997, 272, 18869–18874. [Google Scholar] [CrossRef] [PubMed]
- Massova, I.; Kollman, P.A. Computational Alanine Scanning To Probe Protein−Protein Interactions: A Novel Approach To Evaluate Binding Free Energies. J. Am. Chem. Soc. 1999, 121, 8133–8143. [Google Scholar] [CrossRef]
- Lo Conte, L.; Chothia, C.; Janin, J. The Atomic Structure of Protein-Protein Recognition Sites. J. Mol. Biol. 1999, 285, 2177–2198. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.J.; Lin, S.L.; Wolfson, H.J.; Nussinov, R. Studies of Protein-Protein Interfaces: A Statistical Analysis of the Hydrophobic Effect. Protein Sci. 1997, 6, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Kundrotas, P.J.; Alexov, E. Electrostatic Properties of Protein-Protein Complexes. Biophys. J. 2006, 91, 1724–1736. [Google Scholar] [CrossRef] [PubMed]
- Echalier, A.; Endicott, J.A.; Noble, M.E.M. Recent Developments in Cyclin-Dependent Kinase Biochemical and Structural Studies. Biochim. Biophys. Acta 2010, 1804, 511–519. [Google Scholar] [CrossRef] [PubMed]
- Lolli, G. Structural Dissection of Cyclin Dependent Kinases Regulation and Protein Recognition Properties. Cell Cycle 2010, 9, 1551–1561. [Google Scholar] [CrossRef] [PubMed]
- Flocco, M.M.; Mowbray, S.L. Planar Stacking Interactions of Arginine and Aromatic Side-Chains in Proteins. J. Mol. Biol. 1994, 235, 709–717. [Google Scholar] [CrossRef] [PubMed]
- Wood, D.J.; Endicott, J.A. Structural Insights into the Functional Diversity of the CDK–Cyclin Family. Open Biol. 2018, 8, 180112. [Google Scholar] [CrossRef] [PubMed]
- Baumli, S.; Lolli, G.; Lowe, E.D.; Troiani, S.; Rusconi, L.; Bullock, A.N.; Debreczeni, J.É.; Knapp, S.; Johnson, L.N. The Structure of P-TEFb (CDK9/Cyclin T1), Its Complex with Flavopiridol and Regulation by Phosphorylation. EMBO J. 2008, 27, 1907–1918. [Google Scholar] [CrossRef] [PubMed]
- Heitz, F.; Morris, M.C.; Fesquet, D.; Cavadore, J.-C.; Dorée, M.; Divita, G. Interactions of Cyclins with Cyclin-Dependent Kinases: A Common Interactive Mechanism. Biochemistry 1997, 36, 4995–5003. [Google Scholar] [CrossRef]
- Gondeau, C.; Gerbal-Chaloin, S.; Bello, P.; Aldrian-Herrada, G.; Morris, M.C.; Divita, G. Design of a Novel Class of Peptide Inhibitors of Cyclin-Dependent Kinase/Cyclin Activation. J. Biol. Chem. 2005, 280, 13793–13800. [Google Scholar] [CrossRef]
- Canela, N.; Orzáez, M.; Fucho, R.; Mateo, F.; Gutierrez, R.; Pineda-Lucena, A.; Bachs, O.; Pérez-Payá, E. Identification of an Hexapeptide That Binds to a Surface Pocket in Cyclin A and Inhibits the Catalytic Activity of the Complex Cyclin-Dependent Kinase 2-Cyclin A. J. Biol. Chem. 2006, 281, 35942–35953. [Google Scholar] [CrossRef]
- The UniProt Consortium; Bateman, A.; Martin, M.-J.; Orchard, S.; Magrane, M.; Ahmad, S.; Alpi, E.; Bowler-Barnett, E.H.; Britto, R.; Bye-A-Jee, H.; et al. UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023, 51, D523–D531. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, Scalable Generation of High-Quality Protein Multiple Sequence Alignments Using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Sali, A.; Blundell, T.L. Comparative Protein Modelling by Satisfaction of Spatial Restraints. J. Mol. Biol. 1993, 234, 779–815. [Google Scholar] [CrossRef]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A Program to Check the Stereochemical Quality of Protein Structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Wiederstein, M.; Sippl, M.J. ProSA-Web: Interactive Web Service for the Recognition of Errors in Three-Dimensional Structures of Proteins. Nucleic Acids Res. 2007, 35, W407–W410. [Google Scholar] [CrossRef]
- Case, D.; Babin, V.; Berryman, J.; Betz, R.; Cai, Q.; Cerutti, D.; Cheatham, T.; Darden, T.; Duke, R.; Gohlke, H.; et al. Amber14; University of California: San Francisco, CA, USA, 2014. [Google Scholar]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and Testing of a General Amber Force Field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef]
- Wang, J.; Wang, W.; Kollman, P.A.; Case, D.A. Automatic Atom Type and Bond Type Perception in Molecular Mechanical Calculations. J. Mol. Graph. Model. 2006, 25, 247–260. [Google Scholar] [CrossRef]
- Hall, H.K. Potentiometric Determination of the Base Strength of Amines in Non-Protolytic Solvents. J. Phys. Chem. 1956, 60, 63–70. [Google Scholar] [CrossRef]
- Jakalian, A.; Jack, D.B.; Bayly, C.I. Fast, Efficient Generation of High-Quality Atomic Charges. AM1-BCC Model: II. Parameterization and Validation. J. Comput. Chem. 2002, 23, 1623–1641. [Google Scholar] [CrossRef]
- Olsson, M.H.M.; Søndergaard, C.R.; Rostkowski, M.; Jensen, J.H. PROPKA3: Consistent Treatment of Internal and Surface Residues in Empirical pKa Predictions. J. Chem. Theory Comput. 2011, 7, 525–537. [Google Scholar] [CrossRef]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef]
- Andersen, H.C. Rattle: A “Velocity” Version of the Shake Algorithm for Molecular Dynamics Calculations. J. Comput. Phys. 1983, 52, 24–34. [Google Scholar] [CrossRef]
- Schlitter, J.; Engels, M.; Krüger, P. Targeted Molecular Dynamics: A New Approach for Searching Pathways of Conformational Transitions. J. Mol. Graph. 1994, 12, 84–89. [Google Scholar] [CrossRef]
- Grant, B.J.; Rodrigues, A.P.C.; ElSawy, K.M.; McCammon, J.A.; Caves, L.S.D. Bio3d: An R Package for the Comparative Analysis of Protein Structures. Bioinformatics 2006, 22, 2695–2696. [Google Scholar] [CrossRef]
- Kollman, P.A.; Massova, I.; Reyes, C.; Kuhn, B.; Huo, S.; Chong, L.; Lee, M.; Lee, T.; Duan, Y.; Wang, W.; et al. Calculating Structures and Free Energies of Complex Molecules: Combining Molecular Mechanics and Continuum Models. Acc. Chem. Res. 2000, 33, 889–897. [Google Scholar] [CrossRef]
- Onufriev, A.; Bashford, D.; Case, D.A. Exploring Protein Native States and Large-Scale Conformational Changes with a Modified Generalized Born Model. Proteins 2004, 55, 383–394. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD—Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Bowers, K.; Chow, E.; Xu, H.; Dror, R.; Eastwood, M.; Gregersen, B.; Klepeis, J.; Kolossvary, I.; Moraes, M.; Sacerdoti, F.; et al. Scalable Algorithms for Molecular Dynamics Simulations on Commodity Clusters. In Proceedings of the ACM/IEEE SC 2006 Conference (SC’06), Tampa, FL, USA, 11–17 November 2006; p. 43. [Google Scholar]


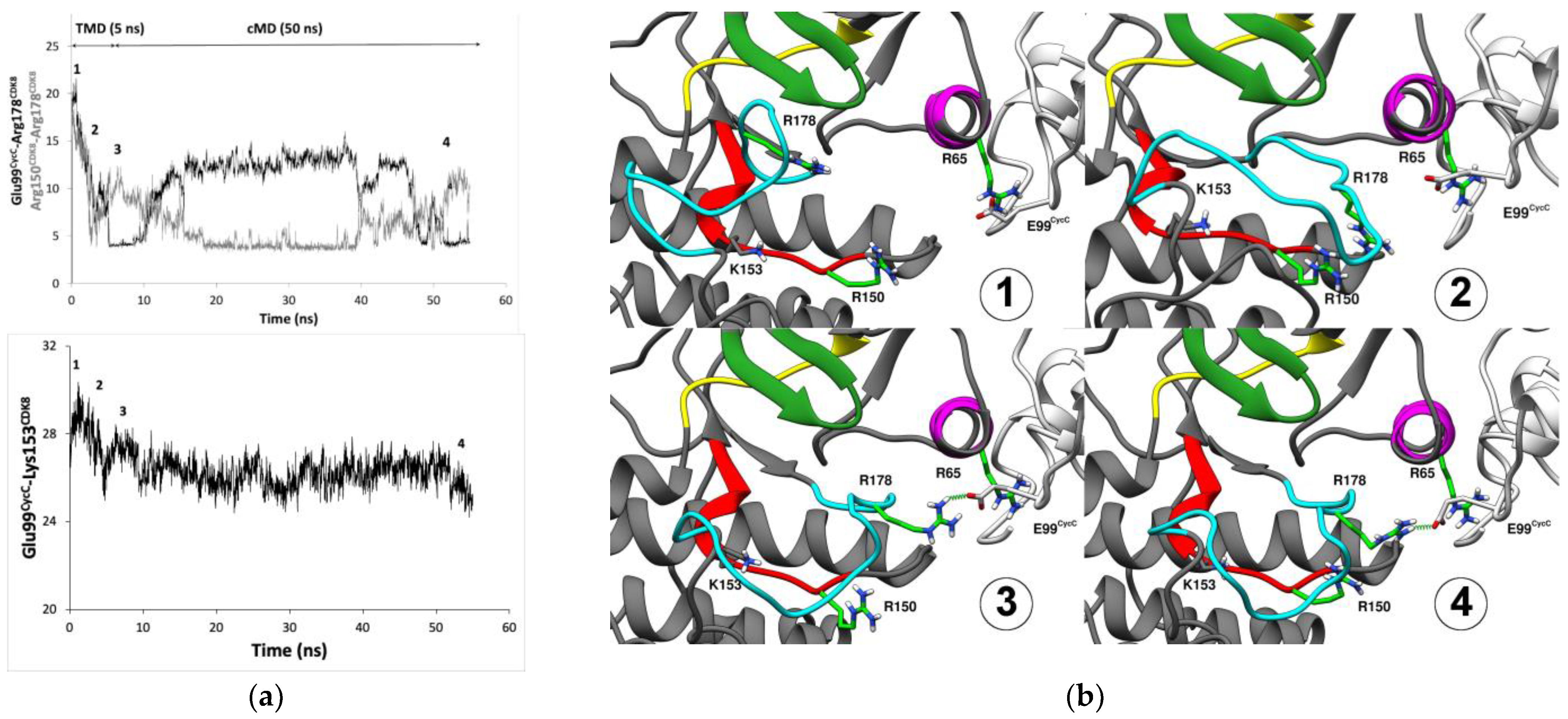
| System ID with Cyclin C | Average RMSD (Å) (±Standard Deviation) | System ID Without Cyclin C | Average RMSD (Å) (±Standard Deviation) |
|---|---|---|---|
| 1a | 3.6 ± 0.2 | 2a | 5.5 ± 1 |
| 1b | 3.9 ± 0.3 | 2b | 5.5 ± 0.3 |
| 3 | 3.8 ± 0.3 | 4 | 5.4 ± 0.9 |
| 6 | 3.2 ± 0.2 | 7 | 5 ± 0.6 |
| 8 | 3.4 ± 0.3 | 9 | 3.7 ± 0.4 |
| Systems’ PDB ID Conformation | System 1a (4F6U) DMG-out | System 1b (4F6U-Replica) DMG-out | System 3 (4F6U-apo) DMG-out | System 5 (4F6UR65A_E66A) DMG-out | System 6 (4F7L) DMG-out | System 8 (4F7S) DMG-in |
|---|---|---|---|---|---|---|
| ΔEVDW | −163.0 ± 0.1 | −160.7 ± 0.1 | −160.0 ± 0.2 | −148.7 ± 0.1 | −156.3 ± 0.1 | −176.5 ± 0.1 |
| ΔEeel | −508.4 ± 1.0 | −436.5 ± 1.0 | −490.0 ± 1.5 | −573.9 ± 1 | −500.6 ± 0.9 | −588.5 ± 0.9 |
| ΔEGB | 554.3 ± 0.9 | 491.1 ± 0.9 | 543.2 ± 1.4 | 613.9 ± 0.9 | 541.0 ± 0.8 | 613.9 ± 0.8 |
| ΔEnp | −23.9 ± 0.0 | −23.1 ± 0.0 | −23.2 ± 0.3 | −22.7 ± 0.0 | −22.9 ± 0.0 | −25.4 ± 0.0 |
| ΔGtotal (Without entropy) | −141.0± 0.2 | −129.2 ± 0.2 | −130.0 ± 0.3 | −131.5 ± 0.2 | −138.8 ± 0.2 | −124.6 ± 0.2 |
| System ID | PDB ID | Ligand Name | DMG Conformation | Manipulation |
|---|---|---|---|---|
| 1a | 4F6U | 0SR | DMG-out | (-) |
| 1b (replica) | 4F6U | 0SR | DMG-out | (-) |
| 2a | 4F6U | 0SR | DMG-out | Removal of CycC |
| 2b (replica) | 4F6U | 0SR | DMG-out | Removal of CycC |
| 3 | 4F6U | (-) | DMG-out | Removal of ligand |
| 4 | 4F6U | (-) | DMG-out | Removal of ligand and CycC |
| 5 | 4F6U | 0SR | DMG-out | CDK8 mutations: E66A, R65A |
| 6 | 4F7L | 0SO | DMG-out | (-) |
| 7 | 4F7L | 0SO | DMG-out | Removal of CycC |
| 8 | 4F7S | 0SW | DMG-in | (-) |
| 9 | 4F7S | 0SW | DMG-in | Removal of CycC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ziada, S.; Diharce, J.; Serillon, D.; Bonnet, P.; Aci-Sèche, S. Highlighting the Major Role of Cyclin C in Cyclin-Dependent Kinase 8 Activity through Molecular Dynamics Simulations. Int. J. Mol. Sci. 2024, 25, 5411. https://doi.org/10.3390/ijms25105411
Ziada S, Diharce J, Serillon D, Bonnet P, Aci-Sèche S. Highlighting the Major Role of Cyclin C in Cyclin-Dependent Kinase 8 Activity through Molecular Dynamics Simulations. International Journal of Molecular Sciences. 2024; 25(10):5411. https://doi.org/10.3390/ijms25105411
Chicago/Turabian StyleZiada, Sonia, Julien Diharce, Dylan Serillon, Pascal Bonnet, and Samia Aci-Sèche. 2024. "Highlighting the Major Role of Cyclin C in Cyclin-Dependent Kinase 8 Activity through Molecular Dynamics Simulations" International Journal of Molecular Sciences 25, no. 10: 5411. https://doi.org/10.3390/ijms25105411
APA StyleZiada, S., Diharce, J., Serillon, D., Bonnet, P., & Aci-Sèche, S. (2024). Highlighting the Major Role of Cyclin C in Cyclin-Dependent Kinase 8 Activity through Molecular Dynamics Simulations. International Journal of Molecular Sciences, 25(10), 5411. https://doi.org/10.3390/ijms25105411







