Genome Variability in Artificial Allopolyploid Hybrids of Avena sativa L. and Avena macrostachya Balansa ex Coss. et Durieu Based on Marker Sequences of Satellite DNA and the ITS1–5.8S rDNA Region
Abstract
:1. Introduction
2. Results
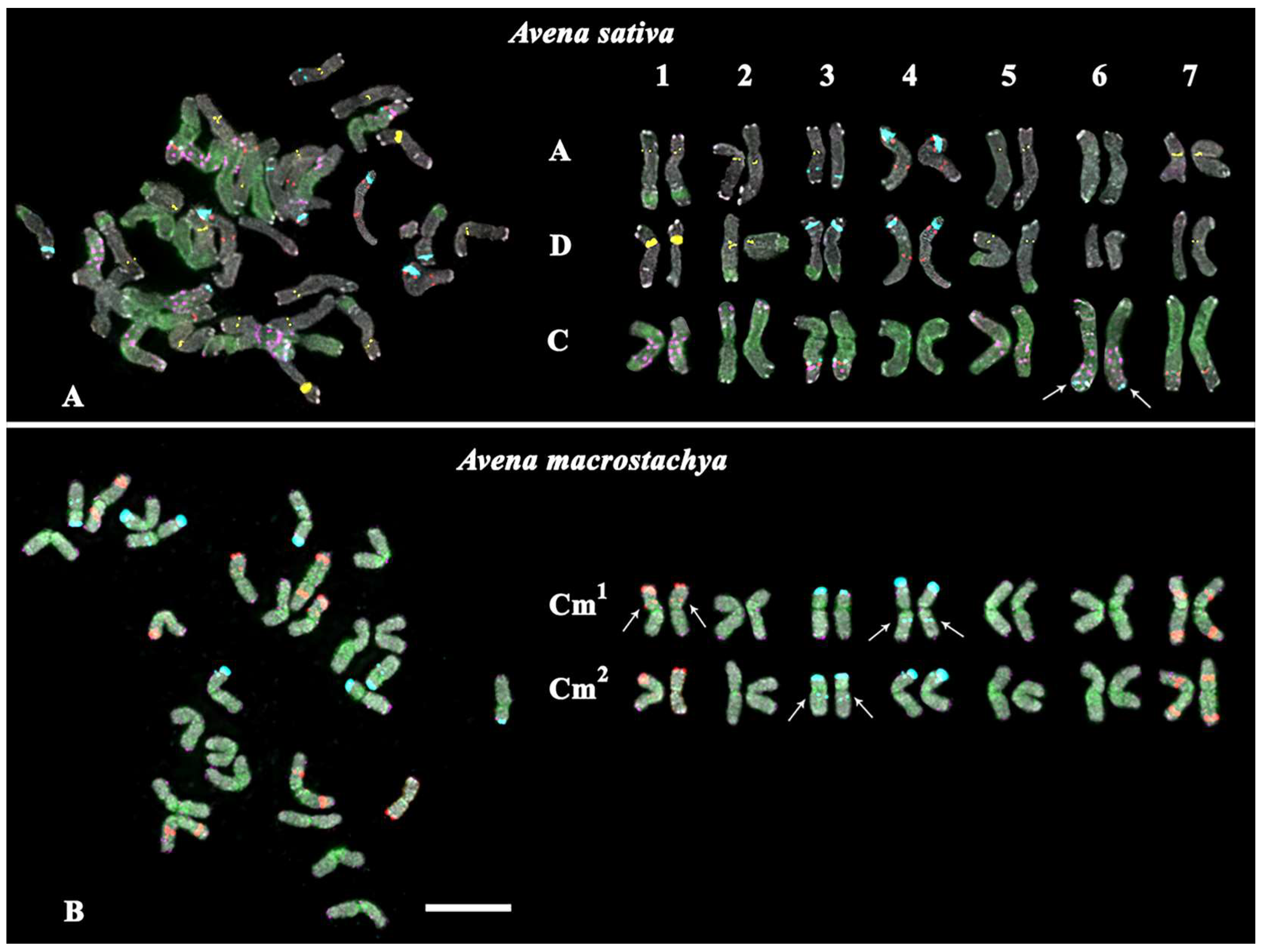
2.1. Chromosomal Structural Variations
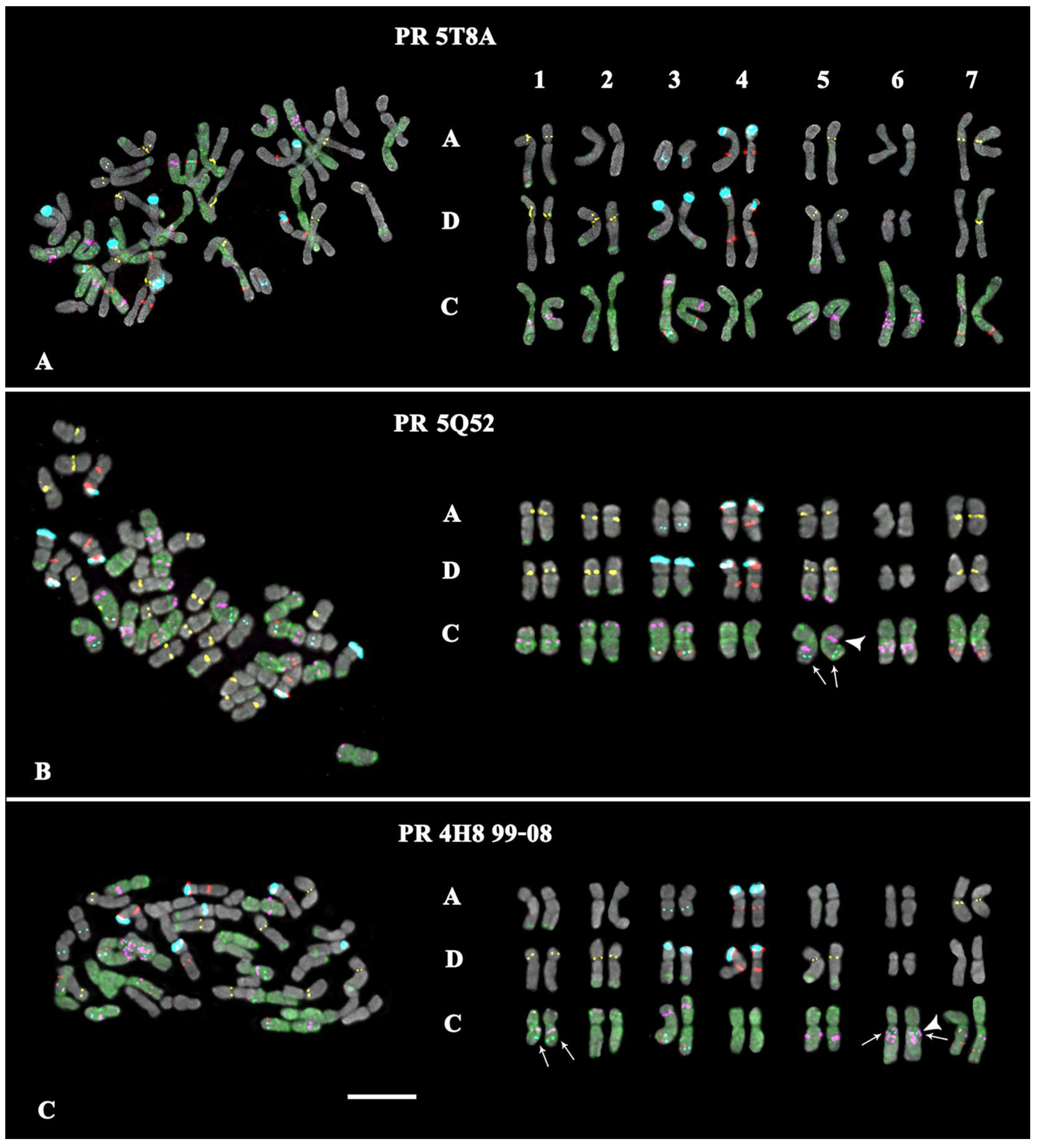
- In the karyotype of PR 5T8A, only constant minor 35S rDNA loci were detected (on chromosome pairs 3A and 3C). On chromosome pairs 2A and 3A, no GTT clusters were revealed, although they were detected in A. sativa; and in pairs 5A and 7D, GTT loci were observed in the hemizygous state (Figure 2A; Supplementary Figures S2 and S5).
- In the karyotype of PR 5Q52, in addition to the constant minor 35S rDNA signals, a polymorphic minor locus was revealed on chromosome pairs 5C. A pericentric inversion occurred in one of the homologs of chromosome pair 5C. On chromosome pair 3A, a GTT locus was not detected (Figure 2B; Supplementary Figures S2 and S5).
- In the karyotype of PR 4H8 99-08, polymorphic minor 35S rDNA loci were revealed on chromosome pairs 1C and 6C. A chromosome inversion occurred within the long arms of chromosome pair 5C. On chromosome pairs 1A, 2A, 3A, 5A, and 7A, no GTT loci were observed (Figure 2C; Supplementary Figures S2 and S5).
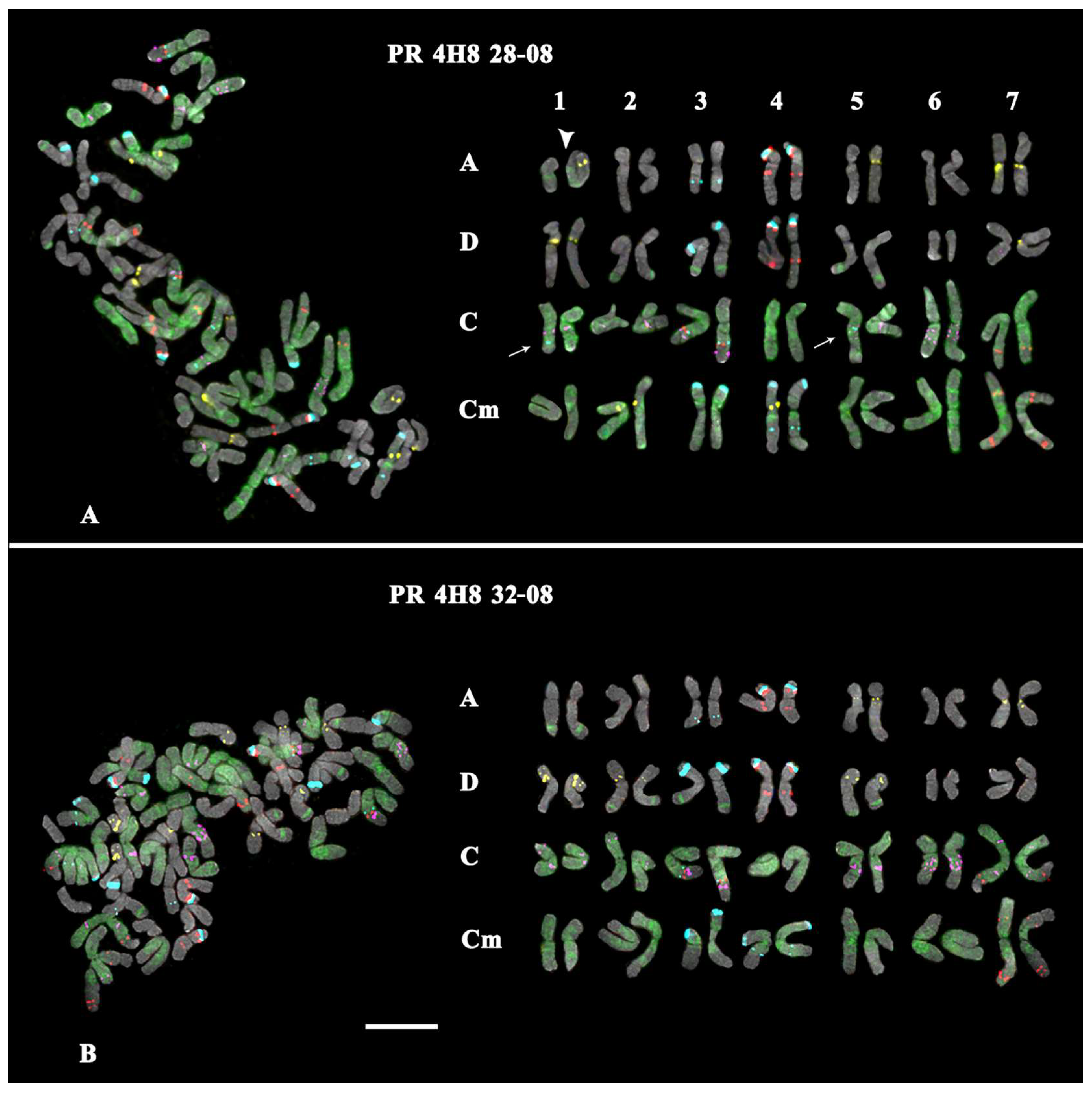
- In the karyotype of PR 4H8 28-08, polymorphic minor 35S rDNA loci were detected on chromosome pairs 1C and 5C (both in the hemizygous state). In pair 1A, a translocation occurred between homologous chromosomes (Figure 3A). No GTT clusters were observed on chromosome pairs 2A, 3A, and 2D. Additional GTT clusters were revealed on chromosome pairs 2Cm and 4Cm (in the hemizygous state), which were not observed in the Cm genome of A. macrostachya (Figure 1B and Figure 3A; Supplementary Figures S3 and S6).
- In the karyotype of PR 4H8 32-08, only constant minor 35S rDNA loci were detected (on chromosome pairs 3A and 3C). On chromosome pairs 1A, 2A, 3A, and 7D, no GTT clusters were revealed (Figure 3B; Supplementary Figures S3 and S6).
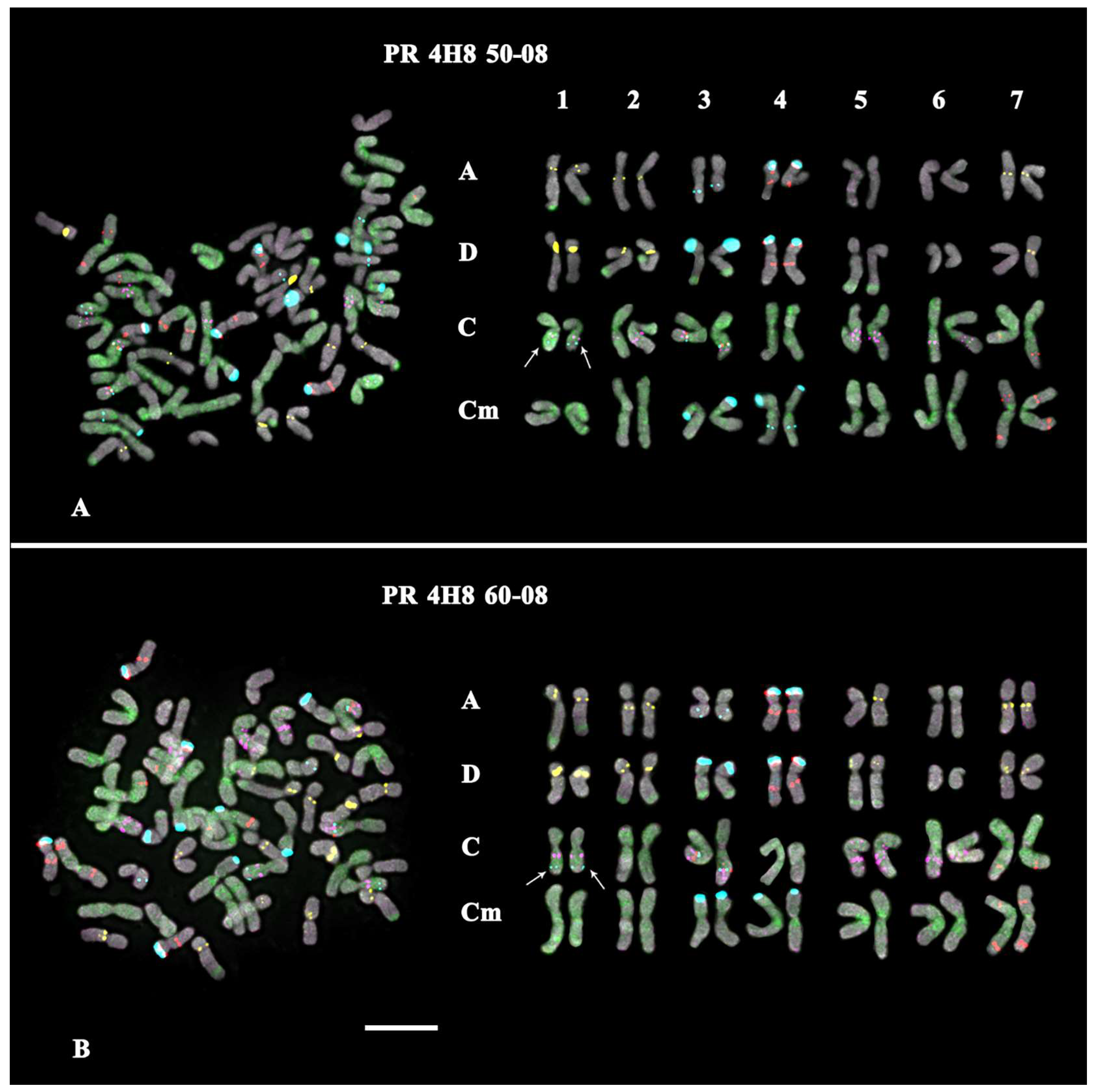
- In the karyotype of PR 4H8 50-08, one polymorphic minor 35S rDNA locus was revealed on chromosome pair 1C. On chromosome pairs 3A, 5A, and 5D, no GTT signals were revealed; and in pair 7D, GTT clusters were observed in the hemizygous state (Figure 4A; Supplementary Figures S4 and S6).
- In the karyotype of PR 4H8 60-08, one polymorphic minor 35S rDNA locus was revealed on chromosome pair 1C. GTT signals were not revealed on chromosome pair 3A; and in pair 5A, GTT clusters were observed in the hemizygous state (Figure 4B; Supplementary Figures S4 and S6).
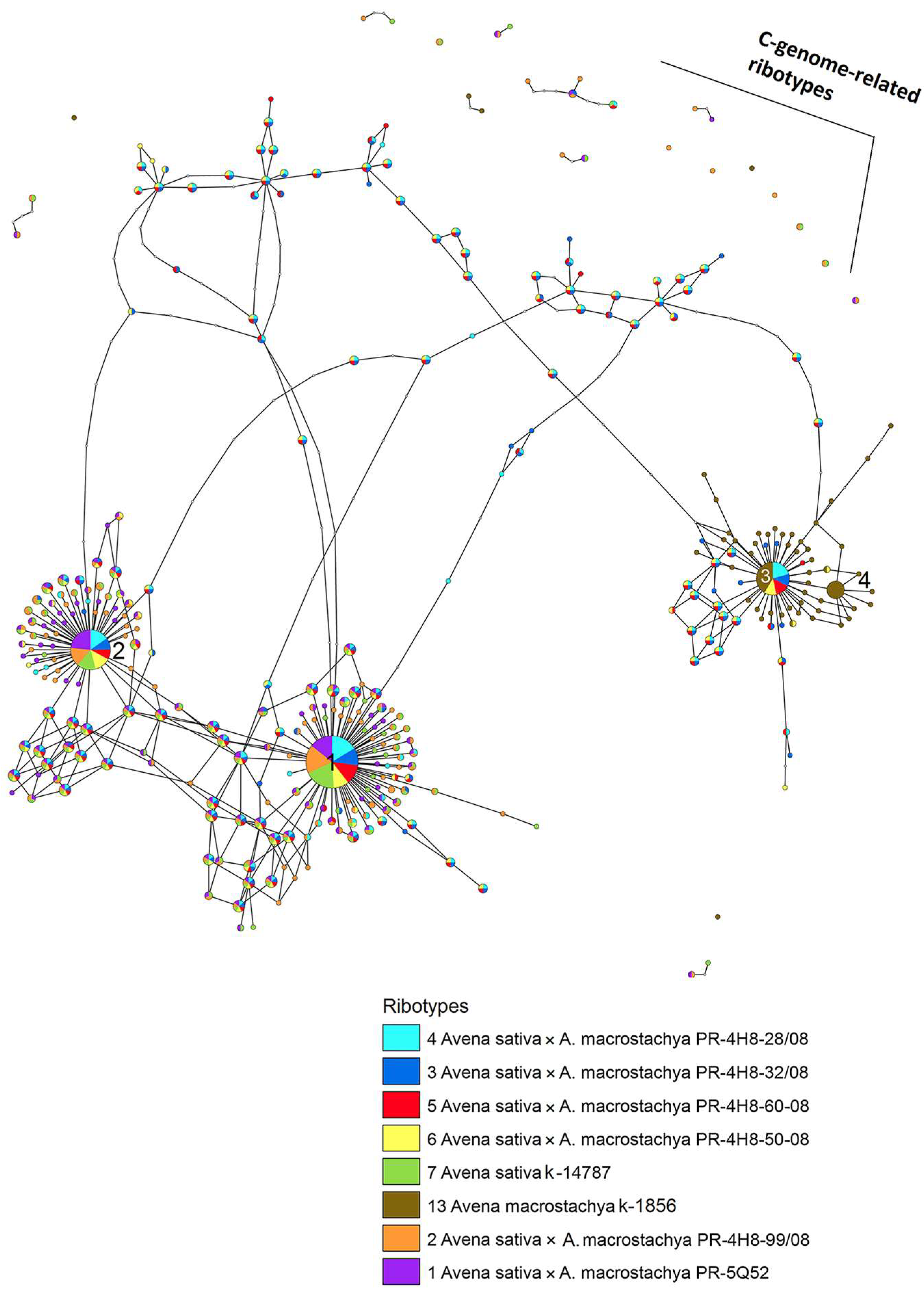
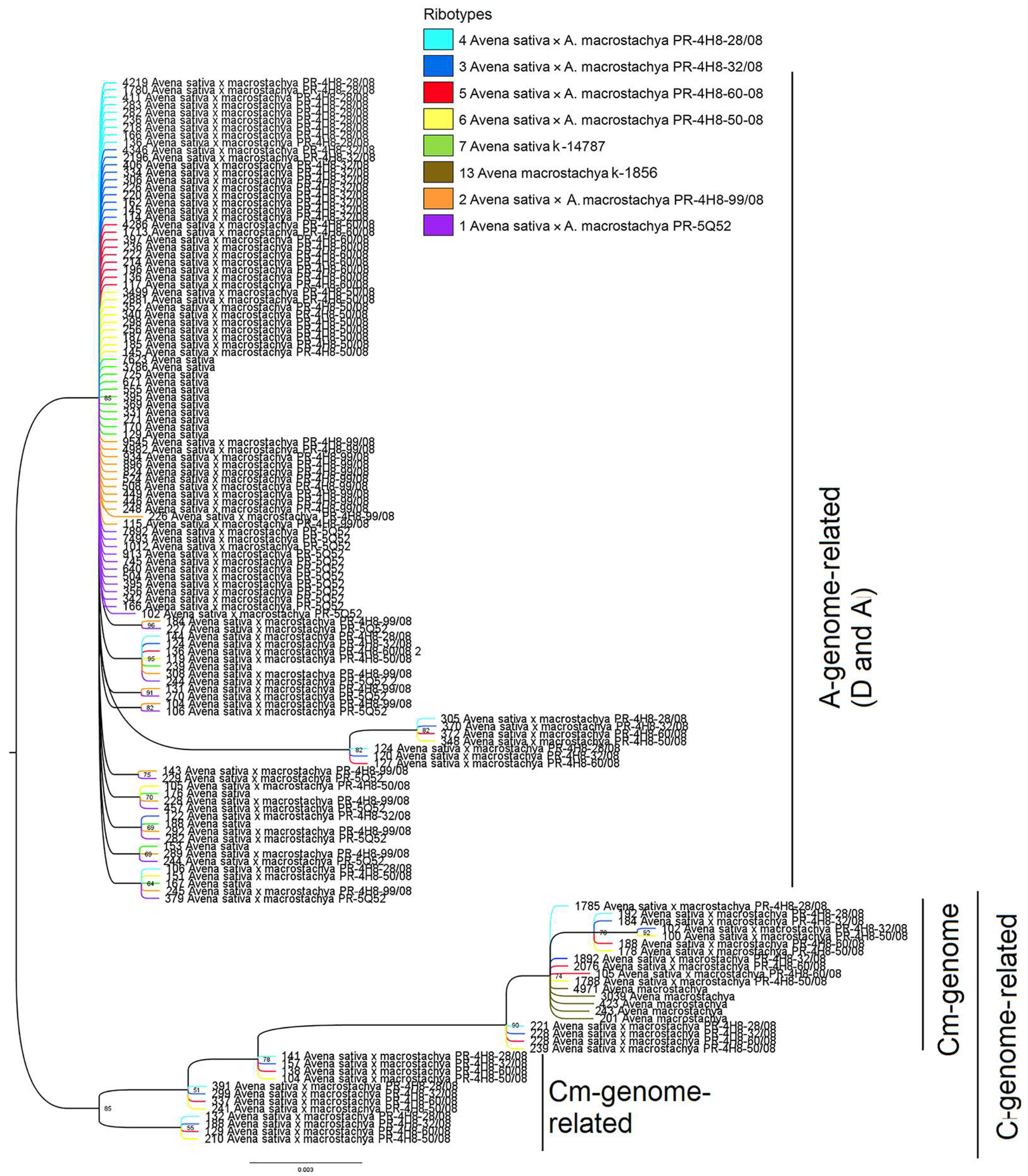
2.2. Molecular Phylogenetic Analysis
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Chromosome Spread Preparation
4.3. Multicolor Fluorescence In Situ Hybridization
4.4. Chromosome Analysis
4.5. Molecular Phylogenetic Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Strychar, R. World Oat Production, Trade, and Usage. In Oats: Chemistry and Technology; Webster, F., Wood, P., Eds.; AACC International Inc.: St. Paul, MN, USA, 2011; pp. 1–10. [Google Scholar]
- Zhang, J.; Li, X.; Wang, J.; Yang, L.; Yang, Q.; Xiang, D.; Wan, Y.; Nevo, E.; Yan, J.; Fan, Y.; et al. Wild oats offer new possibilities for forage because of the higher nutrition content and feed value. Agronomy 2023, 13, 2575. [Google Scholar] [CrossRef]
- Yan, H.; Martin, S.L.; Bekele, W.A.; Latta, R.G.; Diederichsen, A.; Peng, Y.; Tinker, N.A. Genome size variation in the genus Avena. Genome 2016, 59, 209–220. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Yan, H.; Guo, L.; Deng, C.; Wang, C.; Wang, Y.; Kang, L.; Zhou, P.; Yu, K.; Dong, X.; et al. Reference genome assemblies reveal the origin and evolution of allohexaploid oat. Nat. Genet. 2022, 54, 1248–1258. [Google Scholar] [CrossRef] [PubMed]
- Murat, F.; Xu, J.H.; Tannier, E.; Abrouk, M.; Guilhot, N.; Pont, C.; Messing, J.; Salse, J. Ancestral grass karyotype reconstruction unravels new mechanisms of genome shuffling as a source of plant evolution. Genome Res. 2010, 20, 1545–1557. [Google Scholar] [CrossRef] [PubMed]
- Panchy, N.L.; Azodi, C.B.; Winship, E.F.; O’Malley, R.C.; Shiu, S.H. Expression and regulatory asymmetry of retained Arabidopsis thaliana transcription factor genes derived from whole genome duplication. BMC Evol. Biol. 2019, 19, 2294–2316. [Google Scholar] [CrossRef] [PubMed]
- Salse, J. Deciphering the evolutionary interplay between subgenomes following polyploidy: A paleogenomics approach in grasses. Am. J. Bot. 2016, 103, 1167–1174. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Ye, L.; Li, M.; Wang, Z.; Xiong, G.; Ye, Y.; Tu, T.; Schwarzacher, T.; Heslop-Harrison, J.S. Genome-wide expansion and reorganization during grass evolution: From 30 Mb chromosomes in rice and Brachypodium to 550 Mb in Avena. BMC Plant Biol. 2023, 23, 627. [Google Scholar] [CrossRef] [PubMed]
- Thomas, H. Cytogenetics of Avena. In Oat Science and Technology; Marshall, H.G., Sorrells, M.E., Eds.; American Society of Agronomy and Crop Science Society of America: Madison, WI, USA, 1992; pp. 473–507. [Google Scholar]
- Katsiotis, A.; Loukas, M.; Heslop-Harrison, J.S. Repetitive DNA, genome and species relationships in Avena and Arrhenatherum (Poaceae). Ann. Bot. 2000, 86, 1135–1142. [Google Scholar] [CrossRef]
- Loskutov, I.G. On evolutionary pathways of Avena species. Genet. Resour. Crop. Evol. 2008, 55, 211–220. [Google Scholar] [CrossRef]
- Liu, Q.; Lin, L.; Zhou, X.; Peterson, P.M.; Wen, J. Unraveling the evolutionary dynamics of ancient and recent polyploidization events in Avena (Poaceae). Sci. Rep. 2017, 7, 41944. [Google Scholar] [CrossRef]
- Fu, Y.-B.; Williams, D.J. AFLP variation in 25 Avena species. Theor. Appl. Genet. 2008, 117, 333–342. [Google Scholar] [CrossRef] [PubMed]
- Loskutov, I.; Rines, H.W. Avena. In Wild Crop Relatives: Genomic and Breeding Resources, Cereals; Kole, C., Ed.; Springer: New York, NY, USA, 2011; pp. 109–184. [Google Scholar]
- Li, W.T.; Peng, Y.Y.; Wei, Y.M.; Baum, B.R.; Zheng, Y.L. Relationship among Avena species as revealed by consensus chloroplast simple sequence repeat (ccSSR) markers. Genet. Resour. Crop. Evol. 2009, 56, 465–480. [Google Scholar] [CrossRef]
- Tomas, D.; Rodrigues, J.; Varela, A.; Veloso, M.M.; Viegas, W.; Silva, M. Use of repetitive sequences for molecular and cytogenetic characterization of Avena species from Portugal. Int. J. Mol. Sci. 2016, 17, 203. [Google Scholar] [CrossRef] [PubMed]
- Rodionov, A.V.; Tiupa, N.B.; Kim, E.S.; Machs, E.M.; Loskutov, I.G. Genomic structure of the autotetraploid oat species Avena macrostachya inferred from comparative analysis of the ITS1 and ITS2 sequences: On the oat karyotype evolution during the early stages of the Avena species divergence. Genetika 2005, 41, 646–656. [Google Scholar] [CrossRef] [PubMed]
- Nikoloudakis, N.; Skaracis, G.A.; Katsiotis, A. Evolutionary insights inferred by molecular analysis of the ITS1-5.8S-ITS2 and IGS Avena sp. sequences. Mol. Phyl. Evol. 2008, 46, 102–115. [Google Scholar] [CrossRef]
- Peng, Y.Y.; Wei, Y.M.; Baum, B.R.; Yan, Z.H.; Lan, X.J.; Dai, S.F.; Zheng, Y.L. Phylogenetic inferences in Avena based on analysis of FL intron2 sequences. Theor. Appl. Genet. 2010, 121, 985–1000. [Google Scholar] [CrossRef]
- Rodrigues, J.; Viegas, W.; Silva, M. 45S rDNA external transcribed spacer organization reveals new phylogenetic relationships in Avena genus. PLoS ONE 2017, 12, e0176170. [Google Scholar] [CrossRef]
- Gnutikov, A.A.; Nosov, N.N.; Loskutov, I.G.; Machs, E.M.; Blinova, E.V.; Probatova, N.S.; Langdon, T.; Rodionov, A.V. New insights into the genomic structure of the oats (Avena L., Poaceae): Intragenomic polymorphism of ITS1 sequences of rare endemic species Avena bruhnsiana Gruner and its relationship to other species with C-genomes. Euphytica 2022, 218, 3. [Google Scholar] [CrossRef]
- Rajhathy, T.; Thomas, H. Cytogenetics of Oats; Miscellaneous Publications of the Genetics Society of Canada: Ottawa, ON, Canada, 1974; pp. 1–90. [Google Scholar]
- Baum, B.R. Oats: Wild and Cultivated. A Monograph of the Genus Avena L. (Poaceae); Agriculture Canada: Ottawa, ON, Canada, 1977. [Google Scholar]
- Fu, Y.B. Oat evolution revealed in the maternal lineages of 25 Avena species. Sci. Rep. 2018, 8, 4252. [Google Scholar] [CrossRef]
- Irigoyen, M.; Loarce, Y.; Linares, C.; Ferrer, E.; Leggett, M.; Fominaya, A. Discrimination of the closely related A and B genomes in AABB tetraploid species of Avena. Theor. Appl. Genet. 2001, 103, 1160–1166. [Google Scholar] [CrossRef]
- Katsiotis, A.; Hagidimitriou, M.; Heslop-Harrison, J.S. The close relationship between the A and B genomes in Avena L. (Poaceae) determined by molecular cytogenetic analysis of total genomic, tandemly and dispersed repetitive DNA sequences. Ann. Bot. 1997, 79, 103–109. [Google Scholar] [CrossRef]
- Linares, C.; Ferrer, E.; Fominaya, A. Discrimination of the closely related A and D genomes of the hexaploid oat Avena sativa L. Proc. Natl. Acad. Sci. USA 1998, 95, 12450–12455. [Google Scholar] [CrossRef] [PubMed]
- Ananiev, E.V.; Vales, M.I.; Phillips, R.L.; Rines, H.W. Isolation of A/D and C genome specific dispersed and clustered repetitive DNA sequences from Avena sativa. Genome 2002, 45, 431–441. [Google Scholar] [CrossRef] [PubMed]
- Kamal, N.; Tsardakas Renhuldt, N.; Bentzer, J.; Gundlach, H.; Haberer, G.; Juhász, A.; Lux, T.; Bose, U.; Tye-Din, J.A.; Lang, D.; et al. The mosaic oat genome gives insights into a uniquely healthy cereal crop. Nature 2022, 606, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Mark, B.; Andon, J.W. State of the art reviews: The oatmeal-cholesterol connection: 10 years later. Am. J. Lifestyle Med. 2008, 2, 51–57. [Google Scholar]
- Agostoni, C.; Bresson, J.; Fairweather-Tait, S.; Flynn, A.; Golly, I.; Korhonen, H.; Lagiou, P.; Løvik, M.; Marchelli, R.; Martin, A.; et al. Scientific opinion on the substantiation of a health claim related to oat beta glucan and lowering blood cholesterol and reduced risk of (coronary) heart disease pursuant to Article 14 of Regulation (EC) No 1924/2006. EFSA J. 2010, 8, 1885. [Google Scholar] [CrossRef]
- Baum, B.R.; Rajhathy, T. A study of Avena macrostachya. Can. J. Bot. 1976, 54, 2434–2439. [Google Scholar] [CrossRef]
- Bolc, P.; Łapiński, B.; Podyma, W.; Boczkowska, M. Genetic diversity and population structure of Algerian endemic plant species Avena macrostachya Bal. ex Cross. et Durieu. Agronomy 2020, 10, 1984. [Google Scholar] [CrossRef]
- Weibull, J. Resistance in the wild crop relatives Avena macrostachya and Hordeum bogdani to the aphid Rhopalosiphum padi. Entomol. Exp. Appl. 1988, 48, 225–232. [Google Scholar] [CrossRef]
- Yu, J.; Herrmann, M. Inheritance and mapping of a powdery mildew resistance gene introgressed from Avena macrostachya in cultivated oat. Theor. Appl. Genet. 2006, 113, 429–437. [Google Scholar] [CrossRef]
- Łapinski, B.; Kała, M.; Nakielna, Z.; Jellen, R.; Livingston, D. The perennial wild species Avena macrostachya as a genetic source for improvement of winterhardiness in winter oat for cultivation in Poland. In Biotechnology and Plant Breeding—Perspectives; Behl, R.E.A., Ed.; Agrobios (International) Publishers: Jodphur, India, 2013; pp. 51–62. [Google Scholar]
- Hoppe, H.-D.; Pohler, W. Successful hybridization between Avena prostrata and Avena macrostachya. Cereal Res. Comm. 1988, 16, 231–235. [Google Scholar]
- Jia, H.; Livingston, D.P.; Murphy, J.P.; Porter, D.R. Evaluation of freezing tolerance in advanced progeny from a cross of Avena sativa × Avena macrostachya. Cereal Res. Commun. 2006, 34, 2–6. [Google Scholar] [CrossRef]
- Lapinski, B.; Rachwalska, A. Using Avena macrostachya for improvement of oat winterhardiness in Poland. Proc. Appl. Bot. Genet. Breed. 2017, 178, 58–67. [Google Scholar] [CrossRef]
- Pohler, W.; Hoppe, H.-D. Homeology between the chromosomes of Avena macrostachya and the Avena C genome. Plant Breed. 1991, 106, 250–253. [Google Scholar] [CrossRef]
- Leggett, J.M. Further hybrids involving the perennial autotetraploid oat Avena macrostachya. Genome 1992, 35, 273–275. [Google Scholar] [CrossRef]
- Cheng, D.W.; Armstrong, K.C.; Drouin, G.; McElroy, A.; Fedak, G.; Molnar, S.D. Isolation and identification of Triticeae chromosome 1 receptor-like kinase genes (Lrk10) from diploid, tetraploid, and hexaploid species of the genus Avena. Genome 2003, 46, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.-Y.; Wei, Y.-M.; Baum, B.R.; Zheng, Y.-L. Molecular diversity of the 5S rRNA gene and genomic relationships in the genus Avena (Poaceae: Aveneae). Genome 2008, 51, 137–154. [Google Scholar] [CrossRef] [PubMed]
- Leggett, J.M.; Markhand, G.S. The Genomic Configuration of Avena Revealed by GISH, Kew Chromosome Conf. IV; Brandham, P.E., Bennett, M.D., Eds.; Royal Botanic Gardens: London, UK, 1995; pp. 133–139. [Google Scholar]
- Postoyko, J.; Hutchinson, J. The identification of Avena chromosomes by means of C-banding. In Proceedings of the 2nd International Oat Conference, The University College of Wales, Welsh Plant Breeding Station, Aberystwyth, UK, 15–18 July 1985; Lawes, D.A., Thomas, H., Eds.; Martinus Nijhoff: Dordrecht, Switzerland, 1986; pp. 50–51. [Google Scholar]
- Fominaya, A.; Vega, C.; Ferrer, E. Giemsa C-banded karyotypes of Avena species. Genome 1988, 30, 627–632. [Google Scholar] [CrossRef]
- Chen, Q.; Armstrong, K. Genomic in situ hybridization in Avena sativa. Genome 1994, 37, 607–612. [Google Scholar] [CrossRef]
- Badaeva, E.D.; Shelukhina, O.Y.; Diederichsen, A.; Loskutov, I.G.; Pukhalskiy, V.A. Comparative cytogenetic analysis of Avena macrostachya and diploid C-genome Avena species. Genome 2010, 53, 125–137. [Google Scholar] [CrossRef]
- Fominaya, A.; Loarce, Y.; Montes, A.; Ferrer, E. Chromosomal distribution patterns of the (AC)10 microsatellite and other repetitive sequences, and their use in chromosome rearrangement analysis of species of the genus Avena. Genome 2017, 60, 216–227. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.M.; Tinker, N.A.; Zhou, Y.H.; Liu, J.C.; Wan, W.L.; Chen, L. Chromosomal distributions of oligo-Am1 and (TTG)6 trinucleotide and their utilization in genome association analysis of sixteen Avena species. Genet. Resour. Crop. Evol. 2018, 65, 1625–1635. [Google Scholar] [CrossRef] [PubMed]
- Maughan, P.J.; Lee, R.; Walstead, R.; Vickerstaff, R.J.; Fogarty, M.C.; Brouwer, C.R.; Reid, R.R.; Jay, J.J.; Bekele, W.A.; Jackson, E.W.; et al. Genomic insights from the first chromosome-scale assemblies of oat (Avena spp.) diploid species. BMC Biol. 2019, 17, 92. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Li, X.; Zhou, X.; Li, M.; Zhang, F.; Schwarzacher, T.; Heslop-Harrison, J.S. The repetitive DNA landscape in Avena (Poaceae): Chromosome and genome evolution defined by major repeat classes in whole-genome sequence reads. BMC Plant Biol. 2019, 19, 226. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Jiang, C.; Yuan, W.; Zhang, M.; Fang, Z.; Li, Y.; Li, G.; Jia, J.; Yang, Z. A universal karyotypic system for hexaploid and diploid Avena species brings oat cytogenetics into the genomics era. BMC Plant Biol. 2021, 21, 213. [Google Scholar] [CrossRef] [PubMed]
- Sanz, M.J.; Jellen, E.N.; Loarce, Y.; Irigoyen, M.L.; Ferrer, E.; Fominaya, A. A new chromosome nomenclature system for oat (Avena sativa L. and A. byzantine C. Koch) based on FISH analysis of monosomic lines. Theor Appl Genet 2010, 121, 1541–1552. [Google Scholar] [CrossRef] [PubMed]
- Chaffin, A.S.; Huang, Y.F.; Smith, S.; Bekele, W.A.; Babiker, E.; Gnanesh, B.N.; Foresman, B.J.; Blanchard, S.G.; Jay, J.J.; Reid, R.W.; et al. A consensus map in cultivated hexaploid oat reveals conserved grass synteny with substantial subgenome rearrangement. Plant Genome 2016, 9, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Mason, A.S.; Batley, J. Creating new interspecific hybrid and polyploid crops. Trends Biotechnol 2015, 33, 436–441. [Google Scholar] [CrossRef] [PubMed]
- Comai, L.; Madlung, A.; Josefsson, C.; Tyagi, A. Do the different parental ‘heteromes’ cause genomic shock in newly formed allopolyploids? Philos. Trans. R. Soc. B Biol. Sci. 2003, 358, 1149–1155. [Google Scholar] [CrossRef]
- Tomás, C.; Vicient, C.M. The genomic shock hypothesis: Genetic and epigenetic alterations of transposable elements after interspecific hybridization in plants. Epigenomes 2024, 8, 2. [Google Scholar] [CrossRef]
- Warschefsky, E.; Penmetsa, R.V.; Cook, D.R.; von Wettberg, E.J.B. Back to the wilds: Tapping evolutionary adaptations for resilient crops through systematic hybridization with crop wild relatives. Am. J. Bot. 2014, 101, 1791–1800. [Google Scholar] [CrossRef] [PubMed]
- Rajendrakumar, P.; Hariprasanna, K.; Seetharama, N. Prediction of heterosis in crop plants-status and prospects. Am. J. Exp. Agric. 2015, 9, 1–16. [Google Scholar] [CrossRef]
- Flavell, R.B.; O’Dell, M.; Hutchinson, J. Nucleotide sequence organization in plant chromosomes and evidence for sequence translocation during evolution. Cold Spring Harb. Symp. Quant. Biol. 1981, 45, 501–508. [Google Scholar] [CrossRef] [PubMed]
- Biscotti, M.A.; Olmo, E.; Heslop-Harrison, J.S. Repetitive DNA in eukaryotic genomes. Chromosom Res. 2015, 23, 415–420. [Google Scholar] [CrossRef] [PubMed]
- Meštrovi´c, N.; Mravinac, B.; Pavlek, M.; Vojvoda-Zeljko, T.; Šatovi´c, E.; Plohl, M. Structural and functional liaisons between transposable elements and satellite DNAs. Chromosome Res. 2015, 23, 583–596. [Google Scholar] [CrossRef] [PubMed]
- Badaeva, E.D.; Shelukhina, O.Y.; Dedkova, O.S.; Loskutov, I.G.; Pukhalsky, V.A. Comparative cytogenetic analysis of hexaploid Avena L. species. Russ. J. Genet. 2011, 47, 691–702. [Google Scholar] [CrossRef]
- Srivastava, A.K.; Schlessinger, D. Structure and organization of ribosomal DNA. Biochimie 1991, 73, 631–638. [Google Scholar] [CrossRef] [PubMed]
- Garcia, S.; Kovařík, A.; Leitch, A.R.; Garnatje, T. Cytogenetic features of rRNA genes across land plants: Analysis of the plant rDNA database. Plant J. 2017, 89, 1020–1030. [Google Scholar] [CrossRef] [PubMed]
- Rogers, S.O.; Bendich, A.J. Ribosomal RNA genes in plants: Variability in copy number and in intergenic spacer. Plant Mol. Biol. 1987, 9, 509–520. [Google Scholar] [CrossRef]
- Linares, C.; González, J.; Ferrer, E.; Fominaya, A. The use of double fluorescence in situ hybridization to physically map the positions of 5S rDNA genes in relation to the chromosomal location of 18S-5.8S-26S rDNA and a C genome specific DNA sequence in the genus Avena. Genome 1996, 39, 535–542. [Google Scholar] [CrossRef]
- Rodionov, A.V.; Amosova, A.V.; Krainova, L.M.; Machs, E.M.; Mikhailova, Y.V.; Gnutikov, A.A.; Muravenko, O.V.; Loskutov, I.G. Phenomenon of multiple mutations in the 35S rRNA genes of the C subgenome of polyploid Avena L. Rus. J. Gen. 2020, 56, 674–683. [Google Scholar] [CrossRef]
- Winterfeld, G.; Döring, E.; Röser, M. Chromosome evolution in wild oat grasses (Aveneae) revealed by molecular phylogeny. Genome 2009, 52, 361–380. [Google Scholar] [CrossRef] [PubMed]
- Tucker, S.; Vitins, A.; Pikaard, C.S. Nucleolar dominance and ribosomal RNA gene silencing. Curr. Opin. Cell Biol. 2010, 22, 351–356. [Google Scholar] [CrossRef] [PubMed]
- Craig, S.; Pikaard, C.G.; Chandrasekhara, C.; McKinlay, A.; Enganti, R.; Fultz, D. Reaching for the off switch in nucleolar dominance. Plant J. 2023, 115, 1185–1192. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Chen, Y.; Wang, L.; Yuan, Y.; Fang, M.; Shi, L.; Lu, R.; Comes, H.P.; Ma, Y.; Chen, Y.; et al. Pangenome of water caltrop reveals structural variations and asymmetric subgenome divergence after allopolyploidization. Hortic. Res. 2023, 10, uhad203. [Google Scholar] [CrossRef] [PubMed]
- Jellen, E.N.; Phillips, R.L.; Rines, H.W. C-banded karyotypes and polymorphisms in hexaploid oat accessions (Avena spp.) using Wright’s stain. Genome 1993, 36, 1129–1137. [Google Scholar] [CrossRef] [PubMed]
- Jellen, E.N.; Gill, B.S.; Cox, T.S. Genomic in situ hybridization differentiates between A/D- and C-genome chromatin and detects intergenomic translocations in polyploid oat species (genus Avena). Genome 1994, 37, 613–618. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Hanson, L.; Bennett, M.D.; Leitch, I.J. Genome structure and evolution in the allohexaploid weed Avena fatua L. (Poaceae). Genome 1999, 42, 512–518. [Google Scholar] [CrossRef] [PubMed]
- Hayasaki, M.; Morikawa, T.; Tarumoto, I. Intergenomic translocations of polyploid oats (genus Avena) revealed by genomic in situ hybridization. Genes Genet. Syst. 2000, 75, 167–171. [Google Scholar] [CrossRef]
- Belyakov, E.A.; Machs, E.M.; Mikhailova, Y.V.; Rodionov, A.V. The study of hybridization processes within genus Sparganium L. subgenus Xanthosparganium Holmb. Based on data of next generation sequencing (NGS). Ecol. Genet. 2019, 17, 27–35. [Google Scholar] [CrossRef]
- Zhang, M.; Tang, Y.W.; Xu, Y.; Yonezawa, T.; Shao, Y.; Wang, Y.G.; Song, Z.-P.; Yang, J.; Zhang, W.J. Concerted and birth-and-death evolution of 26S ribosomal DNA in Camellia L. Ann. Bot. 2021, 127, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Gnutikov, A.A.; Nosov, N.N.; Loskutov, I.G.; Blinova, E.V.; Shneyer, V.S.; Rodionov, A.V. Origin of wild polyploid Avena species inferred from polymorphism of the ITS1 rDNA in their genomes. Diversity 2023, 15, 717. [Google Scholar] [CrossRef]
- Wang, X.-C.; Liu, C.; Huang, L.; Bengtsson-Palme, J.; Chen, H.; Zhang, J.-H.; Cai, D.; Li, J.-Q. ITS1: A DNA barcode better than ITS2 in eukaryotes? Mol. Ecol. Res. 2015, 15, 573–586. [Google Scholar] [CrossRef] [PubMed]
- Schultz, J.; Maisel, S.; Gerlach, D.; Müller, T.; Wolf, M. A common core of secondary structure of 33 the internal transcribed spacer 2 (ITS2) throughout the Eukaryota. RNA 2005, 11, 361–364. [Google Scholar] [CrossRef] [PubMed]
- Coleman, A.W. Nuclear rRNA transcript processing versus internal transcribed spacer 29 secondary structure. Trends Genet. 2015, 31, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Cao, Y.; Zhang, W.; Simmons, M.P. Adenine· cytosine substitutions are an alternative 13 pathway of compensatory mutation in angiosperm ITS2. RNA 2020, 26, 209–217. [Google Scholar] [CrossRef] [PubMed]
- Brassac, J.; Blattne, r.F.R. Species-level phylogeny and polyploid relationships in Hordeum (Poaceae) inferred by next-generation sequencing and in silico cloning of multiple nuclear loci. Syst. Biol. 2015, 64, 792–808. [Google Scholar] [CrossRef]
- Rodionov, A.V.; Gnutikov, A.A.; Nosov, N.N.; Machs, E.M.; Mikhaylova, Y.V.; Shneyer, V.S.; Punina, E.O. Intragenomic polymorphism of the ITS 1 region of 35S rRNA gene in the group of grasses with two-chromosome species: Different genome composition in closely related Zingeria species. Plants 2020, 9, 1647. [Google Scholar] [CrossRef] [PubMed]
- Amosova, A.V.; Yurkevich, O.Y.; Rodionov, A.V.; Bolsheva, N.L.; Samatadze, T.E.; Zoshchuk, S.A.; Muravenko, O.V. Repeatome Analyses and Satellite DNA Chromosome Patterns in Deschampsia sukatschewii, D. cespitosa, and D. antarctica (Poaceae). Genes 2022, 13, 762. [Google Scholar] [CrossRef]
- Gerlach, W.L.; Bedbrook, J.R. Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucleic Acids Res. 1979, 7, 1869–1885. [Google Scholar] [CrossRef]
- Gerlach, W.L.; Dyer, T.A. Sequence organization of the repeating units in the nucleus of wheat which contain 5S rRNA genes. Nucleic Acids Res. 1980, 8, 4851–4855. [Google Scholar] [CrossRef]
- Ridgway, K.P.; Duck, J.M.; Young, J.P.W. Identification of roots from grass swards using PCR-RFLP and FFLP of the plastid trnL (UAA) intron. BMC Ecol. 2003, 3, 8. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Golosova, O.; Fursov, M.; the UGENE team. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef] [PubMed]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahe, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef] [PubMed]
- Clement, M.; Posada, D.; Crandall, K.A. TCS: A computer program to estimate gene genealogies. Mol. Ecol. 2000, 9, 1657–1660. [Google Scholar] [CrossRef] [PubMed]
- Múrias dos Santos, A.; Cabezas, M.P.; Tavares, A.I.; Xavier, R.; Branco, M. tcsBU: A tool to extend TCS network layout and visualization. Bioinformatics 2016, 32, 627–628. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amosova, A.V.; Gnutikov, A.A.; Rodionov, A.V.; Loskutov, I.G.; Nosov, N.N.; Yurkevich, O.Y.; Samatadze, T.E.; Zoshchuk, S.A.; Muravenko, O.V. Genome Variability in Artificial Allopolyploid Hybrids of Avena sativa L. and Avena macrostachya Balansa ex Coss. et Durieu Based on Marker Sequences of Satellite DNA and the ITS1–5.8S rDNA Region. Int. J. Mol. Sci. 2024, 25, 5534. https://doi.org/10.3390/ijms25105534
Amosova AV, Gnutikov AA, Rodionov AV, Loskutov IG, Nosov NN, Yurkevich OY, Samatadze TE, Zoshchuk SA, Muravenko OV. Genome Variability in Artificial Allopolyploid Hybrids of Avena sativa L. and Avena macrostachya Balansa ex Coss. et Durieu Based on Marker Sequences of Satellite DNA and the ITS1–5.8S rDNA Region. International Journal of Molecular Sciences. 2024; 25(10):5534. https://doi.org/10.3390/ijms25105534
Chicago/Turabian StyleAmosova, Alexandra V., Alexander A. Gnutikov, Alexander V. Rodionov, Igor G. Loskutov, Nikolai N. Nosov, Olga Yu. Yurkevich, Tatiana E. Samatadze, Svyatoslav A. Zoshchuk, and Olga V. Muravenko. 2024. "Genome Variability in Artificial Allopolyploid Hybrids of Avena sativa L. and Avena macrostachya Balansa ex Coss. et Durieu Based on Marker Sequences of Satellite DNA and the ITS1–5.8S rDNA Region" International Journal of Molecular Sciences 25, no. 10: 5534. https://doi.org/10.3390/ijms25105534




