Prediction of Pesticide Interactions with Proteins Involved in Human Reproduction by Using a Virtual Screening Approach: A Case Study of Famoxadone Binding CRBP-III and Izumo
Abstract
1. Introduction
2. Results and Discussion
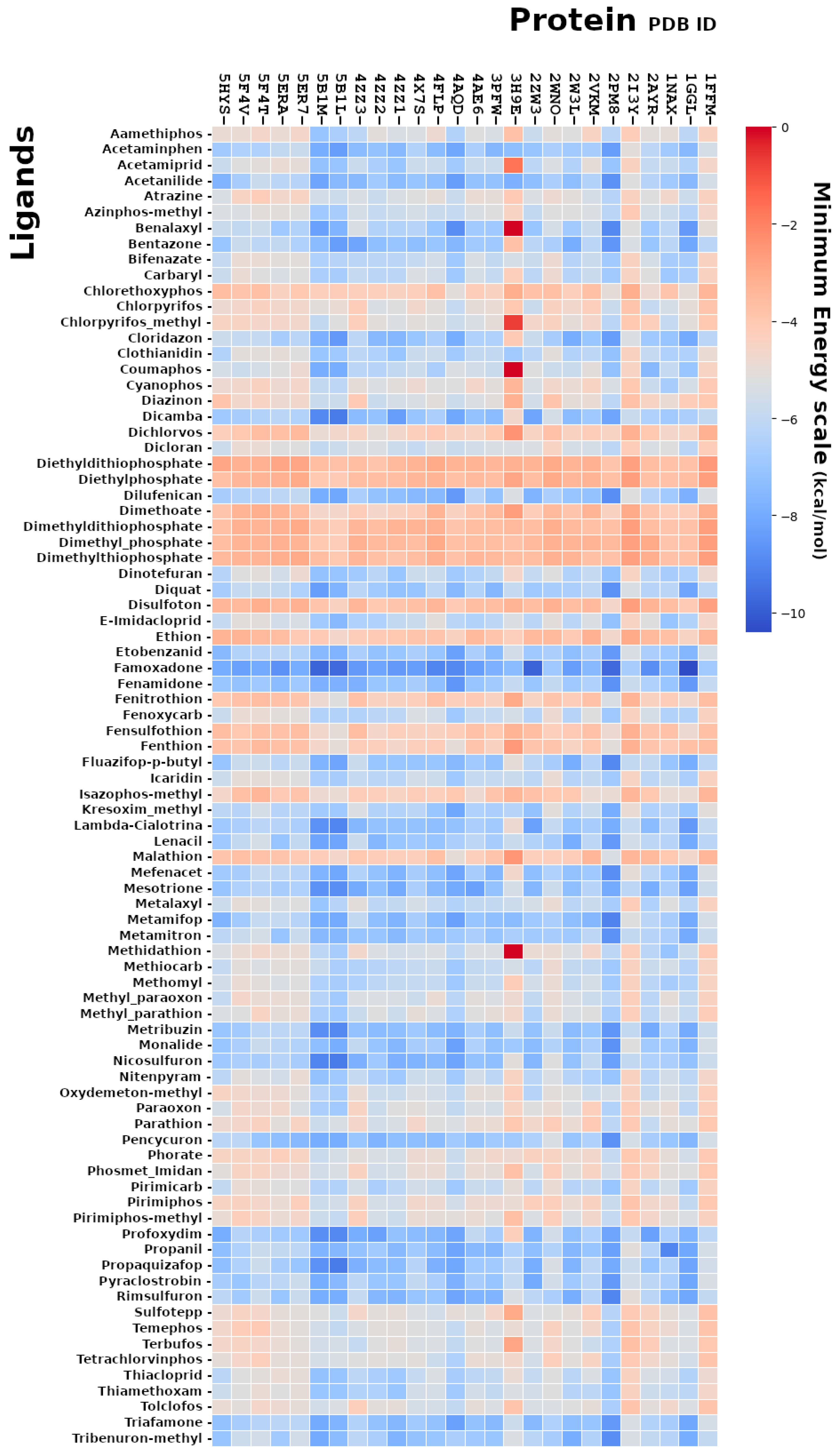
2.1. Molecular Docking Analysis Using AutoDock Vina
2.2. CRBP-III Structure, Sequence, and Functional Role
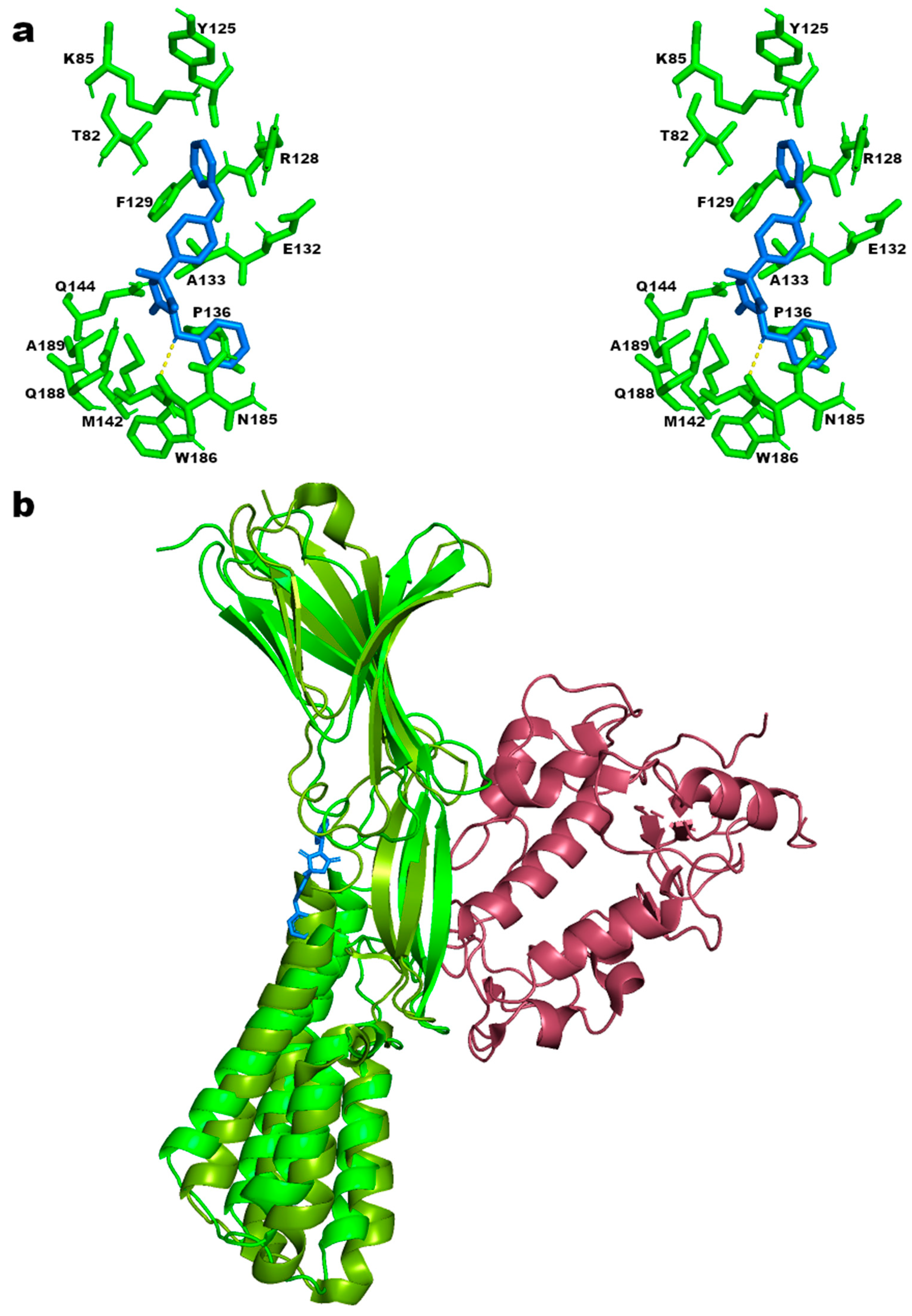
2.3. CRBP-III–Famoxadone Interaction Model
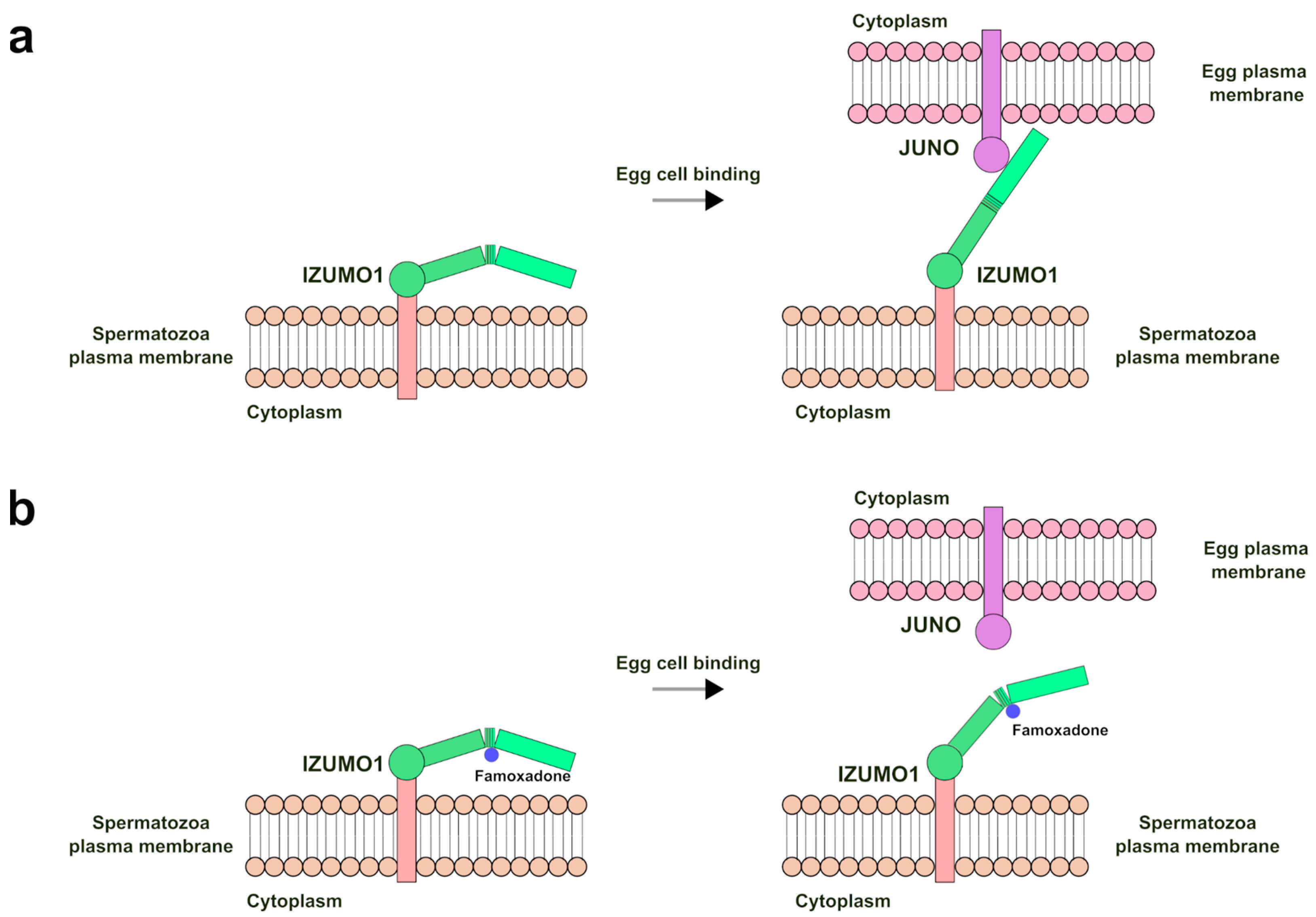
2.4. IZUMO1 Structure, Sequence, and Functional Role
2.5. IZUMO–Famoxadone Interaction Model
3. Materials and Methods
3.1. Ligand Dataset Preparation
3.2. Protein Datasets Preparation
3.3. Molecular Docking Analysis
3.4. Bibliography Research
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shirakawa, H.; Kikuchi, T.; Ito, M. Calcium Signaling in Mammalian Eggs at Fertilization. Curr. Top. Med. Chem. 2016, 16, 2664–2676. [Google Scholar] [CrossRef] [PubMed]
- Zafar, M.I.; Lu, S.; Li, H. Sperm-Oocyte Interplay: An Overview of Spermatozoon’s Role in Oocyte Activation and Current Perspectives in Diagnosis and Fertility Treatment. Cell Biosci. 2021, 11, 4. [Google Scholar] [CrossRef] [PubMed]
- Fainberg, J.; Kashanian, J.A. Recent Advances in Understanding and Managing Male Infertility. F1000Res 2019, 8, F1000 Faculty Rev-670. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, M.L.; Esteves, S.C.; Lamb, D.J.; Hotaling, J.M.; Giwercman, A.; Hwang, K.; Cheng, Y.-S. Male Infertility. Nat. Rev. Dis. Primers. 2023, 9, 49. [Google Scholar] [CrossRef] [PubMed]
- Lotti, F.; Corona, G.; Castellini, G.; Maseroli, E.; Fino, M.G.; Cozzolino, M.; Maggi, M. Semen Quality Impairment Is Associated with Sexual Dysfunction According to Its Severity. Hum. Reprod. 2016, 31, 2668–2680. [Google Scholar] [CrossRef] [PubMed]
- Muratori, M.; Marchiani, S.; Tamburrino, L.; Baldi, E. Sperm DNA Fragmentation: Mechanisms of Origin. In Genetic Damage in Human Spermatozoa; Baldi, E., Muratori, M., Eds.; Advances in Experimental Medicine and Biology; Springer International Publishing: Cham, Switzerland, 2019; pp. 75–85. ISBN 978-3-030-21664-1. [Google Scholar]
- Alvarez, J.G. DNA Fragmentation in Human Spermatozoa: Significance in the Diagnosis and Treatment of Infertility. Minerva Ginecol. 2003, 55, 233–239. [Google Scholar] [PubMed]
- Corsini, C.; Boeri, L.; Candela, L.; Pozzi, E.; Belladelli, F.; Capogrosso, P.; Fallara, G.; Schifano, N.; Cignoli, D.; Ventimiglia, E.; et al. Is There a Relevant Clinical Impact in Differentiating Idiopathic versus Unexplained Male Infertility? World J. Mens. Health 2023, 41, 354–362. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Singh, A.K. Impact of Environmental Factors on Human Semen Quality and Male Fertility: A Narrative Review. Environ. Sci. Eur. 2022, 34, 6. [Google Scholar] [CrossRef]
- Inhorn, M.C.; Patrizio, P. Infertility around the Globe: New Thinking on Gender, Reproductive Technologies and Global Movements in the 21st Century. Hum. Reprod. Update 2015, 21, 411–426. [Google Scholar] [CrossRef]
- Pathak, U.I.; Gabrielsen, J.S.; Lipshultz, L.I. Cutting-Edge Evaluation of Male Infertility. Urologic. Clin. 2020, 47, 129–138. [Google Scholar] [CrossRef]
- Jurewicz, J.; Dziewirska, E.; Radwan, M.; Hanke, W. Air Pollution from Natural and Anthropic Sources and Male Fertility. Reprod. Biol. Endocrinol. 2018, 16, 109. [Google Scholar] [CrossRef] [PubMed]
- González-Montaña, J.-R.; Senís, E.; Alonso, A.-J.; Alonso, M.-E.; Alonso, M.-P.; Domínguez, J.-C. Some Toxic Metals (Al, As, Mo, Hg) from Cow’s Milk Raised in a Possibly Contaminated Area by Different Sources. Environ. Sci. Pollut. Res. 2019, 26, 28909–28918. [Google Scholar] [CrossRef] [PubMed]
- Montano, L.; Pironti, C.; Pinto, G.; Ricciardi, M.; Buono, A.; Brogna, C.; Venier, M.; Piscopo, M.; Amoresano, A.; Motta, O. Polychlorinated Biphenyls (PCBs) in the Environment: Occupational and Exposure Events, Effects on Human Health and Fertility. Toxics 2022, 10, 365. [Google Scholar] [CrossRef] [PubMed]
- Bordajandi, L.R.; Gómez, G.; Abad, E.; Rivera, J.; Fernández-Bastón, M.d.M.; Blasco, J.; González, M.J. Survey of Persistent Organochlorine Contaminants (PCBs, PCDD/Fs, and PAHs), Heavy Metals (Cu, Cd, Zn, Pb, and Hg), and Arsenic in Food Samples From Huelva (Spain): Levels and Health Implications. J. Agric. Food Chem. 2004, 52, 992–1001. [Google Scholar] [CrossRef] [PubMed]
- Mohammad, N.S.; Nazli, R.; Zafar, H.; Fatima, S. Effects of Lipid Based Multiple Micronutrients Supplement on the Birth Outcome of Underweight Pre-Eclamptic Women: A Randomized Clinical Trial. Pak. J. Med. Sci. 2022, 38, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Dickerson, E.H.; Sathyapalan, T.; Knight, R.; Maguiness, S.M.; Killick, S.R.; Robinson, J.; Atkin, S.L. Endocrine Disruptor & Nutritional Effects of Heavy Metals in Ovarian Hyperstimulation. J. Assist. Reprod. Genet. 2011, 28, 1223–1228. [Google Scholar] [CrossRef] [PubMed]
- Lettieri, G.; D’Agostino, G.; Mele, E.; Cardito, C.; Esposito, R.; Cimmino, A.; Giarra, A.; Trifuoggi, M.; Raimondo, S.; Notari, T.; et al. Discovery of the Involvement in DNA Oxidative Damage of Human Sperm Nuclear Basic Proteins of Healthy Young Men Living in Polluted Areas. Int. J. Mol. Sci. 2020, 21, 4198. [Google Scholar] [CrossRef] [PubMed]
- Lettieri, G.; Marra, F.; Moriello, C.; Prisco, M.; Notari, T.; Trifuoggi, M.; Giarra, A.; Bosco, L.; Montano, L.; Piscopo, M. Molecular Alterations in Spermatozoa of a Family Case Living in the Land of Fires. A First Look at Possible Transgenerational Effects of Pollutants. Int. J. Mol. Sci. 2020, 21, 6710. [Google Scholar] [CrossRef]
- Bracke, A.; Peeters, K.; Punjabi, U.; Hoogewijs, D.; Dewilde, S. A Search for Molecular Mechanisms Underlying Male Idiopathic Infertility. Reprod. Biomed. Online 2018, 36, 327–339. [Google Scholar] [CrossRef]
- Foucaut, A.-M.; Faure, C.; Julia, C.; Czernichow, S.; Levy, R.; Dupont, C.; ALIFERT Collaborative Group. Sedentary Behavior, Physical Inactivity and Body Composition in Relation to Idiopathic Infertility among Men and Women. PLoS ONE 2019, 14, e0210770. [Google Scholar] [CrossRef]
- Stucki, A.O.; Barton-Maclaren, T.S.; Bhuller, Y.; Henriquez, J.E.; Henry, T.R.; Hirn, C.; Miller-Holt, J.; Nagy, E.G.; Perron, M.M.; Ratzlaff, D.E.; et al. Use of New Approach Methodologies (NAMs) to Meet Regulatory Requirements for the Assessment of Industrial Chemicals and Pesticides for Effects on Human Health. Front. Toxicol. 2022, 4, 964553. [Google Scholar] [CrossRef]
- Cavasotto, C.N.; Scardino, V. Machine Learning Toxicity Prediction: Latest Advances by Toxicity End Point. ACS Omega 2022, 7, 47536–47546. [Google Scholar] [CrossRef]
- Sakhteman, A.; Failli, M.; Kublbeck, J.; Levonen, A.L.; Fortino, V. A Toxicogenomic Data Space for System-Level Understanding and Prediction of EDC-Induced Toxicity. Environ. Int. 2021, 156, 106751. [Google Scholar] [CrossRef] [PubMed]
- Igarashi, Y.; Kojima, R.; Matsumoto, S.; Iwata, H.; Okuno, Y.; Yamada, H. Developing a GNN-Based AI Model to Predict Mitochondrial Toxicity Using the Bagging Method. J. Toxicol. Sci. 2024, 49, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Matthews, E.J.; Kruhlak, N.L.; Daniel Benz, R.; Ivanov, J.; Klopman, G.; Contrera, J.F. A Comprehensive Model for Reproductive and Developmental Toxicity Hazard Identification: II. Construction of QSAR Models to Predict Activities of Untested Chemicals. Regul. Toxicol. Pharmacol. 2007, 47, 136–155. [Google Scholar] [CrossRef] [PubMed]
- Jenardhanan, P.; Panneerselvam, M.; Mathur, P.P. Computational Methods Involved in Evaluating the Toxicity of the Reproductive Toxicants in Sertoli Cell. In Sertoli Cells: Methods and Protocols; Alves, M.G., Oliveira, P.F., Eds.; Springer: New York, NY, USA, 2018; pp. 253–277. ISBN 978-1-4939-7698-0. [Google Scholar]
- Sternberg, J.A.; Geffken, D.; Adams, J.B.; Pöstages, R.; Sternberg, C.G.; Campbell, C.L.; Moberg, W.K. Famoxadone: The Discovery and Optimisation of a New Agricultural Fungicide. Pest. Manag. Sci. 2001, 57, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Droin, N.M.; Green, D.R. Role of Bcl-2 Family Members in Immunity and Disease. Biochim. Biophys. Acta 2004, 1644, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Warren, C.F.A.; Wong-Brown, M.W.; Bowden, N.A. BCL-2 Family Isoforms in Apoptosis and Cancer. Cell Death Dis. 2019, 10, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Matzuk, M.M.; McKeown, M.R.; Filippakopoulos, P.; Li, Q.; Ma, L.; Agno, J.E.; Lemieux, M.E.; Picaud, S.; Yu, R.N.; Qi, J.; et al. Small-Molecule Inhibition of BRDT for Male Contraception. Cell 2012, 150, 673–684. [Google Scholar] [CrossRef]
- Kappock, T.J.; Ealick, S.E.; Stubbe, J. Modular Evolution of the Purine Biosynthetic Pathway. Curr. Opin. Chem. Biol. 2000, 4, 567–572. [Google Scholar] [CrossRef]
- Hereng, T.H.; Backe, P.H.; Kahmann, J.; Scheich, C.; Bjørås, M.; Skålhegg, B.S.; Rosendal, K.R. Structure and Function of the Human Sperm-Specific Isoform of Protein Kinase A (PKA) Catalytic Subunit Cα2. J. Struct. Biol. 2012, 178, 300–310. [Google Scholar] [CrossRef] [PubMed]
- Folli, C.; Calderone, V.; Ottonello, S.; Bolchi, A.; Zanotti, G.; Stoppini, M.; Berni, R. Identification, Retinoid Binding, and x-Ray Analysis of a Human Retinol-Binding Protein. Proc. Natl. Acad. Sci. USA 2001, 98, 3710–3715. [Google Scholar] [CrossRef] [PubMed]
- Marceau, G.; Gallot, D.; Lemery, D.; Sapin, V. Metabolism of Retinol during Mammalian Placental and Embryonic Development. Vitam. Horm. 2007, 75, 97–115. [Google Scholar] [CrossRef]
- Piantedosi, R.; Ghyselinck, N.; Blaner, W.S.; Vogel, S. Cellular Retinol-Binding Protein Type III Is Needed for Retinoid Incorporation into Milk. J. Biol. Chem. 2005, 280, 24286–24292. [Google Scholar] [CrossRef]
- Vogel, S.; Mendelsohn, C.L.; Mertz, J.R.; Piantedosi, R.; Waldburger, C.; Gottesman, M.E.; Blaner, W.S. Characterization of a New Member of the Fatty Acid-Binding Protein Family That Binds All-Trans-Retinol. J. Biol. Chem. 2001, 276, 1353–1360. [Google Scholar] [CrossRef] [PubMed]
- Spivak, M.; Stone, J.E.; Ribeiro, J.; Saam, J.; Freddolino, P.L.; Bernardi, R.C.; Tajkhorshid, E. VMD as a Platform for Interactive Small Molecule Preparation and Visualization in Quantum and Classical Simulations. J. Chem. Inf. Model. 2023, 63, 4664–4678. [Google Scholar] [CrossRef]
- Cheng, B.; Zhang, H.; Hu, J.; Peng, Y.; Yang, J.; Liao, X.; Liu, F.; Guo, J.; Hu, C.; Lu, H. The Immunotoxicity and Neurobehavioral Toxicity of Zebrafish Induced by Famoxadone-Cymoxanil. Chemosphere 2020, 247, 125870. [Google Scholar] [CrossRef]
- Huang, Y.; Chen, Z.; Meng, Y.; Wei, Y.; Xu, Z.; Ma, J.; Zhong, K.; Cao, Z.; Liao, X.; Lu, H. Famoxadone-Cymoxanil Induced Cardiotoxicity in Zebrafish Embryos. Ecotoxicol. Environ. Saf. 2020, 205, 111339. [Google Scholar] [CrossRef] [PubMed]
- Bournele, D.; Beis, D. Zebrafish Models of Cardiovascular Disease. Heart. Fail. Rev. 2016, 21, 803–813. [Google Scholar] [CrossRef]
- Pascale, A.; Laborde, A. Impact of Pesticide Exposure in Childhood. Rev. Environ. Health 2020, 35, 221–227. [Google Scholar] [CrossRef]
- Aydin, H.; Sultana, A.; Li, S.; Thavalingam, A.; Lee, J.E. Molecular Architecture of the Human Sperm IZUMO1 and Egg JUNO Fertilization Complex. Nature 2016, 534, 562–565. [Google Scholar] [CrossRef] [PubMed]
- Inoue, N.; Hamada, D.; Kamikubo, H.; Hirata, K.; Kataoka, M.; Yamamoto, M.; Ikawa, M.; Okabe, M.; Hagihara, Y. Molecular Dissection of IZUMO1, a Sperm Protein Essential for Sperm-Egg Fusion. Development 2013, 140, 3221–3229. [Google Scholar] [CrossRef]
- Bianchi, E.; Wright, G.J. Find and Fuse: Unsolved Mysteries in Sperm-Egg Recognition. PLoS Biol. 2020, 18, e3000953. [Google Scholar] [CrossRef] [PubMed]
- Hayasaka, S.; Terada, Y.; Inoue, N.; Okabe, M.; Yaegashi, N.; Okamura, K. Positive Expression of the Immunoglobulin Superfamily Protein IZUMO on Human Sperm of Severely Infertile Male Patients. Fertil. Steril. 2007, 88, 214–216. [Google Scholar] [CrossRef]
- Kaji, K.; Oda, S.; Shikano, T.; Ohnuki, T.; Uematsu, Y.; Sakagami, J.; Tada, N.; Miyazaki, S.; Kudo, A. The Gamete Fusion Process Is Defective in Eggs of Cd9-Deficient Mice. Nat. Genet. 2000, 24, 279–282. [Google Scholar] [CrossRef] [PubMed]
- Ohto, U.; Ishida, H.; Krayukhina, E.; Uchiyama, S.; Inoue, N.; Shimizu, T. Structure of IZUMO1-JUNO Reveals Sperm-Oocyte Recognition during Mammalian Fertilization. Nature 2016, 534, 566–569. [Google Scholar] [CrossRef] [PubMed]
- Pacak, P.; Kluger, C.; Vogel, V. Molecular Dynamics of JUNO-IZUMO1 Complexation Suggests Biologically Relevant Mechanisms in Fertilization. Sci. Rep. 2023, 13, 20342. [Google Scholar] [CrossRef] [PubMed]
- Klinovska, K.; Sebkova, N.; Dvorakova-Hortova, K. Sperm-Egg Fusion: A Molecular Enigma of Mammalian Reproduction. Int. J. Mol. Sci. 2014, 15, 10652–10668. [Google Scholar] [CrossRef] [PubMed]
- Miyado, K.; Yoshida, K.; Yamagata, K.; Sakakibara, K.; Okabe, M.; Wang, X.; Miyamoto, K.; Akutsu, H.; Kondo, T.; Takahashi, Y.; et al. The Fusing Ability of Sperm Is Bestowed by CD9-Containing Vesicles Released from Eggs in Mice. Proc. Natl. Acad. Sci. USA 2008, 105, 12921–12926. [Google Scholar] [CrossRef]
- Gaikwad, A.S.; Anderson, A.L.; Merriner, D.J.; O’Connor, A.E.; Houston, B.J.; Aitken, R.J.; O’Bryan, M.K.; Nixon, B. GLIPR1L1 Is an IZUMO-Binding Protein Required for Optimal Fertilization in the Mouse. BMC Biol. 2019, 17, 86. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization and Multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tortora, F.; Guerrera, V.; Lettieri, G.; Febbraio, F.; Piscopo, M. Prediction of Pesticide Interactions with Proteins Involved in Human Reproduction by Using a Virtual Screening Approach: A Case Study of Famoxadone Binding CRBP-III and Izumo. Int. J. Mol. Sci. 2024, 25, 5790. https://doi.org/10.3390/ijms25115790
Tortora F, Guerrera V, Lettieri G, Febbraio F, Piscopo M. Prediction of Pesticide Interactions with Proteins Involved in Human Reproduction by Using a Virtual Screening Approach: A Case Study of Famoxadone Binding CRBP-III and Izumo. International Journal of Molecular Sciences. 2024; 25(11):5790. https://doi.org/10.3390/ijms25115790
Chicago/Turabian StyleTortora, Fabiana, Valentina Guerrera, Gennaro Lettieri, Ferdinando Febbraio, and Marina Piscopo. 2024. "Prediction of Pesticide Interactions with Proteins Involved in Human Reproduction by Using a Virtual Screening Approach: A Case Study of Famoxadone Binding CRBP-III and Izumo" International Journal of Molecular Sciences 25, no. 11: 5790. https://doi.org/10.3390/ijms25115790
APA StyleTortora, F., Guerrera, V., Lettieri, G., Febbraio, F., & Piscopo, M. (2024). Prediction of Pesticide Interactions with Proteins Involved in Human Reproduction by Using a Virtual Screening Approach: A Case Study of Famoxadone Binding CRBP-III and Izumo. International Journal of Molecular Sciences, 25(11), 5790. https://doi.org/10.3390/ijms25115790










