Circadian Rhythm Disruption in Hepatocellular Carcinoma Investigated by Integrated Analysis of Bulk and Single-Cell RNA Sequencing Data
Abstract
:1. Introduction
2. Results
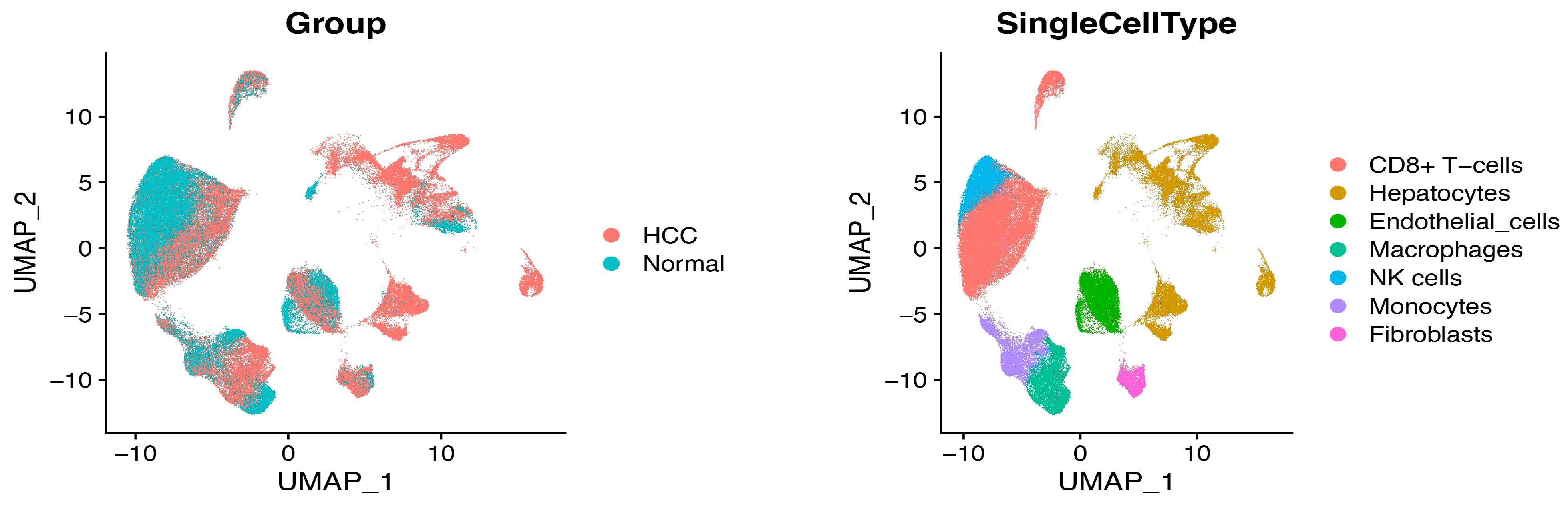
2.1. Single-Cell RNA Sequence Analysis and CRD Scoring in HCC
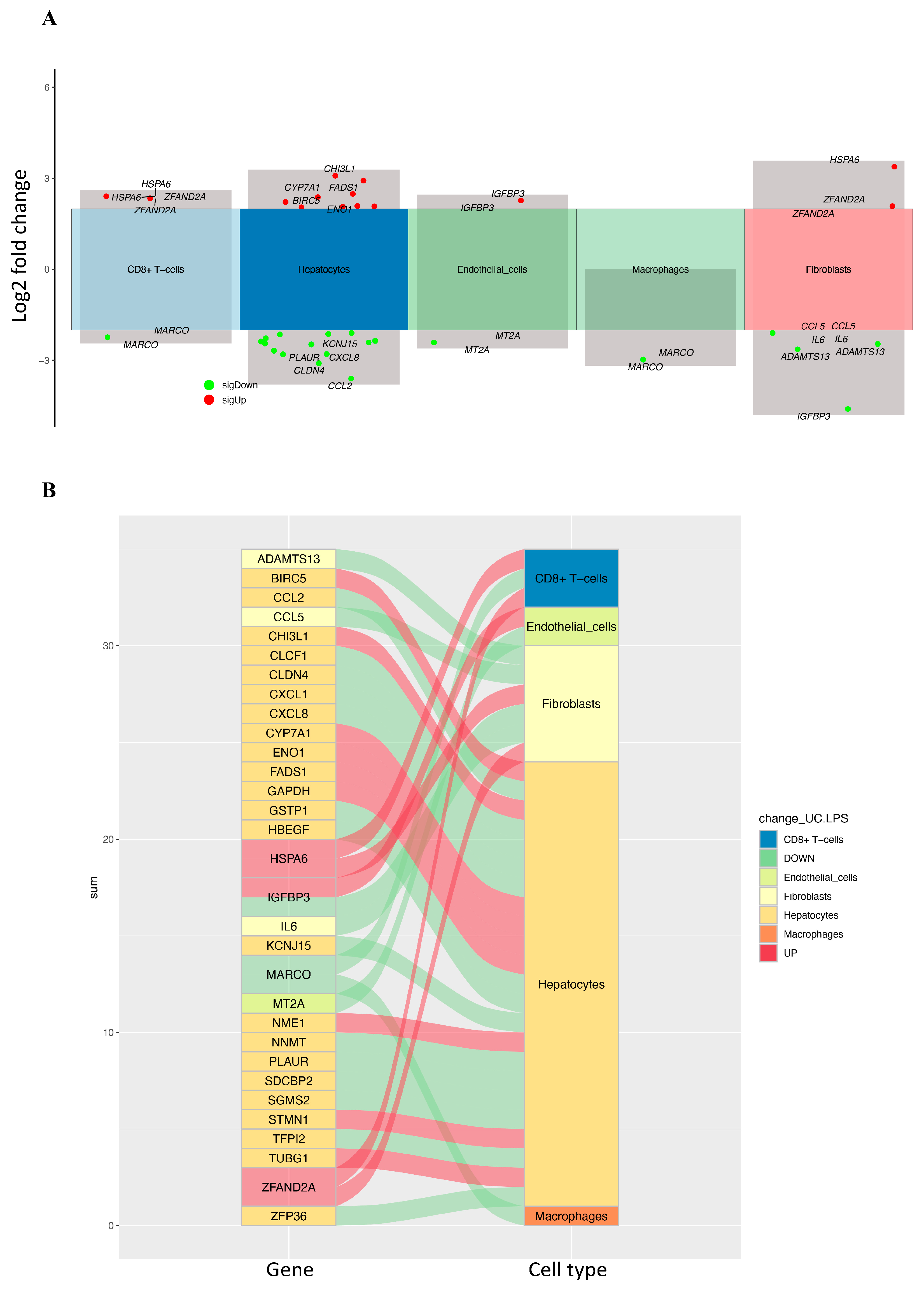
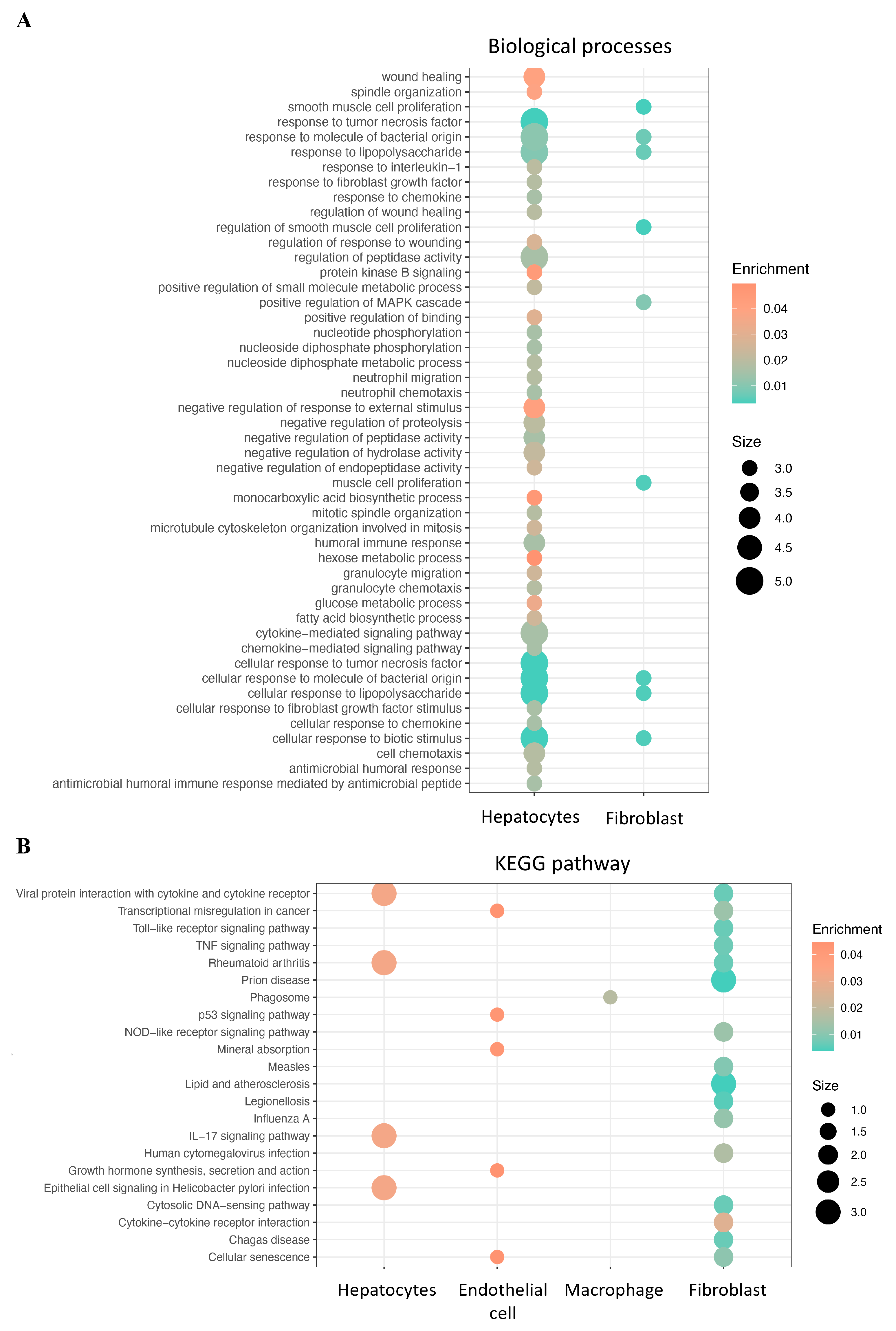
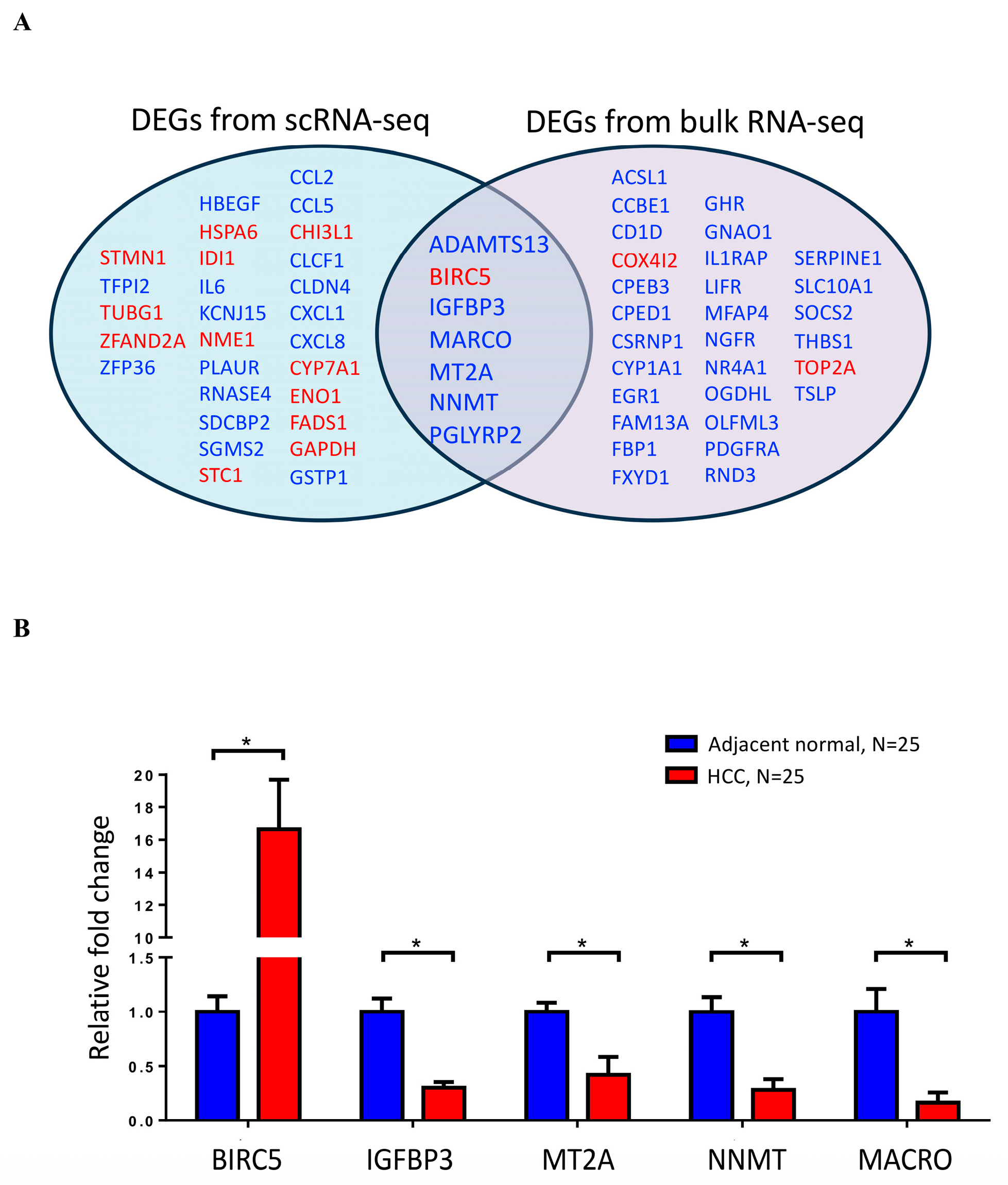
2.2. Identifying Differentially Expressed CRD-Related Genes in HCC by Using scRNA-seq Data Analysis
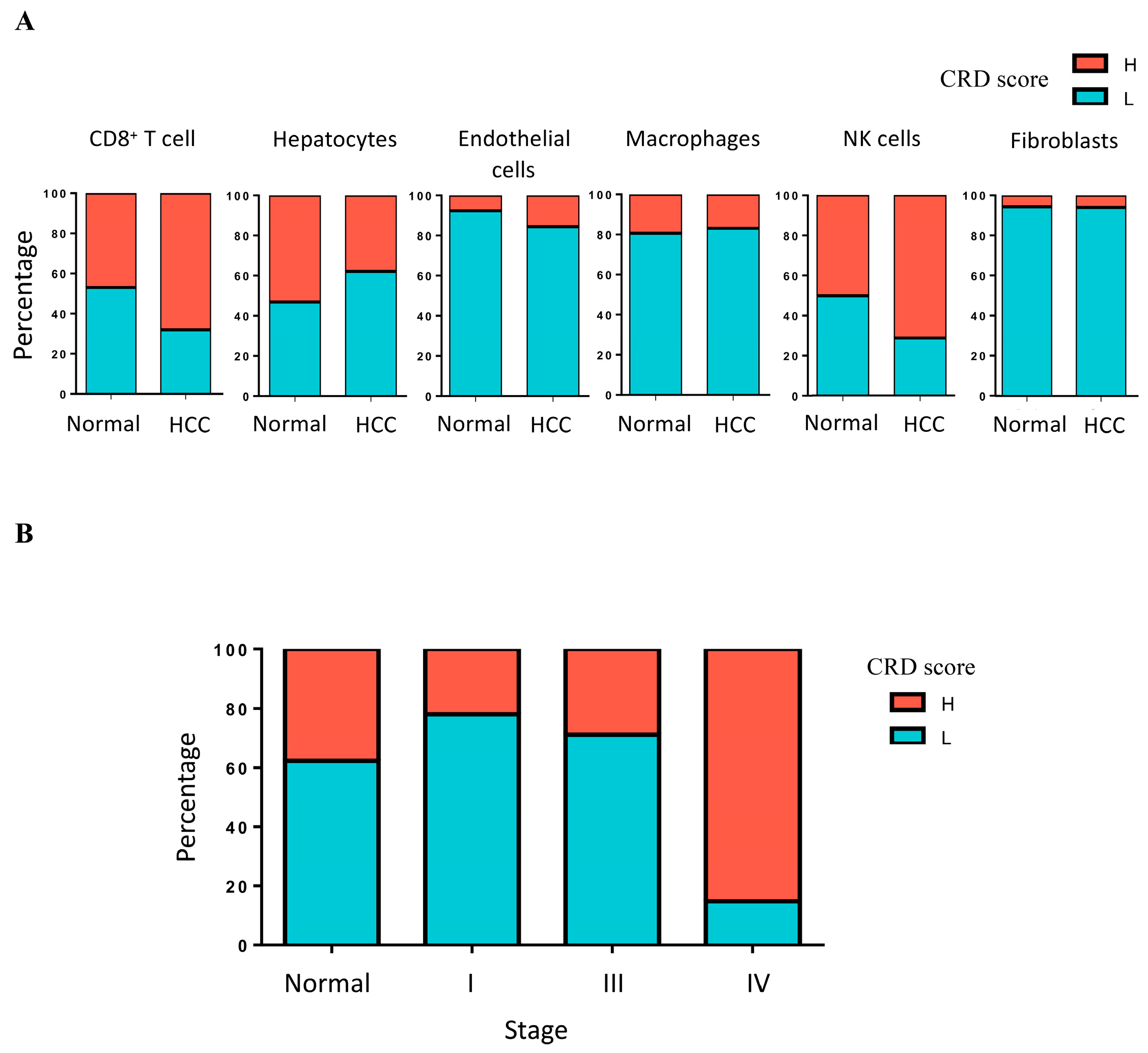
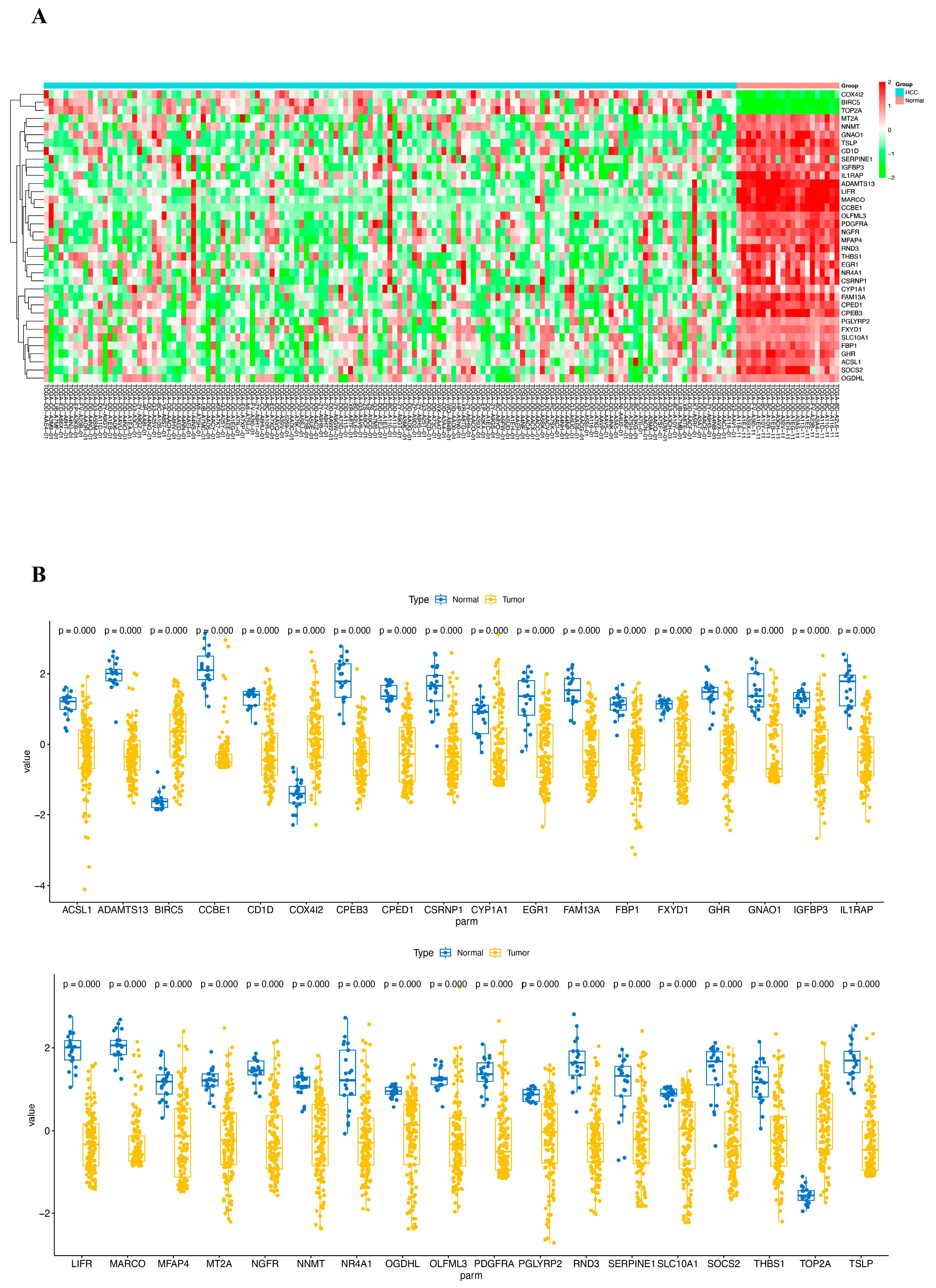
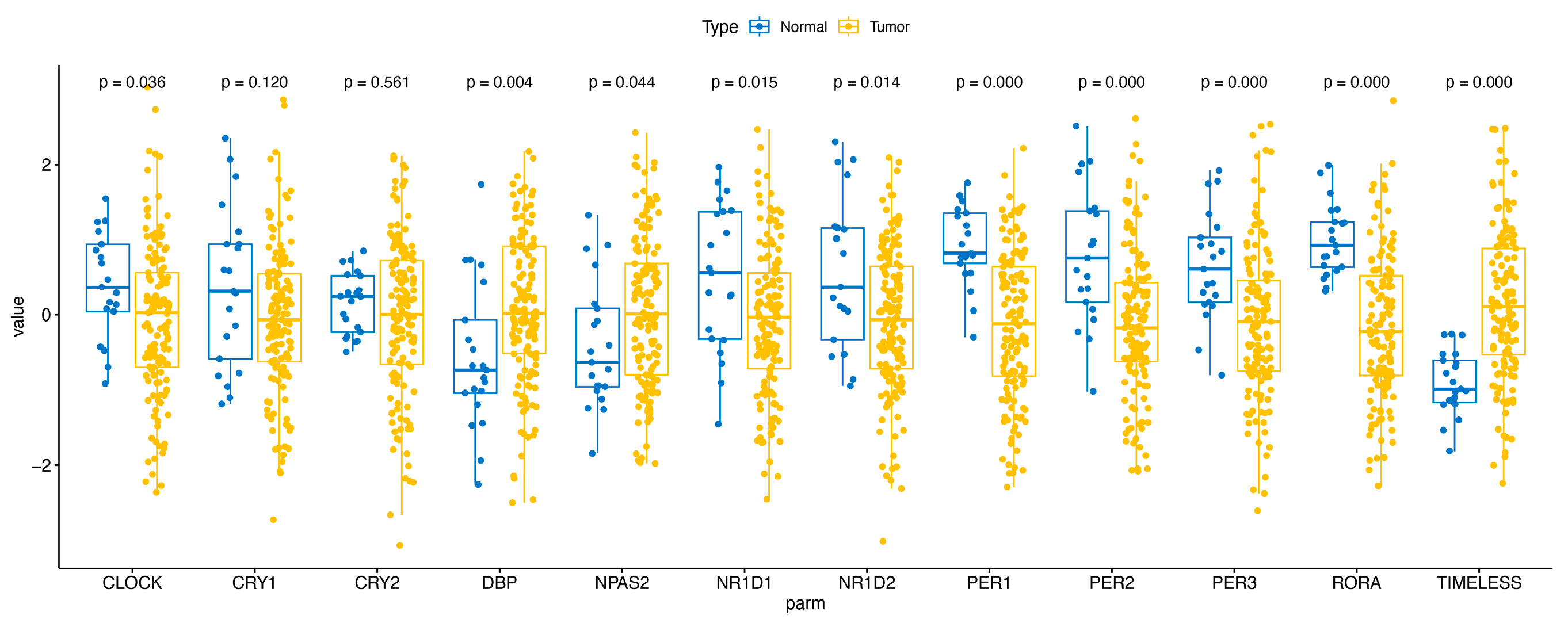
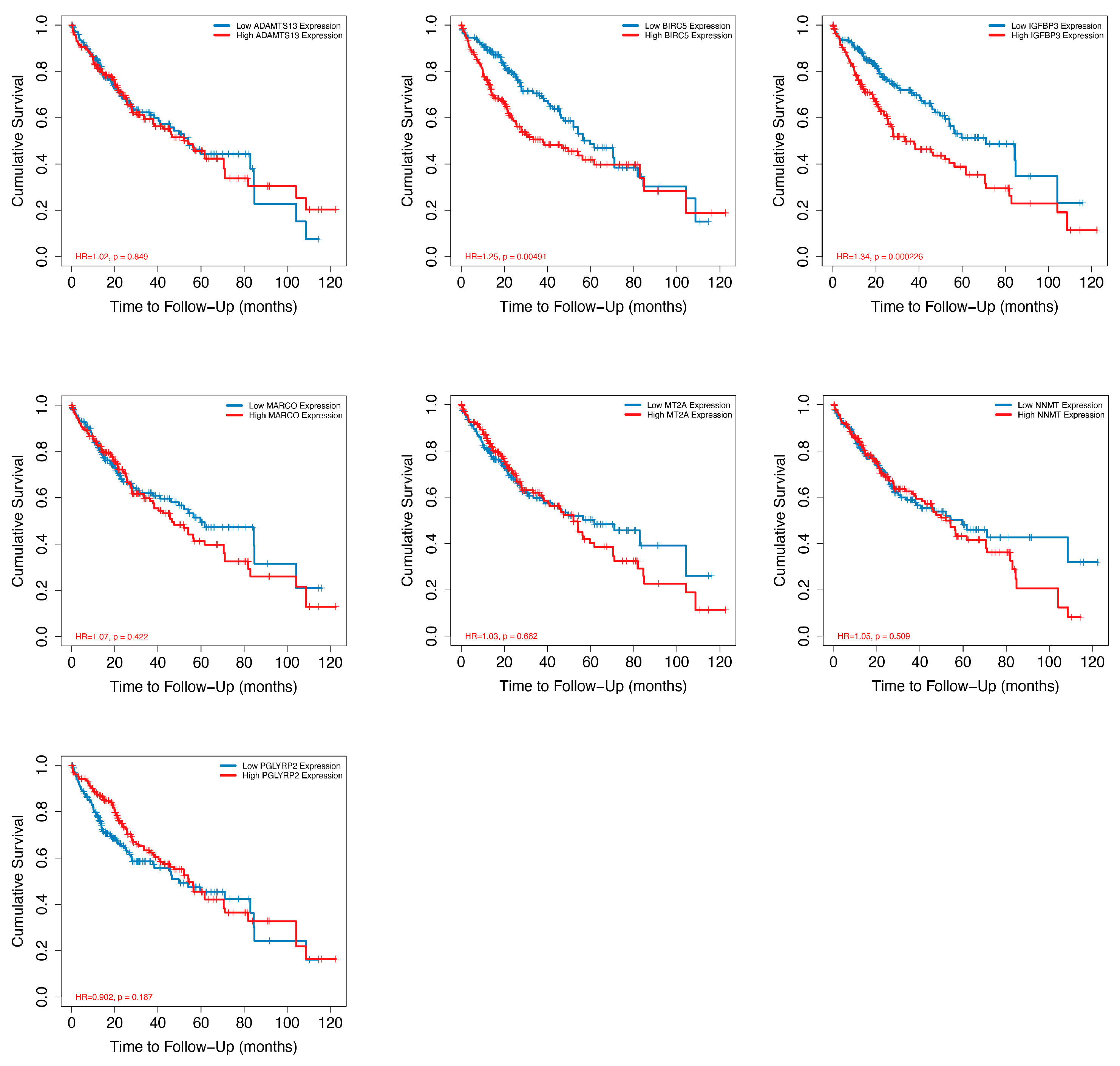
2.3. Analysis of CRD in HCC by Using Bulk RNA-Seq Data Analysis and RT-PCR
3. Materials and Methods
3.1. scRNA-Seq Data Acquisition and Processing
3.2. CRD Score Calculation and Cell Grouping
3.3. Differentially Expressed Genes (DEGs) of CRD
3.4. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Enrichment Analysis
3.5. The Processing of Bulk RNA-Seq Data from TCGA
3.6. Real-Time Quantitative Polymerase Chain Reaction (RT-PCR)
3.7. Kaplan–Meier Survival Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lee, Y. Roles of circadian clocks in cancer pathogenesis and treatment. Exp. Mol. Med. 2021, 53, 1529–1538. [Google Scholar] [CrossRef]
- Hastings, M.H.; Maywood, E.S.; Brancaccio, M. Generation of circadian rhythms in the suprachiasmatic nucleus. Nat. Rev. Neurosci. 2018, 19, 453–469. [Google Scholar] [CrossRef] [PubMed]
- Savvidis, C.; Koutsilieris, M. Circadian rhythm disruption in cancer biology. Mol. Med. 2012, 18, 1249–1260. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Shen, X.; Fasae, M.B.; Zhi, F.; Chai, L.; Ou, Y.; Feng, H.; Liu, S.; Liu, Y.; Yang, S. The Expression and Function of Circadian Rhythm Genes in Hepatocellular Carcinoma. Oxid. Med. Cell Longev. 2021, 2021, 4044606. [Google Scholar] [CrossRef] [PubMed]
- Chellappa, S.L.; Vujovic, N.; Williams, J.S.; Scheer, F. Impact of Circadian Disruption on Cardiovascular Function and Disease. Trends Endocrinol. Metab. 2019, 30, 767–779. [Google Scholar] [CrossRef] [PubMed]
- Leng, Y.; Musiek, E.S.; Hu, K.; Cappuccio, F.P.; Yaffe, K. Association between circadian rhythms and neurodegenerative diseases. Lancet Neurol. 2019, 18, 307–318. [Google Scholar] [CrossRef]
- Zimmet, P.; Alberti, K.; Stern, N.; Bilu, C.; El-Osta, A.; Einat, H.; Kronfeld-Schor, N. The Circadian Syndrome: Is the Metabolic Syndrome and much more! J. Intern. Med. 2019, 286, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Zhang, Z.; Nice, E.; Huang, C.; Zhang, W.; Tang, Y. Circadian rhythms and cancers: The intrinsic links and therapeutic potentials. J. Hematol. Oncol. 2022, 15, 21. [Google Scholar] [CrossRef] [PubMed]
- Ruch, B.; Wagler, J.; Kumm, K.; Zhang, C.; Katariya, N.N.; Garcia-Saenz-de-Sicilia, M.; Giorgakis, E.; Mathur, A.K. Hepatocellular Carcinoma, Alpha Fetoprotein, and Liver Allocation for Transplantation: Past, Present and Future. Curr. Oncol. 2022, 29, 7537–7551. [Google Scholar] [CrossRef]
- Kettner, N.M.; Voicu, H.; Finegold, M.J.; Coarfa, C.; Sreekumar, A.; Putluri, N.; Katchy, C.A.; Lee, C.; Moore, D.D.; Fu, L. Circadian Homeostasis of Liver Metabolism Suppresses Hepatocarcinogenesis. Cancer Cell 2016, 30, 909–924. [Google Scholar] [CrossRef]
- Villanueva, A. Hepatocellular Carcinoma. N. Engl. J. Med. 2019, 380, 1450–1462. [Google Scholar] [CrossRef]
- Siegel, A.B.; Zhu, A.X. Metabolic syndrome and hepatocellular carcinoma: Two growing epidemics with a potential link. Cancer 2009, 115, 5651–5661. [Google Scholar] [CrossRef] [PubMed]
- Krishnaiah, S.Y.; Wu, G.; Altman, B.J.; Growe, J.; Rhoades, S.D.; Coldren, F.; Venkataraman, A.; Olarerin-George, A.O.; Francey, L.J.; Mukherjee, S.; et al. Clock Regulation of Metabolites Reveals Coupling between Transcription and Metabolism. Cell Metab. 2017, 25, 961–974.e964. [Google Scholar] [CrossRef] [PubMed]
- Mteyrek, A.; Filipski, E.; Guettier, C.; Okyar, A.; Levi, F. Clock gene Per2 as a controller of liver carcinogenesis. Oncotarget 2016, 7, 85832–85847. [Google Scholar] [CrossRef] [PubMed]
- Mteyrek, A.; Filipski, E.; Guettier, C.; Oklejewicz, M.; van der Horst, G.T.; Okyar, A.; Levi, F. Critical cholangiocarcinogenesis control by cryptochrome clock genes. Int. J. Cancer 2017, 140, 2473–2483. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lv, K.; Zhao, M.; Chen, H.; Ji, G.; Zhang, Y.; Wang, T.; Cao, H.; Li, Y.; Qu, L. Analysis of miRNA expression profiles in the liver of Clock (Delta19) mutant mice. PeerJ 2019, 7, e8119. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.M.; Chang, J.H.; Yeh, K.T.; Yang, M.Y.; Liu, T.C.; Lin, S.F.; Su, W.W.; Chang, J.G. Disturbance of circadian gene expression in hepatocellular carcinoma. Mol. Carcinog. 2008, 47, 925–933. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Yang, X.; Li, J.; Yang, Y.; Yan, Z.; Zhang, H.; Mu, J. Elevated expression of CLOCK is associated with poor prognosis in hepatocellular carcinoma. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi 2018, 34, 163–168. [Google Scholar]
- Qiu, M.J.; Liu, L.P.; Jin, S.; Fang, X.F.; He, X.X.; Xiong, Z.F.; Yang, S.L. Research on circadian clock genes in common abdominal malignant tumors. Chronobiol. Int. 2019, 36, 906–918. [Google Scholar] [CrossRef]
- Zhang, Z.; Liang, Z.; Gao, W.; Yu, S.; Hou, Z.; Li, K.; Zeng, P. Identification of circadian clock genes as regulators of immune infiltration in Hepatocellular Carcinoma. J. Cancer 2022, 13, 3199–3208. [Google Scholar] [CrossRef]
- Qu, M.; Zhang, G.; Qu, H.; Vu, A.; Wu, R.; Tsukamoto, H.; Jia, Z.; Huang, W.; Lenz, H.J.; Rich, J.N.; et al. Circadian regulator BMAL1::CLOCK promotes cell proliferation in hepatocellular carcinoma by controlling apoptosis and cell cycle. Proc. Natl. Acad. Sci. USA 2023, 120, e2214829120. [Google Scholar] [CrossRef] [PubMed]
- Sia, D.; Villanueva, A.; Friedman, S.L.; Llovet, J.M. Liver Cancer Cell of Origin, Molecular Class, and Effects on Patient Prognosis. Gastroenterology 2017, 152, 745–761. [Google Scholar] [CrossRef]
- Schulze, R.J.; Schott, M.B.; Casey, C.A.; Tuma, P.L.; McNiven, M.A. The cell biology of the hepatocyte: A membrane trafficking machine. J. Cell Biol. 2019, 218, 2096–2112. [Google Scholar] [CrossRef] [PubMed]
- Guan, D.; Xiong, Y.; Trinh, T.M.; Xiao, Y.; Hu, W.; Jiang, C.; Dierickx, P.; Jang, C.; Rabinowitz, J.D.; Lazar, M.A. The hepatocyte clock and feeding control chronophysiology of multiple liver cell types. Science 2020, 369, 1388–1394. [Google Scholar] [CrossRef]
- Seo, H.R. Roles of Tumor Microenvironment in Hepatocelluar Carcinoma. Curr. Med. Chem. 2015, 11, 82–93. [Google Scholar] [CrossRef]
- Kalluri, R.; Zeisberg, M. Fibroblasts in cancer. Nat. Rev. Cancer 2006, 6, 392–401. [Google Scholar] [CrossRef] [PubMed]
- Ping, Q.; Yan, R.; Cheng, X.; Wang, W.; Zhong, Y.; Hou, Z.; Shi, Y.; Wang, C.; Li, R. Cancer-associated fibroblasts: Overview, progress, challenges, and directions. Cancer Gene Ther. 2021, 28, 984–999. [Google Scholar] [CrossRef] [PubMed]
- Xuan, W.; Khan, F.; James, C.D.; Heimberger, A.B.; Lesniak, M.S.; Chen, P. Circadian regulation of cancer cell and tumor microenvironment crosstalk. Trends Cell Biol. 2021, 31, 940–950. [Google Scholar] [CrossRef] [PubMed]
- Nagoshi, E.; Saini, C.; Bauer, C.; Laroche, T.; Naef, F.; Schibler, U. Circadian gene expression in individual fibroblasts: Cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell 2004, 119, 693–705. [Google Scholar] [CrossRef]
- DeYoung, V.; Singh, K.; Kretz, C.A. Mechanisms of ADAMTS13 regulation. J. Thromb. Haemost. 2022, 20, 2722–2732. [Google Scholar] [CrossRef]
- Kamm, D.R.; McCommis, K.S. Hepatic stellate cells in physiology and pathology. J. Physiol. 2022, 600, 1825–1837. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.J.; Niiya, M.; Zheng, X.W.; Shang, D.Z.; Zheng, X.L. Inflammatory cytokines inhibit ADAMTS13 synthesis in hepatic stellate cells and endothelial cells. J. Thromb. Haemost. 2008, 6, 1233–1235. [Google Scholar] [CrossRef]
- Kim, H.S. Role of insulin-like growth factor binding protein-3 in glucose and lipid metabolism. Ann. Pediatr. Endocrinol. Metab. 2013, 18, 9–12. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Yang, X.; Li, L.; Liu, P.; Wu, H.; Liu, Z.; Li, Q.; Liao, G.; Wang, X. Low expression levels of insulin-like growth factor binding protein-3 are correlated with poor prognosis for patients with hepatocellular carcinoma. Oncol. Lett. 2017, 13, 3395–3402. [Google Scholar] [CrossRef] [PubMed]
- Su, C. Survivin in survival of hepatocellular carcinoma. Cancer Lett. 2016, 379, 184–190. [Google Scholar] [CrossRef]
- Zhang, L.; Nie, L.; Cai, S.Y.; Chen, J.; Chen, J. Role of a macrophage receptor with collagenous structure (MARCO) in regulating monocyte/macrophage functions in ayu, Plecoglossus altivelis. Fish. Shellfish Immunol. 2018, 74, 141–151. [Google Scholar] [CrossRef]
- Xiao, Y.; Chen, B.; Yang, K.; Wang, Q.; Liu, P.; Gu, Y.; Zhong, Q.; Liu, Z.; He, Y.; Liu, Q. Down-regulation of MARCO associates with tumor progression in hepatocellular carcinoma. Exp. Cell Res. 2019, 383, 111542. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Wei, Y.; Li, Y.; Jiao, X. Low MARCO Expression is Associated with Poor Survival in Patients with Hepatocellular Carcinoma Following Liver Transplantation. Cancer Manag. Res. 2022, 14, 1935–1944. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.H.; Park, C.W.; Yoon, G.; Hong, S.M.; Choi, K.Y. NNMT depletion contributes to liver cancer cell survival by enhancing autophagy under nutrient starvation. Oncogenesis 2018, 7, 58. [Google Scholar] [CrossRef]
- Kim, J.; Hong, S.J.; Lim, E.K.; Yu, Y.S.; Kim, S.W.; Roh, J.H.; Do, I.G.; Joh, J.W.; Kim, D.S. Expression of nicotinamide N-methyltransferase in hepatocellular carcinoma is associated with poor prognosis. J. Exp. Clin. Cancer Res. 2009, 28, 20. [Google Scholar] [CrossRef]
- Zhang, Y.; van der Fits, L.; Voerman, J.S.; Melief, M.J.; Laman, J.D.; Wang, M.; Wang, H.; Wang, M.; Li, X.; Walls, C.D.; et al. Identification of serum N-acetylmuramoyl-l-alanine amidase as liver peptidoglycan recognition protein 2. Biochim. Biophys. Acta 2005, 1752, 34–46. [Google Scholar] [CrossRef] [PubMed]
- Dziarski, R.; Gupta, D. Review: Mammalian peptidoglycan recognition proteins (PGRPs) in innate immunity. Innate Immun. 2010, 16, 168–174. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Feng, J.; Xiao, L.; Chen, X.; Yao, Y.; Li, Y.; Tang, Y.; Zhang, S.; Lu, M.; Qian, Y.; et al. Tumor-Derived Peptidoglycan Recognition Protein 2 Predicts Survival and Antitumor Immune Responses in Hepatocellular Carcinoma. Hepatology 2020, 71, 1626–1642. [Google Scholar] [CrossRef] [PubMed]
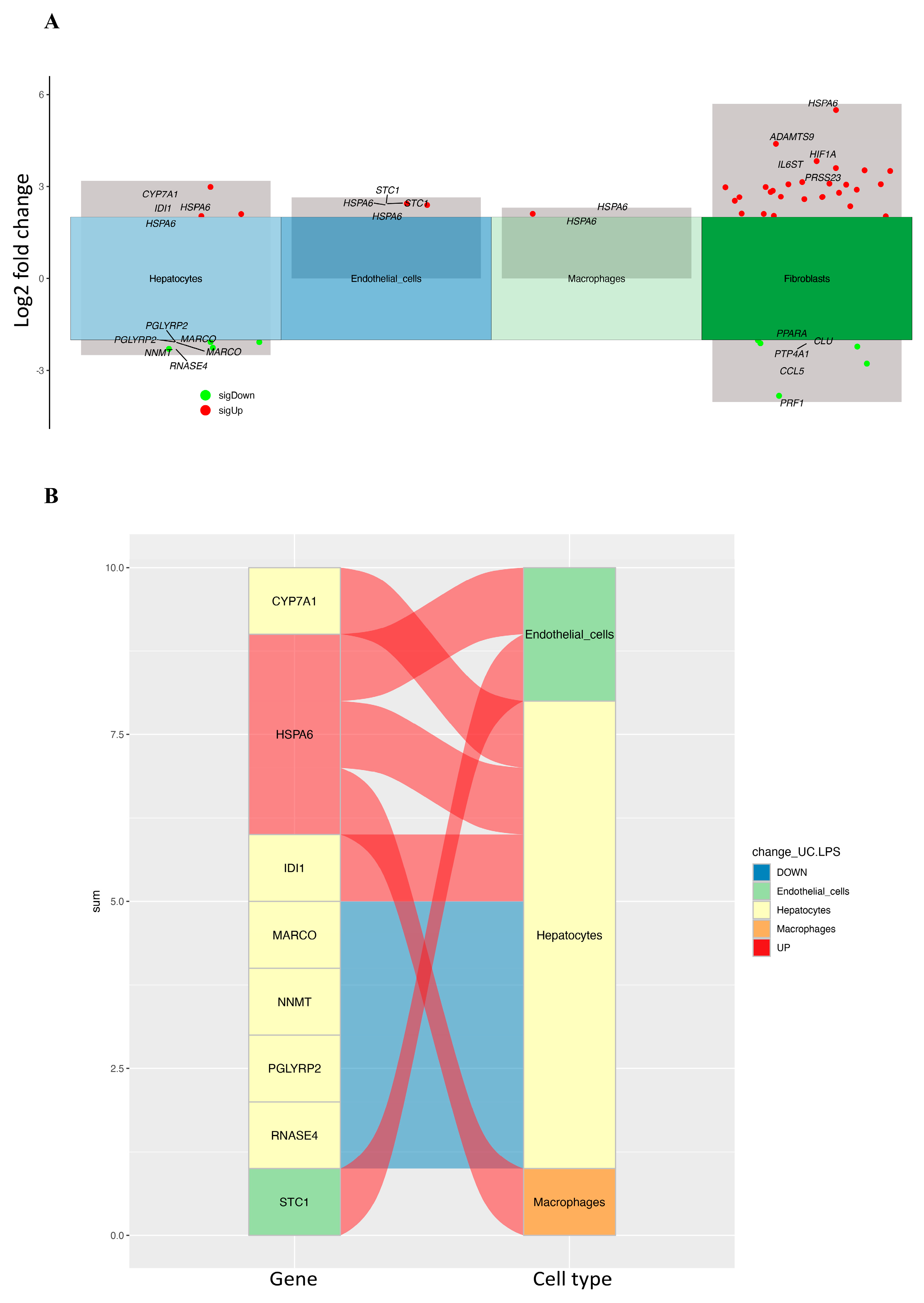

| Gene | Log2 Fold Change | Adjusted p-Value | Cell Type |
|---|---|---|---|
| RNASE4 | −2.303 | p < 0.0001 | Hepatocytes |
| NNMT | −2.267 | p < 0.0001 | Hepatocytes |
| PGLYRP2 | −2.081 | p < 0.0001 | Hepatocytes |
| MARCO | −2.083 | p < 0.0001 | Hepatocytes |
| IDI1 | 2.098 | p < 0.0001 | Hepatocytes |
| CYP7A1 | 2.985 | p < 0.0001 | Hepatocytes |
| STC1 | 2.435 | p < 0.0001 | Endothelial cells |
| HSPA6 | 2.105 | p < 0.0001 | Macrophages |
| HSPA6 | 2.031 | p < 0.0001 | Hepatocytes |
| HSPA6 | 2.389 | p < 0.0001 | Endothelial cells |
| Gene | Log2 Fold Change | Adjusted p-Value | Cell Type |
|---|---|---|---|
| ADAMTS13 | −2.640 | p < 0.0001 | Fibroblasts |
| BIRC5 | 2.380 | p < 0.0001 | Hepatocytes |
| CCL2 | −3.604 | p < 0.0001 | Hepatocytes |
| CCL5 | −2.100 | p < 0.0001 | Fibroblasts |
| CHI3L1 | 3.086 | p < 0.0001 | Hepatocytes |
| CLCF1 | −2.133 | p < 0.0001 | Hepatocytes |
| CLDN4 | −3.097 | p < 0.0001 | Hepatocytes |
| CXCL1 | −2.412 | p < 0.0001 | Hepatocytes |
| CXCL8 | −2.796 | p < 0.0001 | Hepatocytes |
| CYP7A1 | 2.926 | p < 0.0001 | Hepatocytes |
| ENO1 | 2.217 | p < 0.0001 | Hepatocytes |
| FADS1 | 2.486 | p < 0.0001 | Hepatocytes |
| GAPDH | 2.086 | p < 0.0001 | Hepatocytes |
| GSTP1 | −2.273 | p < 0.0001 | Hepatocytes |
| HBEGF | −2.360 | p < 0.0001 | Hepatocytes |
| HSPA6 | 2.408 | p < 0.0001 | CD8+ T-cells |
| HSPA6 | 3.386 | p < 0.0001 | Fibroblasts |
| IGFBP3 | 2.268 | p < 0.0001 | Endothelial cells |
| IGFBP3 | −4.606 | p < 0.0001 | Fibroblasts |
| IL6 | −2.461 | p < 0.0001 | Fibroblasts |
| KCNJ15 | −2.683 | p < 0.0001 | Hepatocytes |
| MARCO | −2.974 | p < 0.0001 | Macrophages |
| MARCO | −2.242 | p < 0.0001 | CD8+ T-cells |
| MT2A | −2.412 | p < 0.0001 | Endothelial cells |
| NME1 | 2.063 | p < 0.0001 | Hepatocytes |
| NNMT | −2.096 | p < 0.0001 | Hepatocytes |
| PLAUR | −2.803 | p < 0.0001 | Hepatocytes |
| SDCBP2 | −2.146 | p < 0.0001 | Hepatocytes |
| SGMS2 | −2.476 | p < 0.0001 | Hepatocytes |
| STMN1 | 2.075 | p < 0.0001 | Hepatocytes |
| TFPI2 | −2.448 | p < 0.0001 | Hepatocytes |
| TUBG1 | 2.044 | p < 0.0001 | Hepatocytes |
| ZFAND2A | 2.079 | p < 0.0001 | Fibroblasts |
| ZFAND2A | 2.339 | p < 0.0001 | CD8+ T-cells |
| ZFP36 | −2.380 | p < 0.0001 | Hepatocytes |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, L.-H.; Huang, C.-Y.; Liu, Y.-W.; Chien, P.-C.; Hsieh, T.-M.; Liu, H.-T.; Lin, H.-P.; Wu, C.-J.; Chuang, P.-C.; Hsieh, C.-H. Circadian Rhythm Disruption in Hepatocellular Carcinoma Investigated by Integrated Analysis of Bulk and Single-Cell RNA Sequencing Data. Int. J. Mol. Sci. 2024, 25, 5748. https://doi.org/10.3390/ijms25115748
Huang L-H, Huang C-Y, Liu Y-W, Chien P-C, Hsieh T-M, Liu H-T, Lin H-P, Wu C-J, Chuang P-C, Hsieh C-H. Circadian Rhythm Disruption in Hepatocellular Carcinoma Investigated by Integrated Analysis of Bulk and Single-Cell RNA Sequencing Data. International Journal of Molecular Sciences. 2024; 25(11):5748. https://doi.org/10.3390/ijms25115748
Chicago/Turabian StyleHuang, Lien-Hung, Chun-Ying Huang, Yueh-Wei Liu, Peng-Chen Chien, Ting-Min Hsieh, Hang-Tsung Liu, Hui-Ping Lin, Chia-Jung Wu, Pei-Chin Chuang, and Ching-Hua Hsieh. 2024. "Circadian Rhythm Disruption in Hepatocellular Carcinoma Investigated by Integrated Analysis of Bulk and Single-Cell RNA Sequencing Data" International Journal of Molecular Sciences 25, no. 11: 5748. https://doi.org/10.3390/ijms25115748






