Exploratory Untargeted Metabolomics of Dried Blood Spot Samples from Newborns with Maple Syrup Urine Disease
Abstract
1. Introduction
2. Results
2.1. Participant Demographics and DBS Sample Selection
2.2. Untargeted Metabolomics Profiling of MSUD Newborns
2.3. Biomarkers Analysis of MSUD
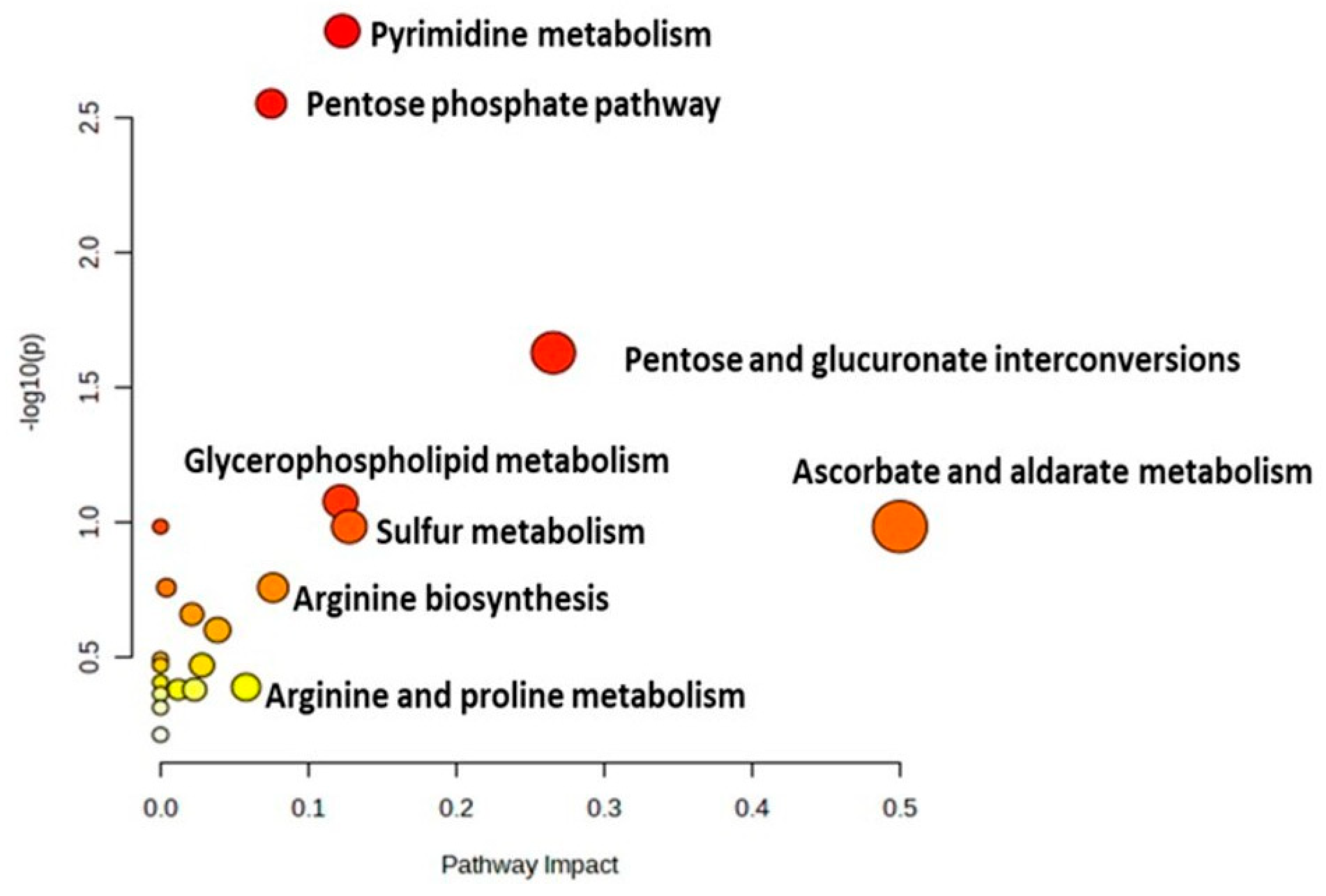
2.4. Metabolomic Pathway Analysis
3. Discussion
3.1. Untargeted Metabolomics as an Additional Diagnostic/Screening Tool for MSUD
3.2. Untargeted Metabolomics Revealed Altered Global Metabolic Profiling of MSUD Newborns
3.3. Alterations in the Amino Acids in MSUD Newborns
3.4. Alterations in the Lipid Species in MSUD Newborns
3.5. Metabolites Involved in Oxidative Events in MSUD Newborns
3.6. New Potential Metabolic Biomarkers/Pathways of MSUD
4. Materials and Methods
4.1. Ethical Approval
4.2. Participants’ Selection Criteria and Sample Collection
4.3. Chemicals
4.4. Sample Preparation
4.5. LC-HRMS Metabolomics Analysis
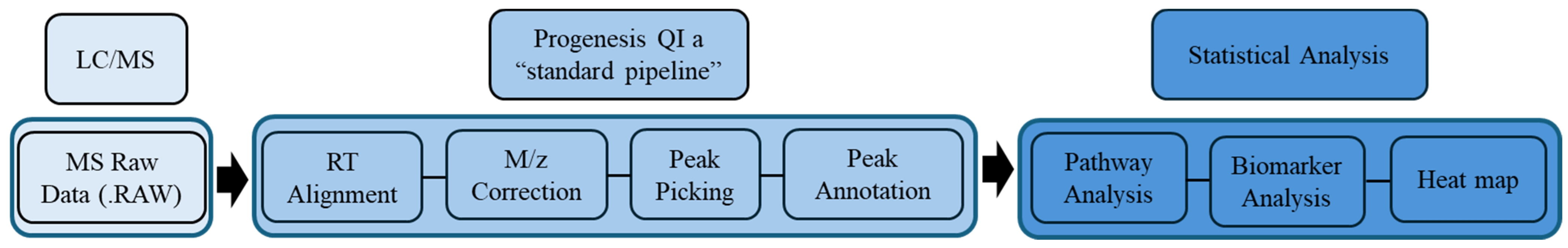
4.6. Metabolomics Data Processing and Statistical Analyses
4.7. Metabolite Identification (Peak Annotation)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bharadwaj, A.; Wahi, N.; Saxena, A. Occurrence of Inborn Errors of Metabolism in Newborns, Diagnosis and Prophylaxis. Endocr. Metab. Immune Disord. Drug Targets 2021, 21, 592–616. [Google Scholar] [CrossRef] [PubMed]
- Guerrero, R.B.; Salazar, D.; Tanpaiboon, P. Laboratory diagnostic approaches in metabolic disorders. Ann. Transl. Med. 2018, 6, 470. [Google Scholar] [CrossRef] [PubMed]
- IEMbase. Inborn Errors of Metabolism Knowledgebase. Available online: http://www.iembase.org/ (accessed on 2 December 2022).
- Pontoizeau, C.; Simon-Sola, M.; Gaborit, C.; Nguyen, V.; Rotaru, I.; Tual, N.; Colella, P.; Girard, M.; Biferi, M.G.; Arnoux, J.B.; et al. Neonatal gene therapy achieves sustained disease rescue of maple syrup urine disease in mice. Nat. Commun. 2022, 13, 3278. [Google Scholar] [CrossRef] [PubMed]
- Imtiaz, F.; Al-Mostafa, A.; Allam, R.; Ramzan, K.; Al-Tassan, N.; Tahir, A.I.; Al-Numair, N.S.; Al-Hamed, M.H.; Al-Hassnan, Z.; Al-Owain, M.; et al. Twenty novel mutations in BCKDHA, BCKDHB and DBT genes in a cohort of 52 Saudi Arabian patients with maple syrup urine disease. Mol. Genet. Metab. Rep. 2017, 11, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Puliyanda, D.P.; Harmon, W.E.; Peterschmitt, M.J.; Irons, M.; Somers, M.J.G. Utility of hemodialysis in maple syrup urine disease. Pediatr. Nephrol. 2002, 17, 239–242. [Google Scholar] [CrossRef]
- Amaral, A.U.; Leipnitz, G.; Fernandes, C.G.; Seminotti, B.; Schuck, P.F.; Wajner, M. α-Ketoisocaproic acid and leucine provoke mitochondrial bioenergetic dysfunction in rat brain. Brain Res. 2010, 1324, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Amaral, A.U.; Wajner, M. Pathophysiology of maple syrup urine disease: Focus on the neurotoxic role of the accumulated branched-chain amino acids and branched-chain α-keto acids. Neurochem. Int. 2022, 157, 105360. [Google Scholar] [CrossRef] [PubMed]
- Strauss, K.A.; Carson, V.J.; Soltys, K.; Young, M.E.; Bowser, L.E.; Puffenberger, E.G.; Brigatti, K.W.; Williams, K.B.; Robinson, D.L.; Hendrickson, C.; et al. Branched-chain α-ketoacid dehydrogenase deficiency (maple syrup urine disease): Treatment, biomarkers, and outcomes. Mol. Genet. Metab. 2020, 129, 193–206. [Google Scholar] [CrossRef]
- Schadewaldt, P.; Bodner-Leidecker, A.; Hammen, H.W.; Wendel, U. Significance of L-alloisoleucine in plasma for diagnosis of maple syrup urine disease. Clin. Chem. 1999, 45, 1734–1740. [Google Scholar] [CrossRef]
- Chen, T.; Lu, D.; Xu, F.; Ji, W.; Zhan, X.; Gao, X.; Qiu, W.; Zhang, H.; Liang, L.; Gu, X.; et al. Newborn screening of maple syrup urine disease and the effect of early diagnosis. Clin. Chim. Acta 2023, 548, 117483. [Google Scholar] [CrossRef]
- Ghosh, A.; Schlecht, H.; Heptinstall, L.E.; Bassett, J.K.; Cartwright, E.; Bhaskar, S.S.; Urquhart, J.; Broomfield, A.; Morris, A.A.; Jameson, E.; et al. Diagnosing childhood-onset inborn errors of metabolism by next-generation sequencing. Arch. Dis. Child. 2017, 102, 1019–1029. [Google Scholar] [CrossRef]
- Fingerhut, R. Recall rate and positive predictive value of MSUD screening is not influenced by hydroxyproline. Eur. J. Pediatr. 2009, 168, 599–604. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Zhu, X.; Feng, Y.; Bai, Y.; Zhao, X.; Liu, N.; Kong, X. Genetic analysis by targeted next-generation sequencing and novel variation identification of maple syrup urine disease in Chinese Han population. Sci. Rep. 2021, 11, 18939. [Google Scholar] [CrossRef] [PubMed]
- Bodian, D.L.; Klein, E.; Iyer, R.K.; Wong, W.S.; Kothiyal, P.; Stauffer, D.; Huddleston, K.C.; Gaither, A.D.; Remsburg, I.; Khromykh, A.J.G.i.M. Utility of whole-genome sequencing for detection of newborn screening disorders in a population cohort of 1696 neonates. Anesth. Analg. 2016, 18, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Tarini, B.A.; Goldenberg, A.J. Ethical Issues with Newborn Screening in the Genomics Era. Annu. Rev. Genom. Hum. Genet. 2012, 13, 381–393. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vázquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N.; et al. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Res. 2018, 46, D608–D617. [Google Scholar] [CrossRef] [PubMed]
- Thistlethwaite, L.R.; Li, X.; Burrage, L.C.; Riehle, K.; Hacia, J.G.; Braverman, N.; Wangler, M.F.; Miller, M.J.; Elsea, S.H.; Milosavljevic, A. Clinical diagnosis of metabolic disorders using untargeted metabolomic profiling and disease-specific networks learned from profiling data. Sci. Rep. 2022, 12, 6556. [Google Scholar] [CrossRef]
- Coene, K.L.M.; Kluijtmans, L.A.J.; van der Heeft, E.; Engelke, U.F.H.; de Boer, S.; Hoegen, B.; Kwast, H.J.T.; van de Vorst, M.; Huigen, M.; Keularts, I.; et al. Next-generation metabolic screening: Targeted and untargeted metabolomics for the diagnosis of inborn errors of metabolism in individual patients. J. Inherit. Metab. Dis. 2018, 41, 337–353. [Google Scholar] [CrossRef] [PubMed]
- Janeckova, H.; Kalivodova, A.; Najdekr, L.; Friedecký, D.; Hron, K.; Bruheim, P.; Adam, T. Untargeted metabolomic analysis of urine samples in the diagnosis of some inherited metabolic disorders. Biomed. Pap. 2015, 159, 582–585. [Google Scholar] [CrossRef]
- Haijes, H.A.; Willemsen, M.; Van der Ham, M.; Gerrits, J.; Pras-Raves, M.L.; Prinsen, H.C.M.T.; Van Hasselt, P.M.; De Sain-van der Velden, M.G.M.; Verhoeven-Duif, N.M.; Jans, J.J.M. Direct Infusion Based Metabolomics Identifies Metabolic Disease in Patients’ Dried Blood Spots and Plasma. Metabolites 2019, 9, 12. [Google Scholar] [CrossRef]
- Adam, M.P.; Feldman, J.; Mirzaa, G.M.; Pagon, R.A.; Wallace, S.E.; Bean, L.J.H.; Gripp, K.W.; Amemiya, A. GeneReviews(®); University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Staufner, C.; Haack, T.B.; Feyh, P.; Gramer, G.; Raga, D.E.; Terrile, C.; Sauer, S.; Okun, J.G.; Fang-Hoffmann, J.; Mayatepek, E.; et al. Genetic cause and prevalence of hydroxyprolinemia. J. Inherit. Metab. Dis. 2016, 39, 625–632. [Google Scholar] [CrossRef] [PubMed]
- Korman, S.H.; Cohen, E.; Preminger, A. Pseudo-maple syrup urine disease due to maternal prenatal ingestion of fenugreek. J. Paediatr. Child. Health 2001, 37, 403–404. [Google Scholar] [CrossRef] [PubMed]
- Burke, W.; Parens, E.; Chung, W.K.; Berger, S.M.; Appelbaum, P.S. The Challenge of Genetic Variants of Uncertain Clinical Significance: A Narrative Review. Ann. Intern. Med. 2022, 175, 994–1000. [Google Scholar] [CrossRef]
- Sajeev, M.; Chin, S.; Ho, G.; Bennetts, B.; Sankaran, B.P.; Gutierrez, B.; Devanapalli, B.; Tolun, A.A.; Wiley, V.; Fletcher, J.; et al. Challenges in Diagnosing Intermediate Maple Syrup Urine Disease by Newborn Screening and Functional Validation of Genomic Results Imperative for Reproductive Family Planning. Int. J. Neonatal Screen. 2021, 7, 25. [Google Scholar] [CrossRef]
- Abadingo, M.E.; Abacan, M.A.R.; Basas, J.R.U.; Padilla, C.D. Pregnancy in an adolescent with maple syrup urine disease: Case report. Mol. Genet. Metab. Rep. 2021, 27, 100745. [Google Scholar] [CrossRef]
- Constantinou, M.A.; Papakonstantinou, E.; Benaki, D.; Spraul, M.; Shulpis, K.; Koupparis, M.A.; Mikros, E. Application of nuclear magnetic resonance spectroscopy combined with principal component analysis in detecting inborn errors of metabolism using blood spots: A metabonomic approach. Anal. Chim. Acta 2004, 511, 303–312. [Google Scholar] [CrossRef]
- Douglas, T.D.; Newby, L.K.; Eckstrand, J.; Wixted, D.; Singh, R.H. Lipid changes in the metabolome of a single case study with maple syrup urine disease (MSUD) after five days of improved diet adherence of controlled branched-chain amino acids (BCAA). Mol. Genet. Metab. Rep. 2020, 25, 100651. [Google Scholar] [CrossRef]
- Miller, M.J.; Kennedy, A.D.; Eckhart, A.D.; Burrage, L.C.; Wulff, J.E.; Miller, L.A.; Milburn, M.V.; Ryals, J.A.; Beaudet, A.L.; Sun, Q.; et al. Untargeted metabolomic analysis for the clinical screening of inborn errors of metabolism. J. Inherit. Metab. Dis. 2015, 38, 1029–1039. [Google Scholar] [CrossRef] [PubMed]
- Minakata, S.; Manabe, S.; Inai, Y.; Ikezaki, M.; Nishitsuji, K.; Ito, Y.; Ihara, Y. Protein C-Mannosylation and C-Mannosyl Tryptophan in Chemical Biology and Medicine. Molecules 2021, 26, 5258. [Google Scholar] [CrossRef]
- Ihara, Y.; Inai, Y.; Ikezaki, M.; Matsui, I.-S.L.; Manabe, S.; Ito, Y. C-Mannosylation: Modification on Tryptophan in Cellular Proteins. In Glycoscience: Biology and Medicine; Taniguchi, N., Endo, T., Hart, G.W., Seeberger, P.H., Wong, C.-H., Eds.; Springer Japan: Tokyo, Japan, 2015; pp. 1091–1099. [Google Scholar]
- Morita, S.; Inai, Y.; Minakata, S.; Kishimoto, S.; Manabe, S.; Iwahashi, N.; Ino, K.; Ito, Y.; Akamizu, T.; Ihara, Y. Quantification of serum C-mannosyl tryptophan by novel assay to evaluate renal function and vascular complications in patients with type 2 diabetes. Sci. Rep. 2021, 11, 1946. [Google Scholar] [CrossRef]
- Schnekenburger, M.; Diederich, M. Chapter 18-Nutritional Epigenetic Regulators in the Field of Cancer: New Avenues for Chemopreventive Approaches. In Epigenetic Cancer Therapy; Gray, S.G., Ed.; Academic Press: Boston, MA, USA, 2015; pp. 393–425. [Google Scholar]
- Qin, D.; Lei, Y.; Xie, W.; Zheng, Q.; Peng, Z.; Liu, Y.; Dai, B.; Ma, T.; Wei, P.; Gao, C.; et al. Methionine sulfoxide suppresses adipogenic differentiation by regulating the mitogen-activated protein kinase signaling pathway. Cell Biol. Int. 2023, 47, 648–659. [Google Scholar] [CrossRef] [PubMed]
- Bennet, S.; Kaufmann, M.; Takami, K.; Sjaarda, C.; Douchant, K.; Moslinger, E.; Wong, H.; Reed, D.E.; Ellis, A.K.; Vanner, S.; et al. Small-molecule metabolome identifies potential therapeutic targets against COVID-19. Sci. Rep. 2022, 12, 10029. [Google Scholar] [CrossRef] [PubMed]
- Barschak, A.G.; Sitta, A.; Deon, M.; Busanello, E.N.; Coelho, D.M.; Cipriani, F.; Dutra-Filho, C.S.; Giugliani, R.; Wajner, M.; Vargas, C.R. Amino acids levels and lipid peroxidation in maple syrup urine disease patients. Clin. Biochem. 2009, 42, 462–466. [Google Scholar] [CrossRef] [PubMed]
- Levine, R.L.; Mosoni, L.; Berlett, B.S.; Stadtman, E.R. Methionine residues as endogenous antioxidants in proteins. Proc. Natl. Acad. Sci. USA 1996, 93, 15036–15040. [Google Scholar] [CrossRef]
- Jobgen, W.S.; Wu, G. L-Arginine increases AMPK phosphorylation and the oxidation of energy substrates in hepatocytes, skeletal muscle cells, and adipocytes. Amino Acids 2022, 54, 1553–1568. [Google Scholar] [CrossRef] [PubMed]
- Dereymaeker, A.; Jansen, K.; Vervisch, J.; Naulaers, G.; Regal, L. P98–3040: Early diagnosis of maple syrup urine disease (MSUD) by means of neonatal EEG and MRI in a newborn with encephalopathy. Eur. J. Paediatr. Neurol. 2015, 19, S121–S122. [Google Scholar] [CrossRef]
- Minamihata, T.; Takano, K.; Moriyama, M.; Nakamura, Y. Lysophosphatidylinositol, an Endogenous Ligand for G Protein-Coupled Receptor 55, Has Anti-inflammatory Effects in Cultured Microglia. Inflammation 2020, 43, 1971–1987. [Google Scholar] [CrossRef]
- Alaamery, M.; Albesher, N.; Aljawini, N.; Alsuwailm, M.; Massadeh, S.; Wheeler, M.A.; Chao, C.C.; Quintana, F.J. Role of sphingolipid metabolism in neurodegeneration. J. Neurochem. 2021, 158, 25–35. [Google Scholar] [CrossRef]
- Calzada, E.; Onguka, O.; Claypool, S.M. Phosphatidylethanolamine Metabolism in Health and Disease. Int. Rev. Cell Mol. Biol. 2016, 321, 29–88. [Google Scholar] [CrossRef]
- Hajeyah, A.A.; Griffiths, W.J.; Wang, Y.; Finch, A.J.; O’Donnell, V.B. The Biosynthesis of Enzymatically Oxidized Lipids. Front. Endocrinol. 2020, 11, 591819. [Google Scholar] [CrossRef]
- Varki, A.; Cummings, R.D.; Esko, J.D.; Stanley, P.; Hart, G.W.; Aebi, M.; Mohnen, D.; Kinoshita, T.; Packer, N.H.; Prestegard, J.H. Essentials of Glycobiology [Internet]; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2022. [Google Scholar]
- Fukuda, M.; Rutishauser, U.; Schnaar, R.L. Neuroglycobiology; Oxford University Press: New York, NY, USA, 2005. [Google Scholar]
- de Chaves, E.P.; Sipione, S. Sphingolipids and gangliosides of the nervous system in membrane function and dysfunction. FEBS Lett. 2010, 584, 1748–1759. [Google Scholar] [CrossRef] [PubMed]
- Svennerholm, L. Gangliosides—A new therapeutic agent against stroke and Alzheimer’s disease. Life Sci. 1994, 55, 2125–2134. [Google Scholar] [CrossRef]
- Dambrova, M.; Makrecka-Kuka, M.; Kuka, J.; Vilskersts, R.; Nordberg, D.; Attwood, M.M.; Smesny, S.; Sen, Z.D.; Guo, A.C.; Oler, E.; et al. Acylcarnitines: Nomenclature, Biomarkers, Therapeutic Potential, Drug Targets, and Clinical Trials. Pharmacol. Rev. 2022, 74, 506–551. [Google Scholar] [CrossRef]
- Sitta, A.; Ribas, G.S.; Mescka, C.P.; Barschak, A.G.; Wajner, M.; Vargas, C.R. Neurological Damage in MSUD: The Role of Oxidative Stress. Cell. Mol. Neurobiol. 2014, 34, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Fairweather-Tait, S.J.; Bao, Y.; Broadley, M.R.; Collings, R.; Ford, D.; Hesketh, J.E.; Hurst, R. Selenium in human health and disease. Antioxid. Redox Signal. 2011, 14, 1337–1383. [Google Scholar] [CrossRef] [PubMed]
- Goff, J.P. Invited review: Mineral absorption mechanisms, mineral interactions that affect acid–base and antioxidant status, and diet considerations to improve mineral status. J. Dairy Sci. 2018, 101, 2763–2813. [Google Scholar] [CrossRef]
- Sharifi-Rad, M.; Anil Kumar, N.V.; Zucca, P.; Varoni, E.M.; Dini, L.; Panzarini, E.; Rajkovic, J.; Tsouh Fokou, P.V.; Azzini, E.; Peluso, I.; et al. Lifestyle, Oxidative Stress, and Antioxidants: Back and Forth in the Pathophysiology of Chronic Diseases. Front. Physiol. 2020, 11, 694. [Google Scholar] [CrossRef]
- Chattoraj, A.; Liu, T.; Zhang, L.S.; Huang, Z.; Borjigin, J. Melatonin formation in mammals: In vivo perspectives. Rev. Endocr. Metab. Disord. 2009, 10, 237–243. [Google Scholar] [CrossRef]
- Smirnoff, N. Ascorbic acid metabolism and functions: A comparison of plants and mammals. Free Radic. Biol. Med. 2018, 122, 116–129. [Google Scholar] [CrossRef]
- Perreault, M.; Białek, A.; Trottier, J.; Verreault, M.; Caron, P.; Milkiewicz, P.; Barbier, O. Role of glucuronidation for hepatic detoxification and urinary elimination of toxic bile acids during biliary obstruction. PLoS ONE 2013, 8, e80994. [Google Scholar] [CrossRef]
- Sebaa, R.; AlMalki, R.H.; Alseraty, W.; Abdel Rahman, A.M. A Distinctive Metabolomics Profile and Potential Biomarkers for Very Long Acylcarnitine Dehydrogenase Deficiency (VLCADD) Diagnosis in Newborns. Metabolites 2023, 13, 725. [Google Scholar] [CrossRef] [PubMed]
- AlMalki, R.H.; Sebaa, R.; Al-Ansari, M.M.; Al-Alwan, M.; Alwehaibi, M.A.; Rahman, A.M.A. E. coli Secretome Metabolically Modulates MDA-MB-231 Breast Cancer Cells’ Energy Metabolism. Int. J. Mol. Sci. 2023, 24, 4219. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Chong, J.; Zhou, G.; de Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.-É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef] [PubMed]
- Worley, B.; Powers, R. Multivariate Analysis in Metabolomics. Curr. Metabolomics 2013, 1, 92–107. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Al Dubayee, M.; Alshahrani, A.; Masood, A.; Benabdelkamel, H.; Zahra, M.; Li, L.; Abdel Rahman, A.M.; Aljada, A. Distinctive Metabolomics Patterns Associated with Insulin Resistance and Type 2 Diabetes Mellitus. Front. Mol. Biosci. 2020, 7, 609806. [Google Scholar] [CrossRef]
- Wishart, D.S.; Guo, A.; Oler, E.; Wang, F.; Anjum, A.; Peters, H.; Dizon, R.; Sayeeda, Z.; Tian, S.; Lee, B.L.; et al. HMDB 5.0: The Human Metabolome Database for 2022. Nucleic Acids Res. 2021, 50, D622–D631. [Google Scholar] [CrossRef]




| Demographic and Clinical Features | MSUD (n = 22) | CTRL (n = 22) | p-Value | ||
|---|---|---|---|---|---|
| Mean | SEM | Mean | SEM | ||
| Age (Day) | 7.63 | 3.07 | 7.72 | 3.18 | 0.9421 |
| Female (%) | 41 | NA | 41 | NA | NA |
| Male (%) | 59 | NA | 59 | NA | NA |
| Xleucine (Cutoff: <245 µM) | 581 | 431.97 | <245 µM | NA | 0.005 ** |
| Valine (Cutoff: <290 µM) | 425.15 | 149.02 | <290 µM | NA | 0.0042 ** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alotaibi, A.Z.; AlMalki, R.H.; Al Mogren, M.; Sebaa, R.; Alanazi, M.; Jacob, M.; Alodaib, A.; Alfares, A.; Abdel Rahman, A.M. Exploratory Untargeted Metabolomics of Dried Blood Spot Samples from Newborns with Maple Syrup Urine Disease. Int. J. Mol. Sci. 2024, 25, 5720. https://doi.org/10.3390/ijms25115720
Alotaibi AZ, AlMalki RH, Al Mogren M, Sebaa R, Alanazi M, Jacob M, Alodaib A, Alfares A, Abdel Rahman AM. Exploratory Untargeted Metabolomics of Dried Blood Spot Samples from Newborns with Maple Syrup Urine Disease. International Journal of Molecular Sciences. 2024; 25(11):5720. https://doi.org/10.3390/ijms25115720
Chicago/Turabian StyleAlotaibi, Abeer Z., Reem H. AlMalki, Maha Al Mogren, Rajaa Sebaa, Mohammad Alanazi, Minnie Jacob, Ahamd Alodaib, Ahmad Alfares, and Anas M. Abdel Rahman. 2024. "Exploratory Untargeted Metabolomics of Dried Blood Spot Samples from Newborns with Maple Syrup Urine Disease" International Journal of Molecular Sciences 25, no. 11: 5720. https://doi.org/10.3390/ijms25115720
APA StyleAlotaibi, A. Z., AlMalki, R. H., Al Mogren, M., Sebaa, R., Alanazi, M., Jacob, M., Alodaib, A., Alfares, A., & Abdel Rahman, A. M. (2024). Exploratory Untargeted Metabolomics of Dried Blood Spot Samples from Newborns with Maple Syrup Urine Disease. International Journal of Molecular Sciences, 25(11), 5720. https://doi.org/10.3390/ijms25115720






