An In Silico Analysis of Genetic Variants and Structural Modeling of the Human Frataxin Protein in Friedreich’s Ataxia
Abstract
:1. Introduction
2. Results
2.1. Protein Sequence and Variant Acquisition
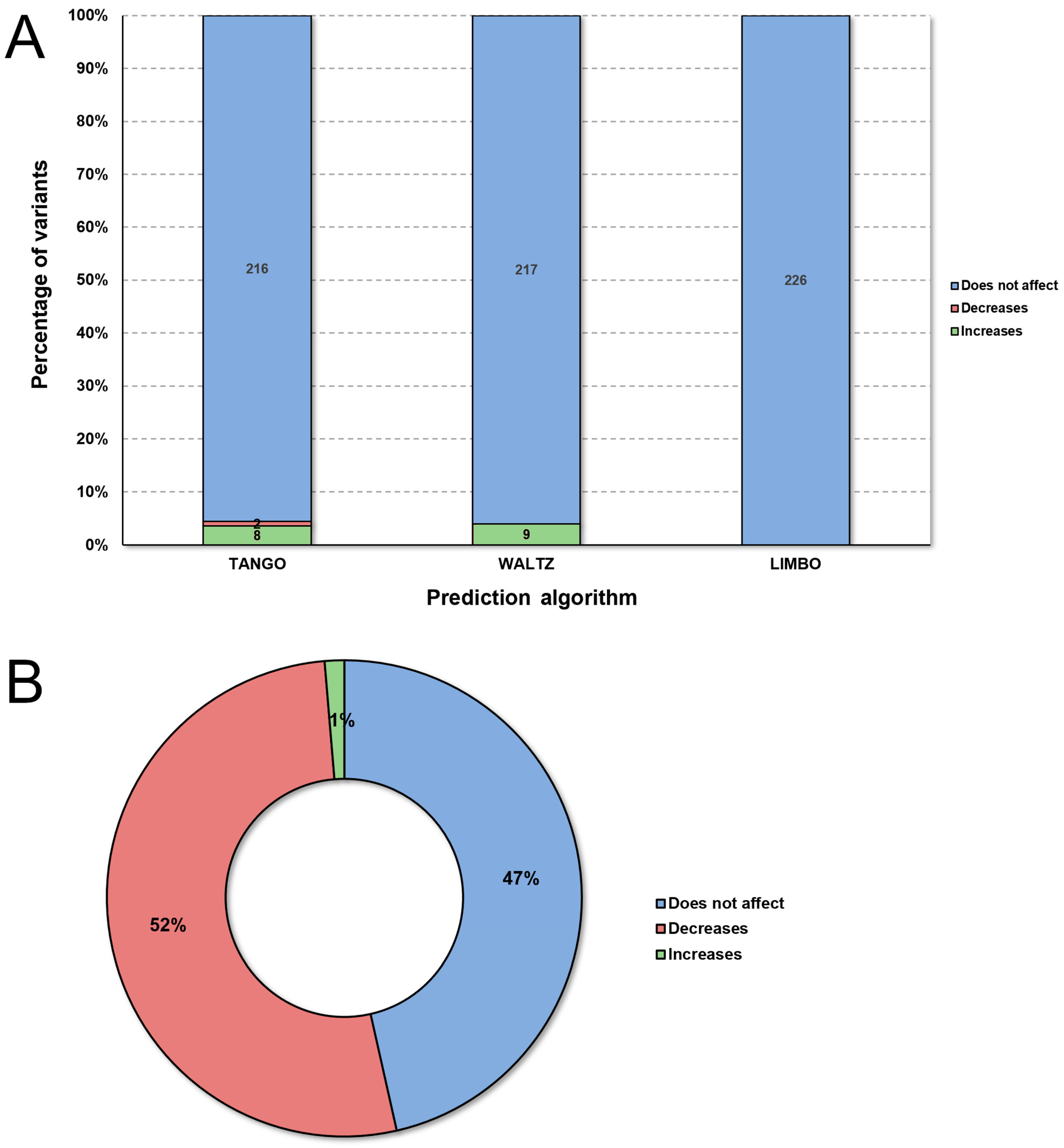
2.2. Predictive Analysis
2.3. Structural Modeling and Validation
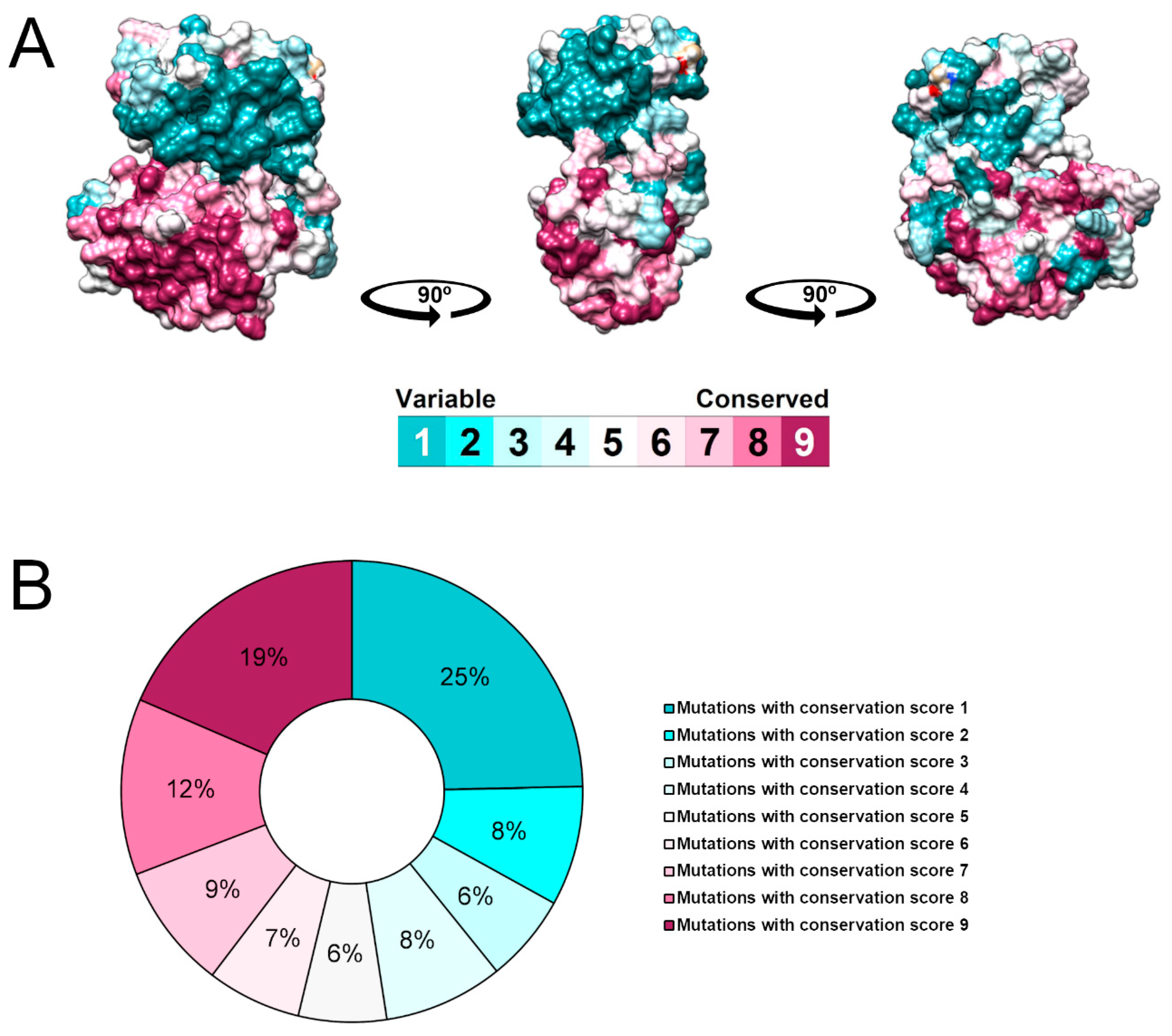
2.4. Evolutionary Conservation Analysis
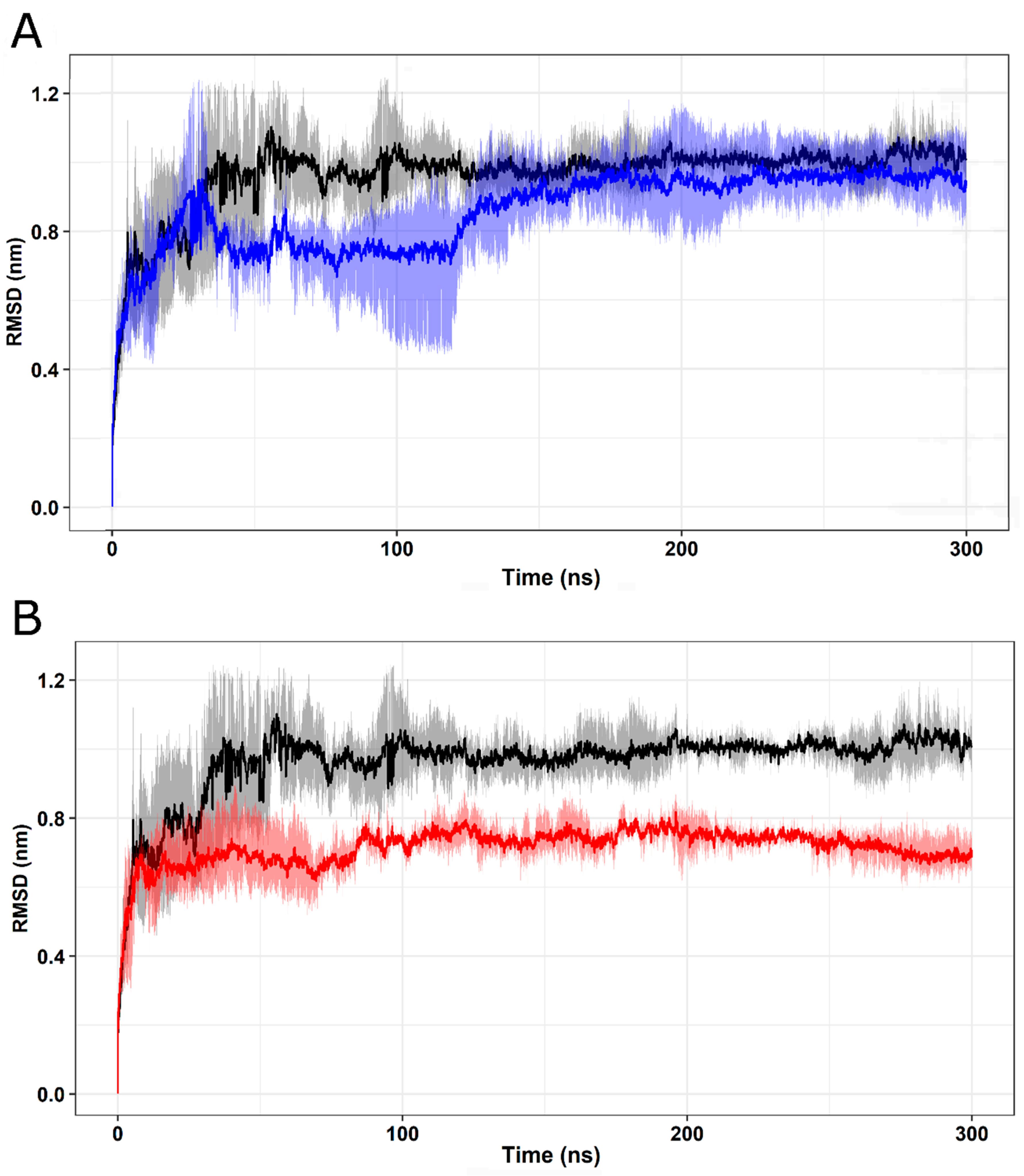
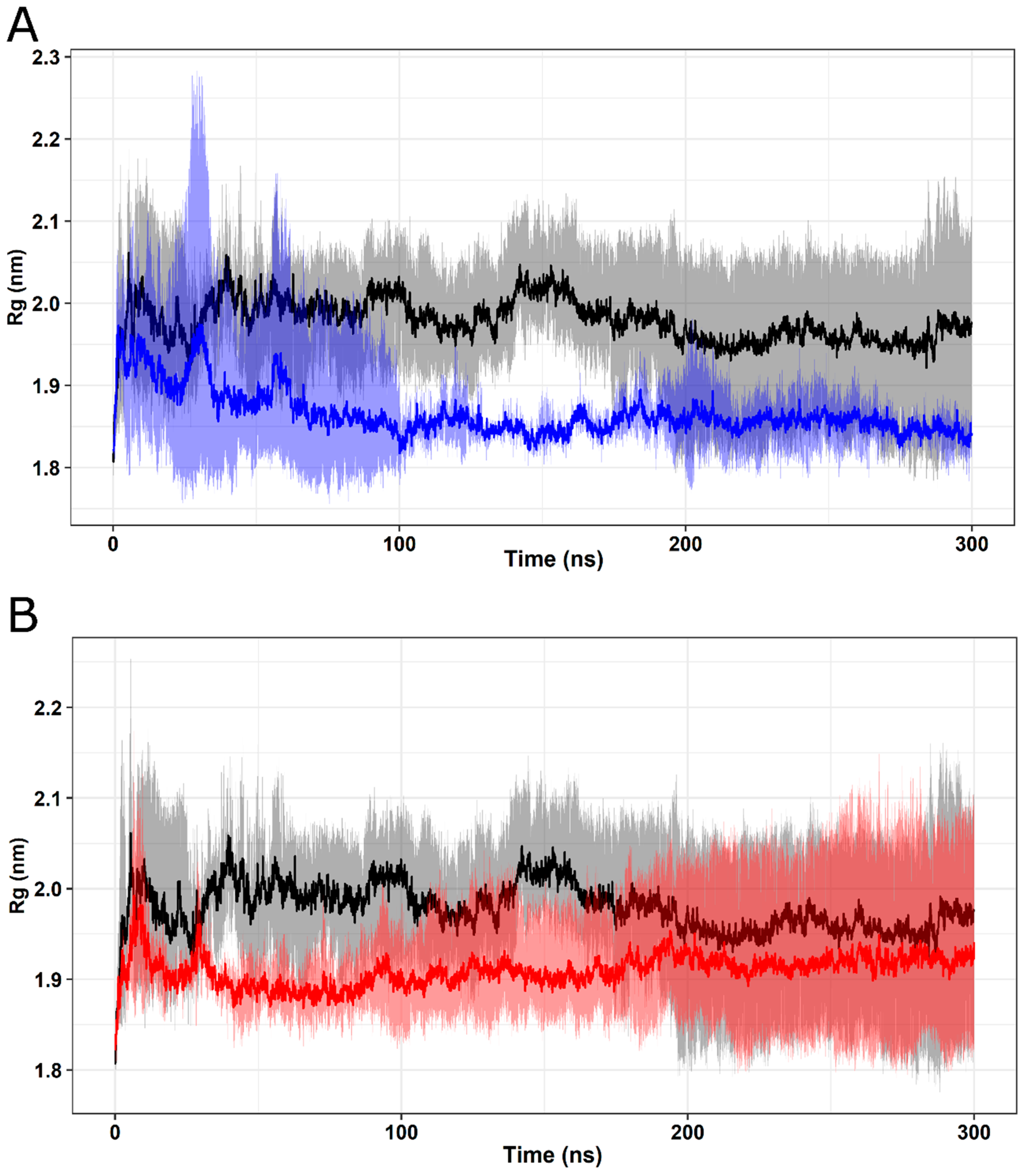
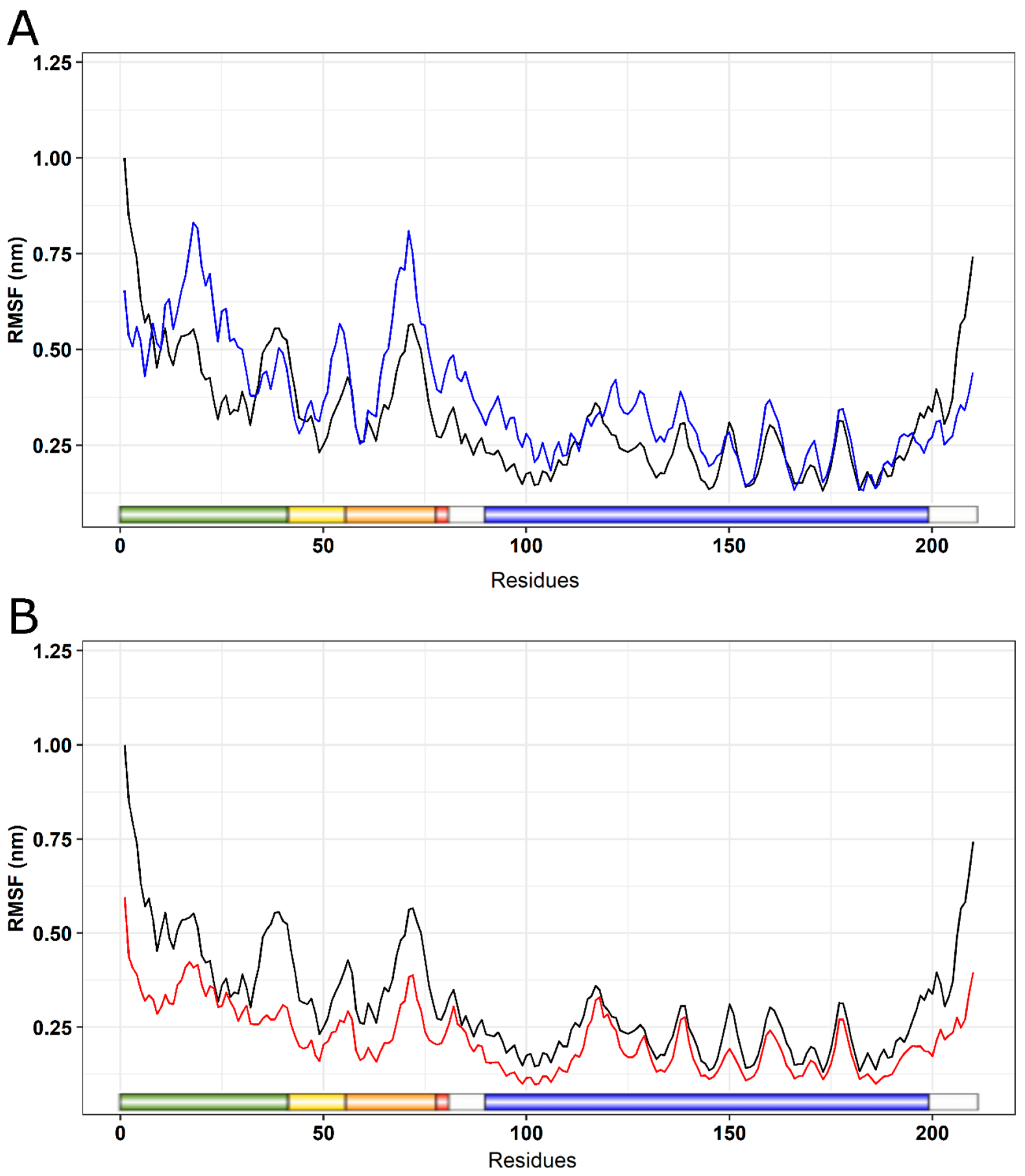
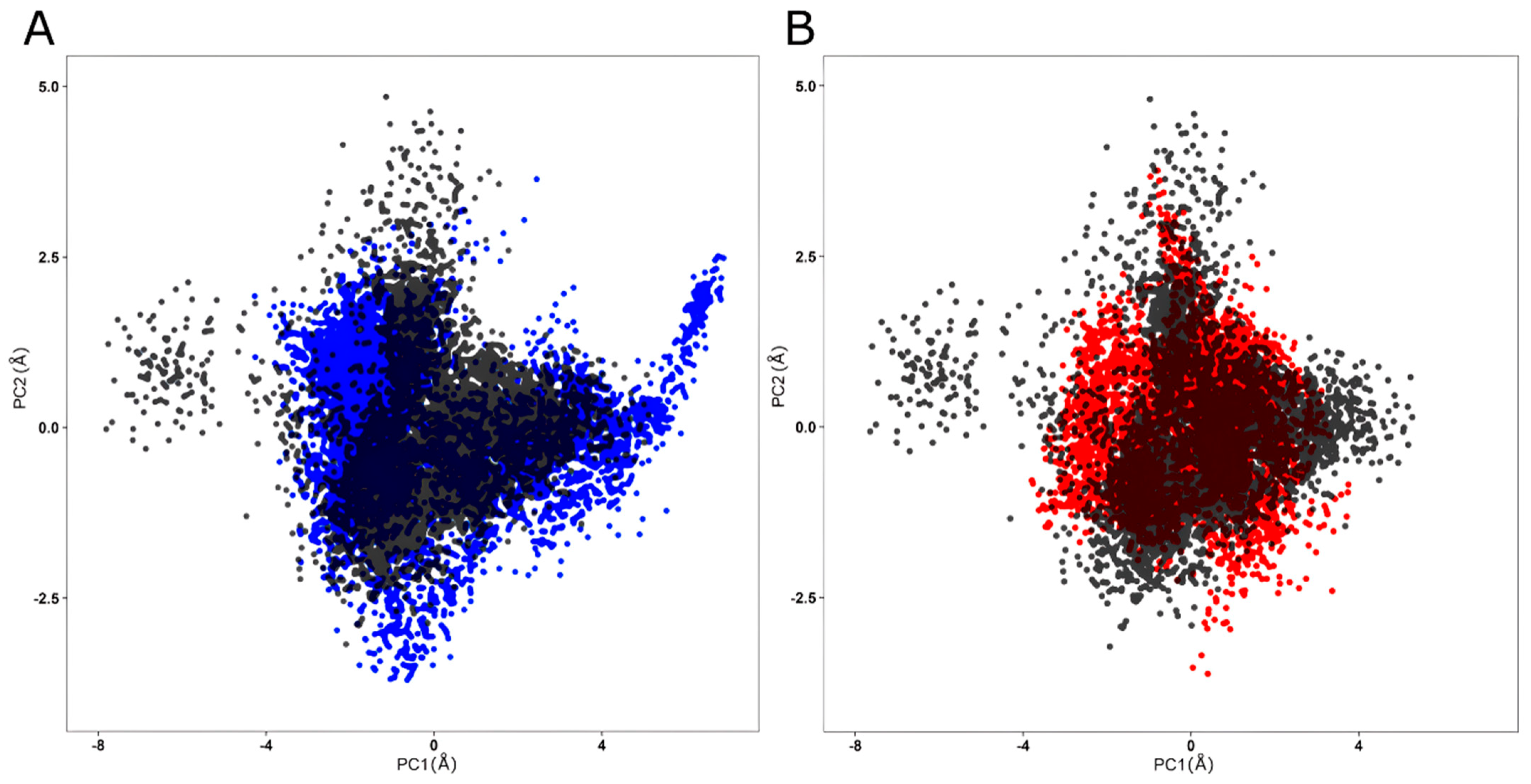
2.5. Molecular Dynamics Simulation
3. Materials and Methods
3.1. Protein Sequence and Variant Acquisition
3.2. Predictive Analysis
3.3. Structural Modeling and Validation
3.4. Evolutionary Conservation Analysis
3.5. Molecular Dynamics Simulation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cossée, M.; Dürr, A.; Schmitt, M.; Dahl, N.; Trouillas, P.; Allinson, P.; Kostrzewa, M.; Nivelon-Chevallier, A.; Gustavson, K.H.; Kohlschütter, A.; et al. Friedreich’s Ataxia: Point Mutations and Clinical Presentation of Compound Heterozygotes. Ann. Neurol. 1999, 45, 200–206. [Google Scholar] [CrossRef] [PubMed]
- Keita, M.; McIntyre, K.; Rodden, L.N.; Schadt, K.; Lynch, D.R. Friedreich ataxia: Clinical features and new developments. Neurodegener. Dis. Manag. 2022, 12, 267–283. [Google Scholar] [CrossRef] [PubMed]
- Maudoux, A.; Teissier, N.; Francois, M.; Abbeele, T.V.D.; Alberti, C.; Husson, I.; Wiener-Vacher, S.R. Vestibular impact of Friedreich ataxia in early onset patients. Cerebellum Ataxias 2020, 7, 6. [Google Scholar] [CrossRef] [PubMed]
- Zeigelboim, B.S.; Teive, H.A.G.; da Rosa, M.R.; Malisky, J.S.; Fonseca, V.R.; Marques, J.M.; Liberalesso, P.B. The importance of central auditory evaluation in Friedreich’s ataxia. Arq. Neuro-Psiquiatr. 2018, 76, 170–176. [Google Scholar] [CrossRef]
- Lynch, D.R.; Chin, M.P.; Delatycki, M.B.; Subramony, S.H.; Corti, M.; Hoyle, J.C.; Boesch, S.; Nachbauer, W.; Mariotti, C.; Mathews, K.D.; et al. Safety and Efficacy of Omaveloxolone in Friedreich Ataxia (MOXIe Study). Ann. Neurol. 2021, 89, 212–225. [Google Scholar] [CrossRef] [PubMed]
- Abeti, R.; Baccaro, A.; Esteras, N.; Giunti, P. Novel Nrf2-Inducer Prevents Mitochondrial Defects and Oxidative Stress in Friedreich’s Ataxia Models. Front. Cell. Neurosci. 2018, 12, 188. [Google Scholar] [CrossRef]
- Luz, G.D.S.; Da Silva, M.R.S.; DeMontigny, F. Priority needs referred by families of rare disease patients. Texto Context.—Enferm. 2016, 25. [Google Scholar] [CrossRef]
- Schoenfeld, R.A.; Napoli, E.; Wong, A.; Zhan, S.; Reutenauer, L.; Morin, D.; Buckpitt, A.R.; Taroni, F.; Lonnerdal, B.; Ristow, M.; et al. Frataxin deficiency alters heme pathway transcripts and decreases mitochondrial heme metabolites in mammalian cells. Hum. Mol. Genet. 2005, 14, 3787–3799. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Carlson, B.E.; Lacis, A.A. Application of spectral analysis techniques in the intercomparison of aerosol data: 1. An EOF approach to analyze the spatial-temporal variability of aerosol optical depth using multiple remote sensing data sets. J. Geophys. Res. Atmos. 2013, 118, 8640–8648. [Google Scholar] [CrossRef]
- Clark, E.; Strawser, C.; Schadt, K.; Lynch, D.R. Identification of a novel missense mutation in Friedreich’s ataxia–FXNW168R. Ann. Clin. Transl. Neurol. 2019, 6, 812–816. [Google Scholar] [CrossRef]
- da Silva, A.N.R.; Pereira, G.R.C.; Bonet, L.F.S.; Outeiro, T.F.; De Mesquita, J.F. In silico analysis of alpha-synuclein protein variants and posttranslational modifications related to Parkinson’s disease. J. Cell. Biochem. 2024, 125, e30523. [Google Scholar] [CrossRef] [PubMed]
- Tavares, G.D.B.; Pereira, G.R.C.; Borges, G.F.; de Mesquita, J.F. Predictive analysis of Tryptophan Hydroxylase 2 (TPH2) missense mutations in psychiatric disorders. Braz. J. Dev. 2022, 8, 61944–61970. [Google Scholar] [CrossRef]
- Pereira, G.R.C.; Abrahim-Vieira, B.d.A.; de Mesquita, J.F. In Silico Analyses of a Promising Drug Candidate for the Treatment of Amyotrophic Lateral Sclerosis Targeting Superoxide Dismutase I Protein. Pharmaceutics 2023, 15, 1095. [Google Scholar] [CrossRef] [PubMed]
- Pereira, G.R.C.; Tellini, G.H.A.S.; Mesquita, J.F. De In Silico Analysis of PFN1 Related to Amyotrophic Lateral Sclerosis. PLoS ONE 2019, 14, e0215723. [Google Scholar] [CrossRef] [PubMed]
- Rose, Y.; Duarte, J.M.; Lowe, R.; Segura, J.; Bi, C.; Bhikadiya, C.; Chen, L.; Rose, A.S.; Bittrich, S.; Burley, S.K.; et al. RCSB Protein Data Bank: Architectural Advances Towards Integrated Searching and Efficient Access to Macromolecular Structure Data from the PDB Archive. J. Mol. Biol. 2020, 433, 166704. [Google Scholar] [CrossRef] [PubMed]
- Batool, M.; Ahmad, B.; Choi, S. A Structure-Based Drug Discovery Paradigm. Int. J. Mol. Sci. 2019, 20, 2783. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, C.C.S.; Pereira, G.R.C.; De Alcantara, J.Y.S.; Antunes, D.; Caffarena, E.R.; Mesquita, J.F. De In Silico Analysis of the V66M Variant of Human BDNF in Psychiatric Disorders: An Approach to Precision Medicine. PLoS ONE 2019, 14, e0215508. [Google Scholar] [CrossRef] [PubMed]
- Loureiro, J.P.; Pereira, G.R.C.; Bloise, L.C.d.S.; Salerno, J.A.d.C.; Silvestre, V.A.; De Mesquita, J.F. Comprehensive in silico analysis of the TDP-43 protein variants related to Amyotrophic Lateral Sclerosis and Frontotemporal Dementia. Braz. J. Dev. 2022, 8, 57746–57775. [Google Scholar] [CrossRef]
- Pereira, G.R.C.; Da Silva, A.N.R.; Nascimento, S.S.D.; De Mesquita, J.F. In silico analysis and molecular dynamics simulation of human superoxide dismutase 3 (SOD3) genetic variants. J. Cell. Biochem. 2018, 120, 3583–3598. [Google Scholar] [CrossRef]
- Sapoval, N.; Aghazadeh, A.; Nute, M.G.; Antunes, D.A.; Balaji, A.; Baraniuk, R.; Barberan, C.J.; Dannenfelser, R.; Dun, C.; Edrisi, M.; et al. Current progress and open challenges for applying deep learning across the biosciences. Nat. Commun. 2022, 13, 1728. [Google Scholar] [CrossRef]
- Filipek, S. Homology Modeling; Springer Science and Business Media LLC.: Dordrecht, The Netherlands, 2012; ISBN 1592592112. [Google Scholar]
- Krebs, B.B.; De Mesquita, J.F. Amyotrophic Lateral Sclerosis Type 20—In Silico Analysis and Molecular Dynamics Simulation of hnRNPA1. PLoS ONE 2016, 11, e0158939. [Google Scholar] [CrossRef]
- Moreira, L.G.A.; Pereira, L.C.; Drummond, P.R.; De Mesquita, J.F. Structural and Functional Analysis of Human SOD1 in Amyotrophic Lateral Sclerosis. PLoS ONE 2013, 8, e81979. [Google Scholar] [CrossRef] [PubMed]
- Bendl, J.; Stourac, J.; Salanda, O.; Pavelka, A.; Wieben, E.D.; Zendulka, J.; Brezovsky, J.; Damborsky, J. PredictSNP: Robust and Accurate Consensus Classifier for Prediction of Disease-Related Mutations. PLOS Comput. Biol. 2014, 10, e1003440. [Google Scholar] [CrossRef]
- Wiederstein, M.; Sippl, M.J. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007, 35, W407–W410. [Google Scholar] [CrossRef]
- Coordinators, N.R. Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2016, 45, D12–D17. [Google Scholar] [CrossRef] [PubMed]
- Schmucker, S.; Argentini, M.; Carelle-Calmels, N.; Martelli, A.; Puccio, H. The in vivo mitochondrial two-step maturation of human frataxin. Hum. Mol. Genet. 2008, 17, 3521–3531. [Google Scholar] [CrossRef] [PubMed]
- Dhe-Paganon, S.; Shigeta, R.; Chi, Y.-I.; Ristow, M.; Shoelson, S.E. Crystal Structure of Human Frataxin. J. Biol. Chem. 2000, 275, 30753–30756. [Google Scholar] [CrossRef]
- Huang, J.; Dizin, E.; Cowan, J.A. Mapping iron binding sites on human frataxin: Implications for cluster assembly on the ISU Fe–S cluster scaffold protein. JBIC J. Biol. Inorg. Chem. 2008, 13, 825–836. [Google Scholar] [CrossRef] [PubMed]
- Cherubini, F.; Serio, D.; Guccini, I.; Fortuni, S.; Arcuri, G.; Condò, I.; Rufini, A.; Moiz, S.; Camerini, S.; Crescenzi, M.; et al. Src inhibitors modulate frataxin protein levels. Hum. Mol. Genet. 2015, 24, 4296–4305. [Google Scholar] [CrossRef]
- De Baets, G.; Van Durme, J.; Reumers, J.; Maurer-Stroh, S.; Vanhee, P.; Dopazo, J.; Schymkowitz, J.; Rousseau, F. SNPeffect 4.0: On-line prediction of molecular and structural effects of protein-coding variants. Nucleic Acids Res. 2011, 40, D935–D939. [Google Scholar] [CrossRef]
- Pereira, G.R.C.; Vieira, B.d.A.A.; De Mesquita, J.F. Comprehensive in silico analysis and molecular dynamics of the superoxide dismutase 1 (SOD1) variants related to amyotrophic lateral sclerosis. PLoS ONE 2021, 16, e0247841. [Google Scholar] [CrossRef] [PubMed]
- Capriotti, E.; Fariselli, P.; Casadio, R. I-Mutant2.0: Predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res. 2005, 33, W306–W310. [Google Scholar] [CrossRef] [PubMed]
- Sherry, S.T.; Ward, M.-H.; Kholodov, M.; Baker, J.; Phan, L.; Smigielski, E.M.; Sirotkin, K. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001, 29, 308–311. [Google Scholar] [CrossRef]
- López-Ferrando, V.; Gazzo, A.; de la Cruz, X.; Orozco, M.; Gelpí, J.L. PMut: A web-based tool for the annotation of pathological variants on proteins, 2017 update. Nucleic Acids Res. 2017, 45, W222–W228. [Google Scholar] [CrossRef] [PubMed]
- Bromberg, Y.; Rost, B. SNAP: Predict effect of non-synonymous polymorphisms on function. Nucleic Acids Res. 2007, 35, 3823–3835. [Google Scholar] [CrossRef] [PubMed]
- Tavares, L.G.; Lopes, H.S.; Lima, C.R.E. Estudo Comparativo de Métodos de Aprendizado de Máquina Na Detecçaoo de Regiões Promotoras de Genes de Escherichia coli. Anais Simpósio Bras. De Inteligência Comput. 2007, 8–11. [Google Scholar]
- Batista, G.E.A.P.A.; Monard, M.C. An analysis of four missing data treatment methods for supervised learning. Appl. Artif. Intell. 2003, 17, 519–533. [Google Scholar] [CrossRef]
- Sanavia, T.; Birolo, G.; Montanucci, L.; Turina, P.; Capriotti, E.; Fariselli, P. Limitations and challenges in protein stability prediction upon genome variations: Towards future applications in precision medicine. Comput. Struct. Biotechnol. J. 2020, 18, 1968–1979. [Google Scholar] [CrossRef] [PubMed]
- Zafar, M.; Khan, H.; Rauf, A.; Khan, A.; Lodhi, M.A. In Silico Study of Alkaloids as α-Glucosidase Inhibitors: Hope for the Discovery of Effective Lead Compounds. Front. Endocrinol. 2016, 7, 153. [Google Scholar] [CrossRef]
- Filho, O.A.S.; de Alencastro, R.B. Modelagem de proteínas por homologia. Quim. Nova 2003, 26, 253–259. [Google Scholar] [CrossRef]
- de Roy, A.; Forano, C.; Besse, J.P. Layered Double Hydroxides: Synthesis and Post-Synthesis Modification. In Layered Double Hydroxides: Present and Future; Nova Science Publisher, Inc.: New York, NY, USA, 2001. [Google Scholar] [CrossRef]
- Silva, L.X.; Bastos, L.L.; Santos, L.H. Modelagem Computacional de Proteínas. Revista Brasileira de Bioinformáticaa, 20 July. [CrossRef]
- Dong, R.; Peng, Z.; Zhang, Y.; Yang, J. mTM-align: An algorithm for fast and accurate multiple protein structure alignment. Bioinformatics 2018, 34, 1719–1725. [Google Scholar] [CrossRef] [PubMed]
- Benkert, P.; Künzli, M.; Schwede, T. QMEAN server for protein model quality estimation. Nucleic Acids Res. 2009, 37, W510–W514. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; Rullmann, J.A.C.; MacArthur, M.W.; Kaptein, R.; Thornton, J.M. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 1996, 8, 477–486. [Google Scholar] [CrossRef] [PubMed]
- Colovos, C.; Yeates, T.O. Verification of protein structures: Patterns of nonbonded atomic interactions. Protein Sci. 1993, 2, 1511–1519. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, D.; Lüthy, R.; Bowie, J.U. VERIFY3D: Assessment of Protein Models with Three-Dimensional Profiles. Methods Enzymol. 1997, 277, 396–404. [Google Scholar] [PubMed]
- Olechnovi, K.; Venclovas, C. VoroMQA: Assessment of Protein Structure Quality Using Interatomic Contact Areas. Proteins Struct. Funct. Bioinforma. 2017, 14, 46. [Google Scholar] [CrossRef]
- Kleywegt, G.J.; Jones, T.A. Model Building and Refinement Practice. Methods Enzymol. 1997, 277, 208–230. [Google Scholar] [CrossRef] [PubMed]
- Karplus, P.A.; Diederichs, K. Linking Crystallographic Model and Data Quality. Science 2012, 336, 1030–1033. [Google Scholar] [CrossRef]
- Choi, M.J.; Torralba, A.; Willsky, A.S. Context models and out-of-context objects. Pattern Recognit. Lett. 2012, 33, 853–862. [Google Scholar] [CrossRef]
- Pereira, G.R.C.; Gonçalves, L.M.; Abrahim-Vieira, B.d.A.; De Mesquita, J.F. In silico analyses of acetylcholinesterase (AChE) and its genetic variants in interaction with the anti-Alzheimer drug Rivastigmine. J. Cell. Biochem. 2022, 123, 1259–1277. [Google Scholar] [CrossRef]
- Layer, G.; Choudens, S.O.-D.; Sanakis, Y.; Fontecave, M. Iron-Sulfur Cluster Biosynthesis. J. Biol. Chem. 2006, 281, 16256–16263. [Google Scholar] [CrossRef] [PubMed]
- Vivas, E.; Skovran, E.; Downs, D.M. Salmonella enterica Strains Lacking the Frataxin Homolog CyaY Show Defects in Fe-S Cluster Metabolism In Vivo. J. Bacteriol. 2006, 188, 1175–1179. [Google Scholar] [CrossRef] [PubMed]
- Knapp, B.; Frantal, S.; Cibena, M.; Schreiner, W.; Bauer, P. Is an Intuitive Convergence Definition of Molecular Dynamics Simulations Solely Based on the Root Mean Square Deviation Possible? J. Comput. Biol. 2011, 18, 997–1005. [Google Scholar] [CrossRef] [PubMed]
- Alvi, S.S.; Iqbal, D.; Ahmad, S.; Khan, M.S. Molecular rationale delineating the role of lycopene as a potent HMG-CoA reductase inhibitor: In vitro and in silico study. Nat. Prod. Res. 2015, 30, 2111–2114. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.-L.; Bridwell-Rabb, J.; Barondeau, D.P. Friedreich’s Ataxia Variants I154F and W155R Diminish Frataxin-Based Activation of the Iron–Sulfur Cluster Assembly Complex. Biochemistry 2011, 50, 6478–6487. [Google Scholar] [CrossRef] [PubMed]
- Koutnikova, H.; Campuzano, V.; Koenig, M. Maturation of wild-type and mutated frataxin by the mitochondrial processing peptidase. Hum. Mol. Genet. 1998, 7, 1485–1489. [Google Scholar] [CrossRef] [PubMed]
- Bahar, I.; Chennubhotla, C.; Tobi, D. Intrinsic dynamics of enzymes in the unbound state and relation to allosteric regulation. Curr. Opin. Struct. Biol. 2007, 17, 633–640. [Google Scholar] [CrossRef] [PubMed]
- Shaw, D.E.; Maragakis, P.; Lindorff-Larsen, K.; Piana, S.; Dror, R.O.; Eastwood, M.P.; Bank, J.A.; Jumper, J.M.; Salmon, J.K.; Shan, Y.; et al. Atomic-Level Characterization of the Structural Dynamics of Proteins. Science 2010, 330, 341–346. [Google Scholar] [CrossRef]
- Martínez, L. Automatic Identification of Mobile and Rigid Substructures in Molecular Dynamics Simulations and Fractional Structural Fluctuation Analysis. PLoS ONE 2015, 10, e0119264. [Google Scholar] [CrossRef]
- Sinha, S.; Verma, S.; Singh, A.; Somvanshi, P.; Grover, A. Simulation Based Investigation of Deleterious nsSNPs in ATXN2 Gene and Its Structural Consequence Toward Spinocerebellar Ataxia. J. Cell. Biochem. 2017, 119, 499–510. [Google Scholar] [CrossRef]
- Wang, Q.; Mehmood, A.; Wang, H.; Xu, Q.; Xiong, Y.; Wei, D.-Q. Computational Screening and Analysis of Lung Cancer Related Non-Synonymous Single Nucleotide Polymorphisms on the Human Kirsten Rat Sarcoma Gene. Molecules 2019, 24, 1951. [Google Scholar] [CrossRef] [PubMed]
- Craveur, P.; Joseph, A.P.; Esque, J.; Narwani, T.J.; Noã«L, F.; Shinada, N.; Goguet, M.; Leonard, S.; Poulain, P.; Bertrand, O.; et al. Protein flexibility in the light of structural alphabets. Front. Mol. Biosci. 2015, 2, 20. [Google Scholar] [CrossRef] [PubMed]
- Condò, I.; Ventura, N.; Malisan, F.; Rufini, A.; Tomassini, B.; Testi, R. In vivo maturation of human frataxin. Hum. Mol. Genet. 2007, 16, 1534–1540. [Google Scholar] [CrossRef] [PubMed]
- Puccio, H. Recent advances in the molecular pathogenesis of Friedreich ataxia. Hum. Mol. Genet. 2000, 9, 887–892. [Google Scholar] [CrossRef] [PubMed]
- Branda, S.S.; Cavadini, P.; Adamec, J.; Kalousek, F.; Taroni, F.; Isaya, G. Yeast and Human Frataxin Are Processed to Mature Form in Two Sequential Steps by the Mitochondrial Processing Peptidase. J. Biol. Chem. 1999, 274, 22763–22769. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, H.A.; Gakh, O.; Isaya, G. Supramolecular Assemblies of Human Frataxin are Formed via Subunit–Subunit Interactions Mediated by a Non-conserved Amino-terminal Region. J. Mol. Biol. 2004, 345, 433–439. [Google Scholar] [CrossRef] [PubMed]
- Gakh, O.; Park, S.; Liu, G.; Macomber, L.; Imlay, J.A.; Ferreira, G.C.; Isaya, G. Mitochondrial iron detoxification is a primary function of frataxin that limits oxidative damage and preserves cell longevity. Hum. Mol. Genet. 2005, 15, 467–479. [Google Scholar] [CrossRef] [PubMed]
- Aloria, K.; Schilke, B.; Andrew, A.; A Craig, E. Iron-induced oligomerization of yeast frataxin homologue Yfh1 is dispensable in vivo. Embo Rep. 2004, 5, 1096–1101. [Google Scholar] [CrossRef] [PubMed]
- Gordon, D.M.; Shi, Q.; Dancis, A.; Pain, D. Maturation of Frataxin Within Mammalian and Yeast Mitochondria: One-Step Processing by Matrix Processing Peptidase. Hum. Mol. Genet. 1999, 8, 2255–2262. [Google Scholar] [CrossRef]
- Kempf, J.G.; Loria, J.P. Protein Dynamics from Solution NMR. Cell Biochem. Biophys. 2002, 37, 187–212. [Google Scholar] [CrossRef]
- Spyrakis, F.; BidonChanal, A.; Barril, X.; Luque, F.J. Protein Flexibility and Ligand Recognition: Challenges for Molecular Modeling. Curr. Top. Med. Chem. 2011, 11, 192–210. [Google Scholar] [CrossRef] [PubMed]
- Forrey, C.; Douglas, J.F.; Gilson, M.K. The fundamental role of flexibility on the strength of molecular binding. Soft Matter. 2012, 8, 6385–6392. [Google Scholar] [CrossRef] [PubMed]
- Musco, G.; Stier, G.; Kolmerer, B.; Adinolfi, S.; Martin, S.; Frenkiel, T.; Gibson, T.; Pastore, A. Towards a structural understanding of Friedreich’s ataxia: The solution structure of frataxin. Structure 2000, 8, 695–707. [Google Scholar] [CrossRef] [PubMed]
- Correia, A.R.; Adinolfi, S.; Pastore, A.; Gomes, C.M. Conformational stability of human frataxin and effect of Friedreich’s ataxia-related mutations on protein folding. Biochem. J. 2006, 398, 605–611. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Gakh, O.; Smith, D.Y.; Ranatunga, W.K.; Isaya, G. Missense Mutations Linked to Friedreich Ataxia Have Different but Synergistic Effects on Mitochondrial Frataxin Isoforms. J. Biol. Chem. 2013, 288, 4116–4127. [Google Scholar] [CrossRef] [PubMed]
- UniProt Consortium. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef] [PubMed]
- Amberger, J.S.; Bocchini, C.A.; Scott, A.F.; Hamosh, A. OMIM.org: Leveraging knowledge across phenotype–gene relationships. Nucleic Acids Res. 2019, 47, D1038–D1043. [Google Scholar] [CrossRef] [PubMed]
- Ashkenazy, H.; Abadi, S.; Martz, E.; Chay, O.; Mayrose, I.; Pupko, T.; Ben-Tal, N. ConSurf 2016: An improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016, 44, W344–W350. [Google Scholar] [CrossRef] [PubMed]
- Gajula, K.S.; Huwe, P.J.; Mo, C.Y.; Crawford, D.J.; Stivers, J.T.; Radhakrishnan, R.; Kohli, R.M. High-throughput mutagenesis reveals functional determinants for DNA targeting by activation-induced deaminase. Nucleic Acids Res. 2014, 42, 9964–9975. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Vieira, I.H.P.; Botelho, E.B.; Gomes, T.J.d.S.; Kist, R.; Caceres, R.A.; Zanchi, F.B. Visual dynamics: A WEB application for molecular dynamics simulation using GROMACS. BMC Bioinform. 2023, 24, 107. [Google Scholar] [CrossRef]
- Frezza, E.; Martin, J.; Lavery, R. A molecular dynamics study of adenylyl cyclase: The impact of ATP and G-protein binding. PLoS ONE 2018, 13, e0196207. [Google Scholar] [CrossRef] [PubMed]
- Yang, M. Molecular dynamics simulations: Chemical advances and applications. J. Phys. Conf. Ser. 2023, 2608, 012044. [Google Scholar] [CrossRef]
- Namba, A.M.; da Silva, V.B.; da Silva, C.H.T.P. Dinâmica molecular: Teoria e aplicações em planejamento de fármacos. Eclet. Quim. 2008, 33, 13–24. [Google Scholar] [CrossRef]
- Grant, B.J.; Rodrigues, A.P.; ElSawy, K.M.; McCammon, J.A.; Caves, L.S. Bio3d: An R package for the comparative analysis of protein structures. Bioinformatics 2006, 22, 2695–2696. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. ggplot2. Wiley Interdiscip. Rev. Comput. Stat. 2011, 3, 180–185. [Google Scholar]
- Goddard, T.D.; Huang, C.C.; Ferrin, T.E. Software Extensions to UCSF Chimera for Interactive Visualization of Large Molecular Assemblies. Structure 2005, 13, 473–482. [Google Scholar] [CrossRef]
- Zhang, Y.; Skolnick, J. TM-Align: A protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005, 33, 2302–2309. [Google Scholar] [CrossRef] [PubMed]
- Capriotti, E.; Calabrese, R.; Fariselli, P.; Martelli, P.L.; Altman, R.B.; Casadio, R. WS-SNPs&GO: A web server for predicting the deleterious effect of human protein variants using functional annotation. BMC Genom. 2013, 14 (Suppl. S3), S6. [Google Scholar] [CrossRef]
- Kumar, P.; Henikoff, S.; Ng, P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009, 4, 1073–1081. [Google Scholar] [CrossRef]
- Adzhubei, I.; Jordan, D.M.; Sunyaev, S.R. Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet. 2013, 76, 7–20. [Google Scholar] [CrossRef] [PubMed]
- Capriotti, E.; Calabrese, R.; Casadio, R. Predicting the insurgence of human genetic diseases associated to single point protein mutations with support vector machines and evolutionary information. Bioinformatics 2006, 22, 2729–2734. [Google Scholar] [CrossRef] [PubMed]
- Pejaver, V.; Urresti, J.; Lugo-Martinez, J.; Pagel, K.A.; Lin, G.N.; Nam, H.-J.; Mort, M.; Cooper, D.N.; Sebat, J.; Iakoucheva, L.M.; et al. Inferring the molecular and phenotypic impact of amino acid variants with MutPred2. Nat. Commun. 2020, 11, 5918. [Google Scholar] [CrossRef]
- Shan, Y.; Guan, D.; Liu, J.; Mi, Z.; Liu, Z.; Liu, J.; Schroeder, H.; Cai, B.; Chen, Y.; Shao, S.; et al. Methodology and applications of city level CO2 emission accounts in China. J. Clean. Prod. 2017, 161, 1215–1225. [Google Scholar] [CrossRef]









| Algorithm | Model | Model Size | Folding |
|---|---|---|---|
| Rosetta | 1 | 210 | complete |
| Rosetta | 2 | 210 | complete |
| Rosetta | 3 | 210 | complete |
| Rosetta | 4 | 210 | complete |
| Rosetta | 5 | 210 | complete |
| I-Tasser * | 1 | 210 | incomplete |
| I-Tasser * | 2 | 210 | incomplete |
| I-Tasser * | 3 | 210 | incomplete |
| I-Tasser * | 4 | 210 | incomplete |
| I-Tasser * | 5 | 210 | incomplete |
| Raptor-X * | 1 | 210 | incomplete |
| MholLine * | 1 | 210 | incomplete |
| Swiss Model * | 1 | 168 | complete |
| Algorithm | Model | RMSD | TM Score |
|---|---|---|---|
| Rosetta | 1 | 0.59 | 0.98055 |
| Rosetta | 2 | 0.67 | 0.97807 |
| Rosetta | 3 | 0.59 | 0.98093 |
| Rosetta | 4 | 1.53 | 0.90771 |
| Rosetta | 5 | 0.70 | 0.97564 |
| Model | ERRAT 1 | PROCHECK 2 | Verify-3D 3 | Prosa-Web 4 | QMEAN 5 | VoroMQA 6 |
|---|---|---|---|---|---|---|
| Rosetta1 | 99 | 86 | 80 | NMR | high resolution | 0.43 |
| Rosetta2 | 99 | 84 | 77 | NMR | high resolution | 0.43 |
| Rosetta3 | 95 | 87 | 86 | NMR | high resolution | 0.42 |
| Rosetta4 | 96 | 91 | 80 | NMR | high resolution | 0.40 |
| Rosetta5 | 98 | 86 | 83 | NMR | high resolution | 0.42 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Da Conceição, L.M.A.; Cabral, L.M.; Pereira, G.R.C.; De Mesquita, J.F. An In Silico Analysis of Genetic Variants and Structural Modeling of the Human Frataxin Protein in Friedreich’s Ataxia. Int. J. Mol. Sci. 2024, 25, 5796. https://doi.org/10.3390/ijms25115796
Da Conceição LMA, Cabral LM, Pereira GRC, De Mesquita JF. An In Silico Analysis of Genetic Variants and Structural Modeling of the Human Frataxin Protein in Friedreich’s Ataxia. International Journal of Molecular Sciences. 2024; 25(11):5796. https://doi.org/10.3390/ijms25115796
Chicago/Turabian StyleDa Conceição, Loiane Mendonça Abrantes, Lucio Mendes Cabral, Gabriel Rodrigues Coutinho Pereira, and Joelma Freire De Mesquita. 2024. "An In Silico Analysis of Genetic Variants and Structural Modeling of the Human Frataxin Protein in Friedreich’s Ataxia" International Journal of Molecular Sciences 25, no. 11: 5796. https://doi.org/10.3390/ijms25115796
APA StyleDa Conceição, L. M. A., Cabral, L. M., Pereira, G. R. C., & De Mesquita, J. F. (2024). An In Silico Analysis of Genetic Variants and Structural Modeling of the Human Frataxin Protein in Friedreich’s Ataxia. International Journal of Molecular Sciences, 25(11), 5796. https://doi.org/10.3390/ijms25115796




