Development of a Normal Porcine Cell Line Growing in a Heme-Supplemented, Serum-Free Condition for Cultured Meat
Abstract
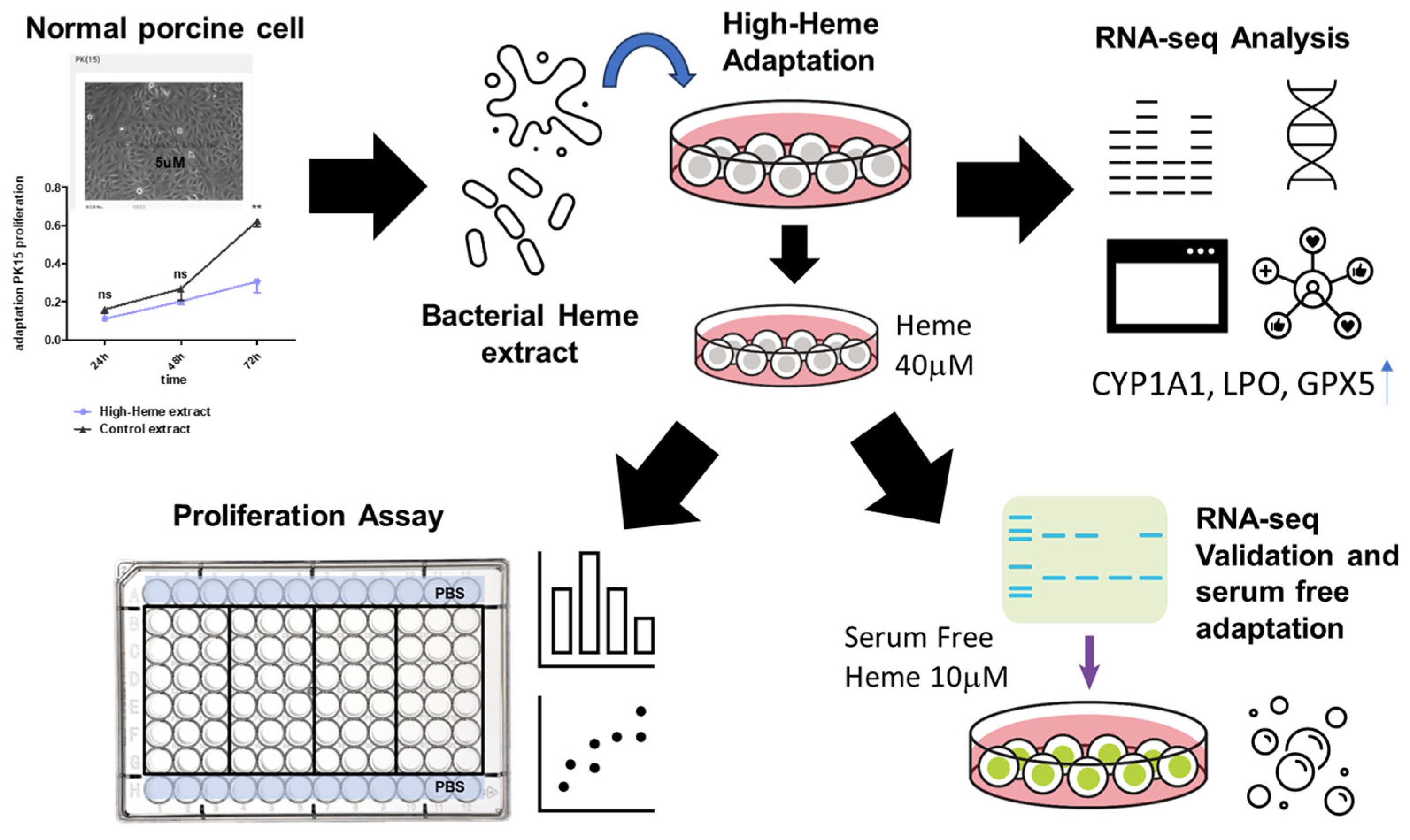
1. Introduction
2. Results
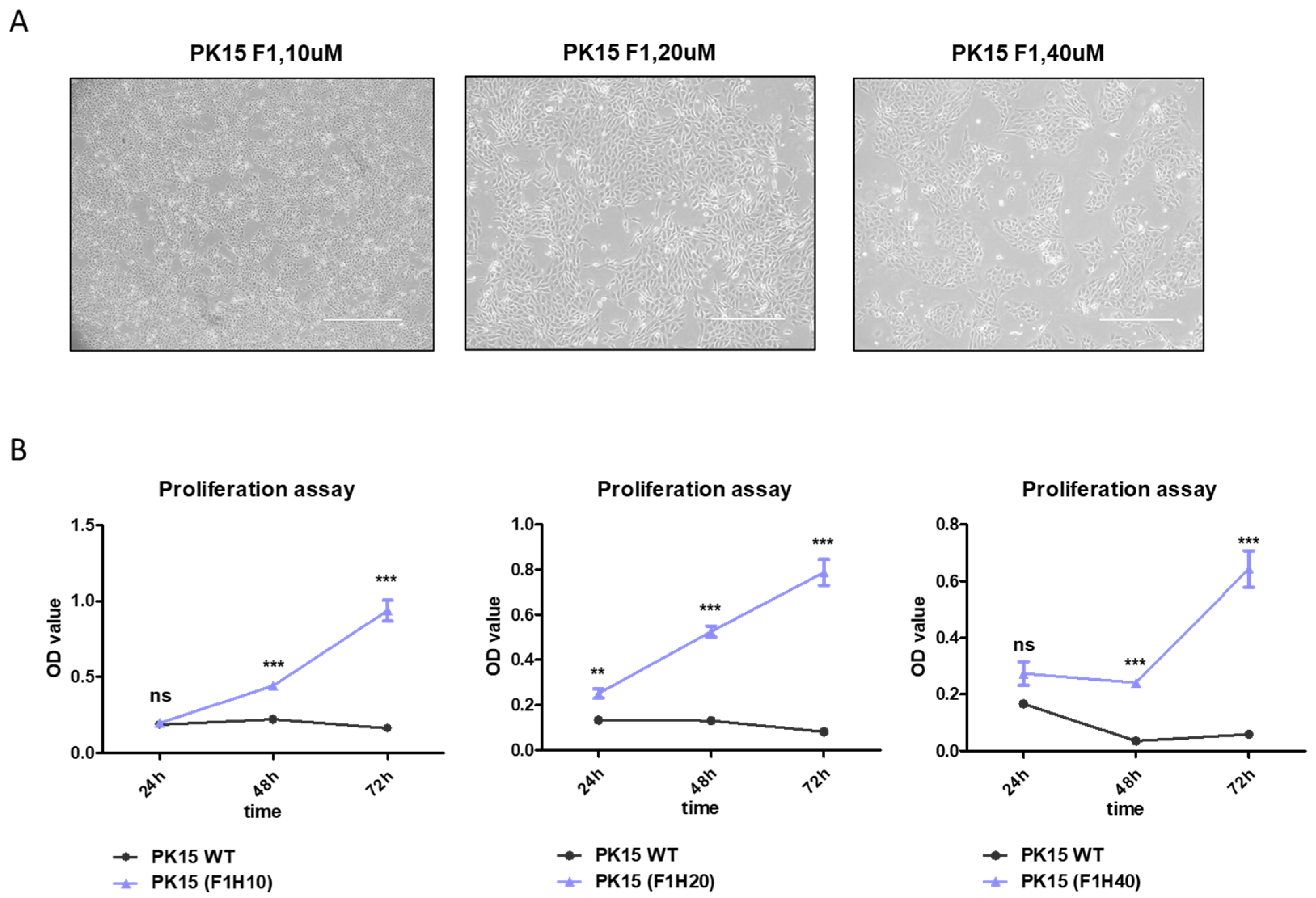
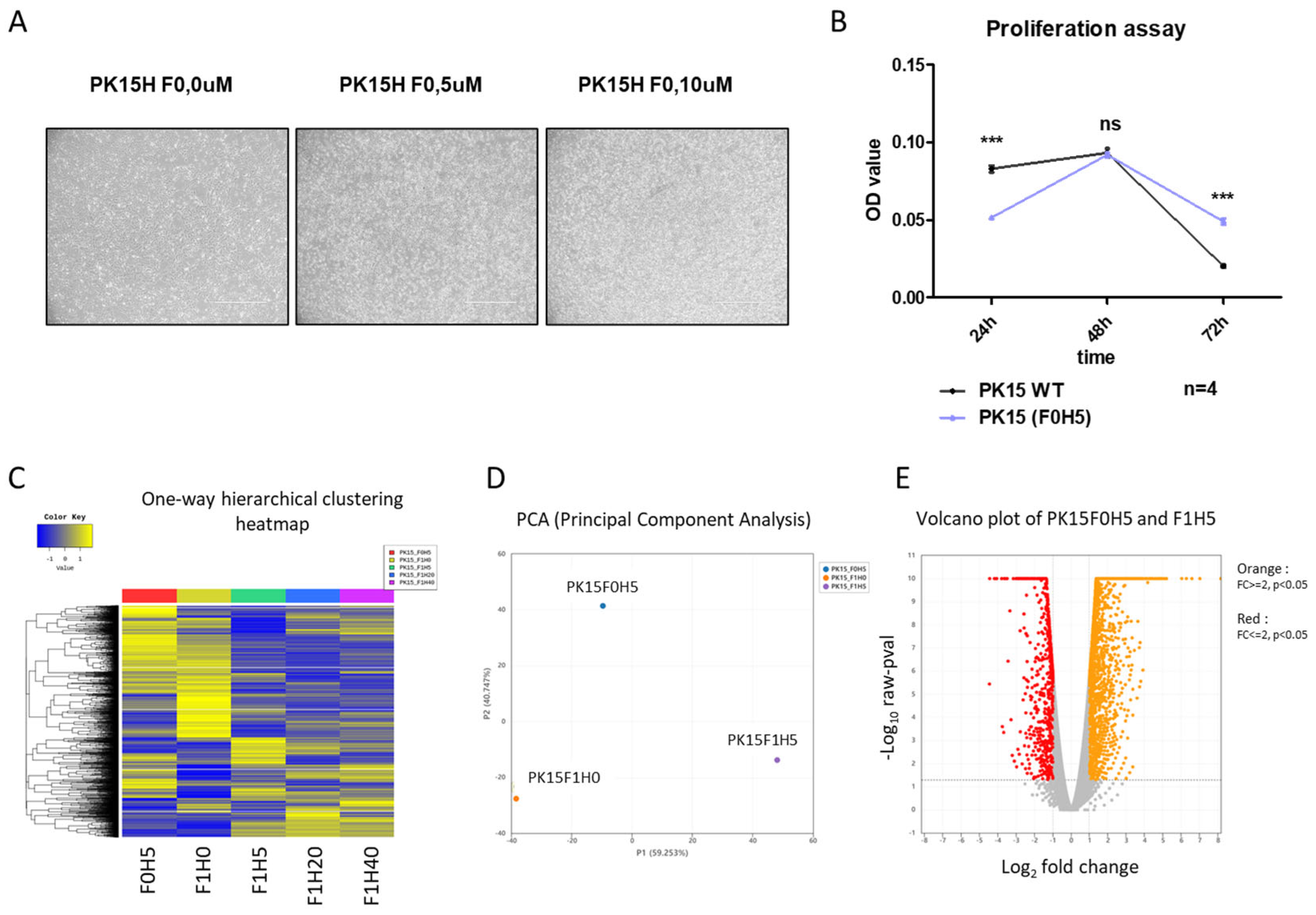
2.1. Culture of PK15 Cells under a Serial Increase of Heme Extract Concentrations Generates a Heme-Adapted Subculture
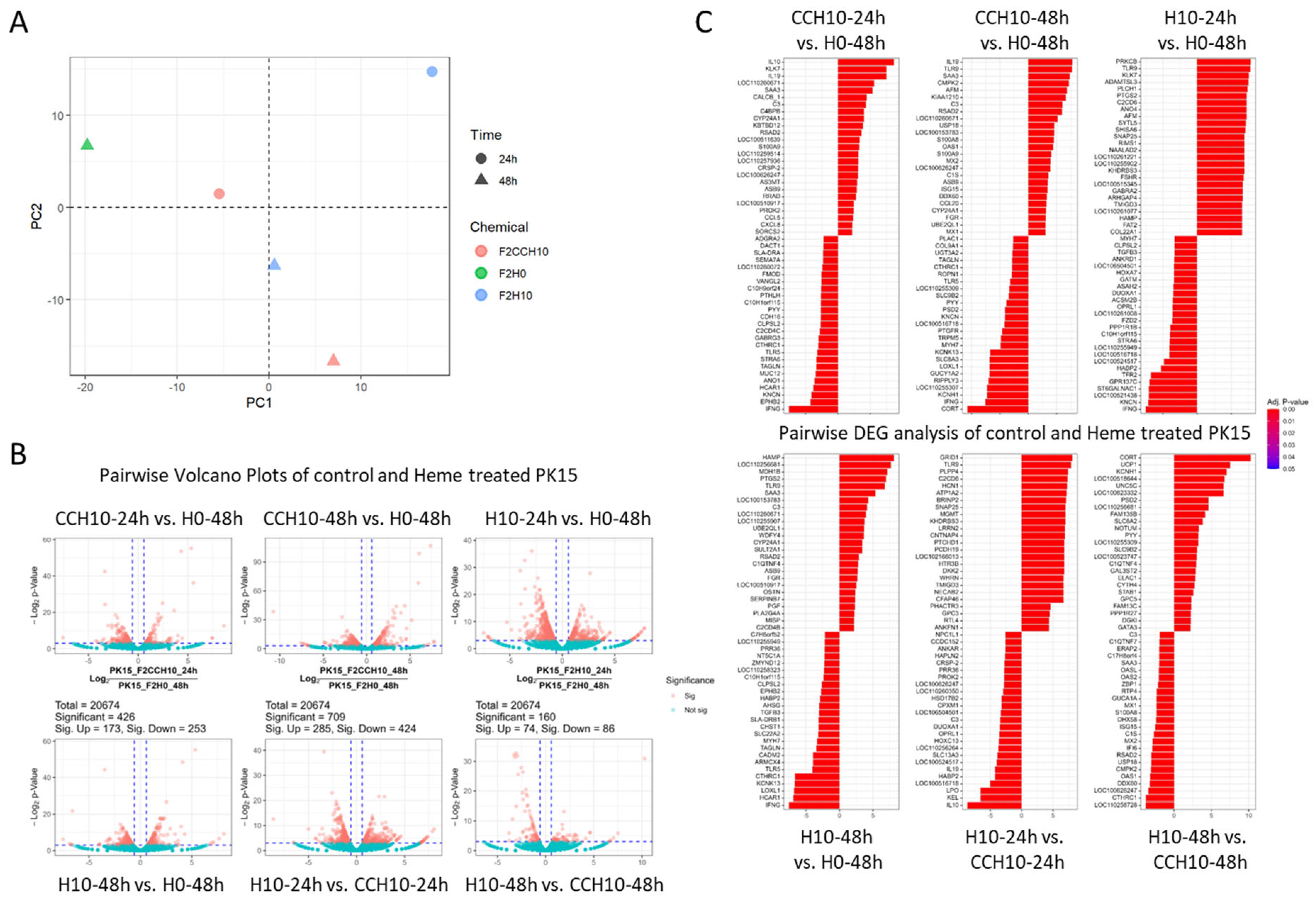
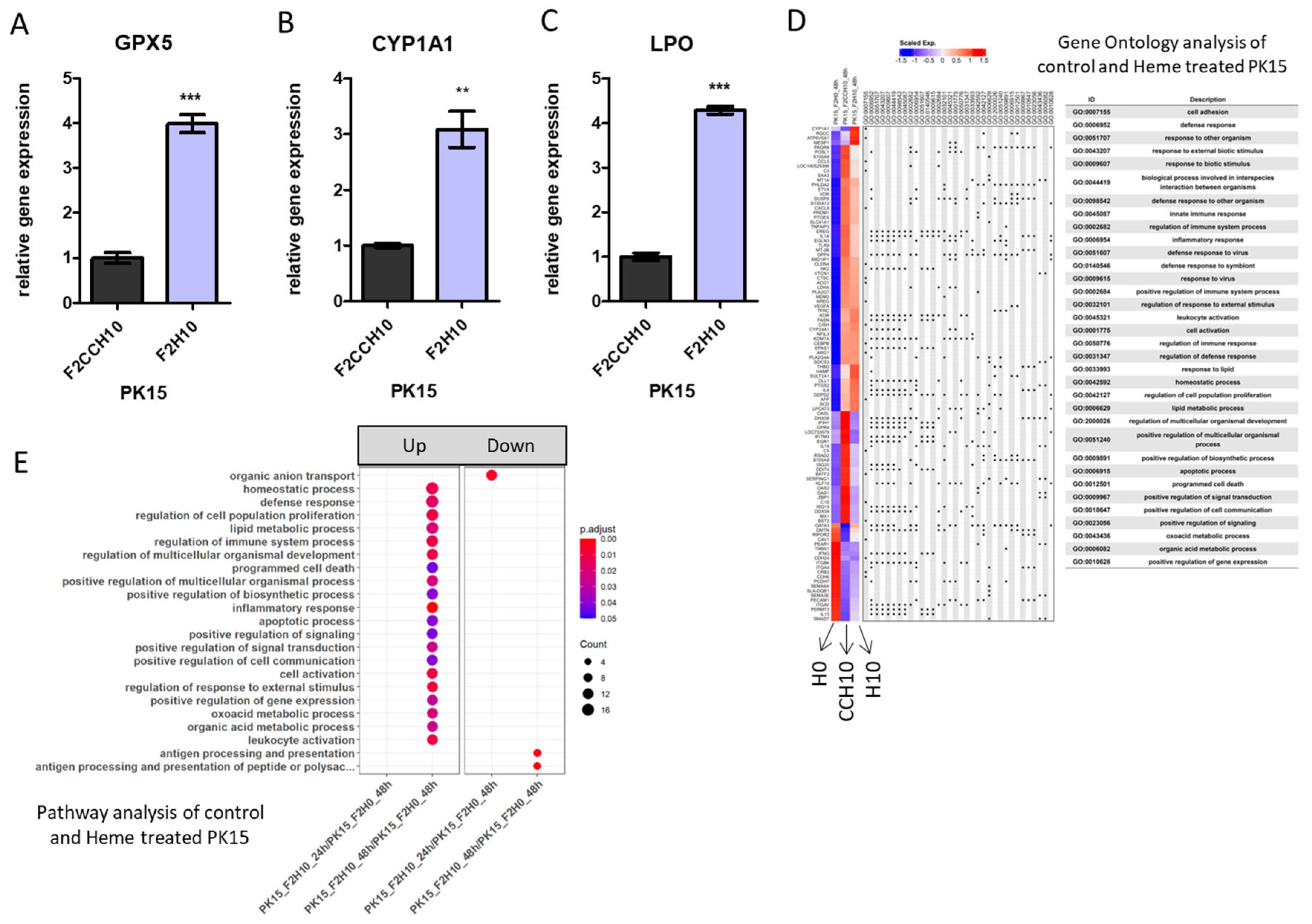
2.2. RNA-seq Analysis Reveals an Increased Apoptotic Process in Heme Extract-Treated PK15 Cells
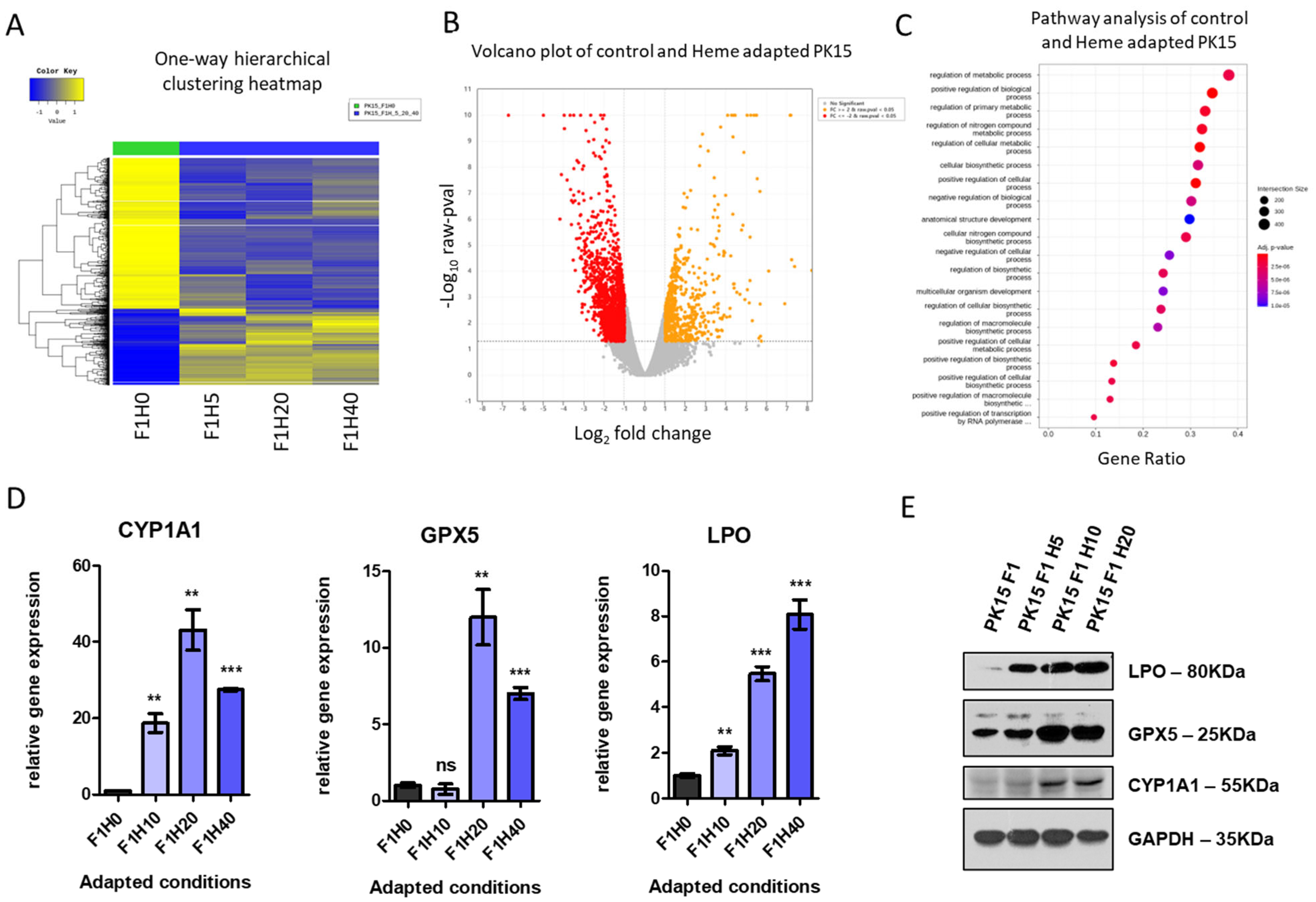
2.3. Heme-Adapted PK15 Cells Display a Distinct Expression Signature
2.4. Bacterial Extract with High-Heme Content Transiently Supports PK15 Cell Growth in Serum-Free Media
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Media
4.2. Heme and Serum-Free Adaptation Culture
4.3. Cell Proliferation Assay
4.4. Heme Sonication and Quantitation
4.5. Protein Extraction and Western Blotting
4.6. RNA Preparation and RT-PCR
4.7. Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hong, T.K.; Shin, D.M.; Choi, J.; Do, J.T.; Han, S.G. Current Issues and Technical Advances in Cultured Meat Production: A Review. Food Sci. Anim. Resour. 2021, 41, 355–372. [Google Scholar] [CrossRef] [PubMed]
- Pakseresht, A.; Ahmadi Kaliji, S.; Canavari, M. Review of factors affecting consumer acceptance of cultured meat. Appetite 2022, 170, 105829. [Google Scholar] [CrossRef] [PubMed]
- Humbird, D. Scale-up economics for cultured meat. Biotechnol. Bioeng. 2021, 118, 3239–3250. [Google Scholar] [CrossRef] [PubMed]
- Gomez Romero, S.; Boyle, N. Systems biology and metabolic modeling for cultivated meat: A promising approach for cell culture media optimization and cost reduction. Compr. Rev. Food Sci. Food Saf. 2023, 22, 3422–3443. [Google Scholar] [CrossRef]
- Yamanaka, K.; Haraguchi, Y.; Takahashi, H.; Kawashima, I.; Shimizu, T. Development of serum-free and grain-derived-nutrient-free medium using microalga-derived nutrients and mammalian cell-secreted growth factors for sustainable cultured meat production. Sci. Rep. 2023, 13, 498. [Google Scholar] [CrossRef] [PubMed]
- Stout, A.J.; Mirliani, A.B.; Rittenberg, M.L.; Shub, M.; White, E.C.; Yuen, J.S.K., Jr.; Kaplan, D.L. Simple and effective serum-free medium for sustained expansion of bovine satellite cells for cell cultured meat. Commun. Biol. 2022, 5, 466. [Google Scholar] [CrossRef]
- Defendi-Cho, G.; Gould, T.M. In vitro culture of bovine fibroblasts using select serum-free media supplemented with Chlorella vulgaris extract. BMC Biotechnol. 2023, 23, 4. [Google Scholar] [CrossRef] [PubMed]
- Stout, A.J.; Rittenberg, M.L.; Shub, M.; Saad, M.K.; Mirliani, A.B.; Dolgin, J.; Kaplan, D.L. A Beefy-R culture medium: Replacing albumin with rapeseed protein isolates. Biomaterials 2023, 296, 122092. [Google Scholar] [CrossRef] [PubMed]
- Fraeye, I.; Kratka, M.; Vandenburgh, H.; Thorrez, L. Sensorial and Nutritional Aspects of Cultured Meat in Comparison to Traditional Meat: Much to Be Inferred. Front. Nutr. 2020, 7, 35. [Google Scholar] [CrossRef]
- Simsa, R.; Yuen, J.; Stout, A.; Rubio, N.; Fogelstrand, P.; Kaplan, D.L. Extracellular Heme Proteins Influence Bovine Myosatellite Cell Proliferation and the Color of Cell-Based Meat. Foods 2019, 8, 521. [Google Scholar] [CrossRef]
- Kumar, S.; Bandyopadhyay, U. Free heme toxicity and its detoxification systems in human. Toxicol. Lett. 2005, 157, 175–188. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.I.; Park, J.; Kim, P. Heme Derived from Corynebacterium glutamicum: A Potential Iron Additive for Swine and an Electron Carrier Additive for Lactic Acid Bacterial Culture. J. Microbiol. Biotechnol. 2017, 27, 500–506. [Google Scholar] [CrossRef] [PubMed]
- Ko, Y.J.; Kim, M.; You, S.K.; Shin, S.K.; Chang, J.; Choi, H.J.; Jeong, W.Y.; Lee, M.E.; Hwang, D.H.; Han, S.O. Animal-free heme production for artificial meat in Corynebacterium glutamicum via systems metabolic and membrane engineering. Metab. Eng. 2021, 66, 217–228. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Choi, A.; Park, K.H.; Lee, S.; Yoon, H.; Kim, P. Single-cell hemoprotein (heme-SCP) exerts the prebiotic potential to establish a healthy gut microbiota in small pet dogs. Food Sci. Biotechnol. 2023, 32, 489–496. [Google Scholar] [CrossRef] [PubMed]
- Dulac, G.C.; Afshar, A. Porcine circovirus antigens in PK-15 cell line (ATCC CCL-33) and evidence of antibodies to circovirus in Canadian pigs. Can. J. Vet. Res. 1989, 53, 431–433. [Google Scholar]
- Wang, D.D.; Liu, Y.; Li, N.; Zhang, Y.; Jin, Q.; Hao, D.C.; Piao, H.L.; Dai, Z.R.; Ge, G.B.; Yang, L. Induction of CYP1A1 increases gefitinib-induced oxidative stress and apoptosis in A549 cells. Toxicol Vitr. 2017, 44, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Taylor, A.; Robson, A.; Houghton, B.C.; Jepson, C.A.; Ford, W.C.; Frayne, J. Epididymal specific, selenium-independent GPX5 protects cells from oxidative stress-induced lipid peroxidation and DNA mutation. Hum. Reprod. 2013, 28, 2332–2342. [Google Scholar] [CrossRef] [PubMed]
- Nasralla, S.N.; Ghoneim, A.I.; Khalifa, A.E.; Gad, M.Z.; Abdel-Naim, A.B. Lactoperoxidase catalyzes in vitro activation of acrylonitrile to cyanide. Toxicol. Lett. 2009, 191, 347–352. [Google Scholar] [CrossRef] [PubMed]
- Kaliman, P.A.; Barannik, T.B. Heme metabolism and oxidative stress. Ukr. Kyi Biokhimichnyi Zhurnal 2001, 73, 5–15. [Google Scholar]
- Jang, M.; Pete, E.S.; Bruheim, P. The impact of serum-free culture on HEK293 cells: From the establishment of suspension and adherent serum-free adaptation cultures to the investigation of growth and metabolic profiles. Front. Bioeng. Biotechnol. 2022, 10, 964397. [Google Scholar] [CrossRef]
- Ozturk, S.; Kaseko, G.; Mahaworasilpa, T.; Coster, H.G. Adaptation of cell lines to serum-free culture medium. Hybrid. Hybridomics 2003, 22, 267–272. [Google Scholar] [CrossRef] [PubMed]
- Abraham, N.G.; Lavrovsky, Y.; Schwartzman, M.L.; Stoltz, R.A.; Levere, R.D.; Gerritsen, M.E.; Shibahara, S.; Kappas, A. Transfection of the human heme oxygenase gene into rabbit coronary microvessel endothelial cells: Protective effect against heme and hemoglobin toxicity. Proc. Natl. Acad. Sci. USA 1995, 92, 6798–6802. [Google Scholar] [CrossRef] [PubMed]
- Ingoglia, G.; Sag, C.M.; Rex, N.; De Franceschi, L.; Vinchi, F.; Cimino, J.; Petrillo, S.; Wagner, S.; Kreitmeier, K.; Silengo, L.; et al. Hemopexin counteracts systolic dysfunction induced by heme-driven oxidative stress. Free Radic Biol. Med. 2017, 108, 452–464. [Google Scholar] [CrossRef] [PubMed]
- Chassaing, S.; Collin, F.; Dorlet, P.; Gout, J.; Hureau, C.; Faller, P. Copper and heme-mediated Abeta toxicity: Redox chemistry, Abeta oxidations and anti-ROS compounds. Curr. Top. Med. Chem. 2012, 12, 2573–2595. [Google Scholar] [CrossRef] [PubMed]
- Spink, D.C.; Katz, B.H.; Hussain, M.M.; Spink, B.C.; Wu, S.J.; Liu, N.; Pause, R.; Kaminsky, L.S. Induction of CYP1A1 and CYP1B1 in T-47D human breast cancer cells by benzo[a]pyrene is diminished by arsenite. Drug Metab. Dispos. 2002, 30, 262–269. [Google Scholar] [CrossRef]
- Seubert, J.M.; Webb, C.D.; Bend, J.R. Acute sodium arsenite treatment induces Cyp2a5 but not Cyp1a1 in the C57Bl/6 mouse in a tissue (kidney) selective manner. J. Biochem. Mol. Toxicol. 2002, 16, 96–106. [Google Scholar] [CrossRef] [PubMed]
- Elbekai, R.H.; El-Kadi, A.O. Transcriptional activation and posttranscriptional modification of Cyp1a1 by arsenite, cadmium, and chromium. Toxicol. Lett. 2007, 172, 106–119. [Google Scholar] [CrossRef] [PubMed]
- Barouki, R.; Morel, Y. Repression of cytochrome P450 1A1 gene expression by oxidative stress: Mechanisms and biological implications. Biochem. Pharmacol. 2001, 61, 511–516. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.L.; Li, H.; Wu, D.C.; Wang, X.W.; Chen, X.Y.; Kong, Q.Y.; Ma, J.X.; Gao, Y.; Liu, J. CYP1A1 and CYP1B1 expressions in medulloblastoma cells are AhR-independent and have no direct link with resveratrol-induced differentiation and apoptosis. Neurosci. Lett. 2005, 384, 33–37. [Google Scholar] [CrossRef]
- Lingappan, K.; Jiang, W.; Wang, L.; Wang, G.; Couroucli, X.I.; Shivanna, B.; Welty, S.E.; Barrios, R.; Khan, M.F.; Nebert, D.W.; et al. Mice deficient in the gene for cytochrome P450 (CYP)1A1 are more susceptible than wild-type to hyperoxic lung injury: Evidence for protective role of CYP1A1 against oxidative stress. Toxicol. Sci. 2014, 141, 68–77. [Google Scholar] [CrossRef]
- Reszka, K.J.; Matuszak, Z.; Chignell, C.F.; Dillon, J. Oxidation of biological electron donors and antioxidants by a reactive lactoperoxidase metabolite from nitrite (NO2−): An EPR and spin trapping study. Free Radic Biol. Med. 1999, 26, 669–678. [Google Scholar] [CrossRef]
- Hanstrom, L.; Johansson, A.; Carlsson, J. Lactoperoxidase and thiocyanate protect cultured mammalian cells against hydrogen peroxide toxicity. Med. Biol. 1983, 61, 268–274. [Google Scholar] [PubMed]
- Di Rocco, G.; Bernini, F.; Battistuzzi, G.; Ranieri, A.; Bortolotti, C.A.; Borsari, M.; Sola, M. Hydrogen peroxide induces heme degradation and protein aggregation in human neuroglobin: Roles of the disulfide bridge and hydrogen-bonding in the distal heme cavity. FEBS J. 2023, 290, 148–161. [Google Scholar] [CrossRef] [PubMed]
- Pasitka, L.; Cohen, M.; Ehrlich, A.; Gildor, B.; Reuveni, E.; Ayyash, M.; Wissotsky, G.; Herscovici, A.; Kaminker, R.; Niv, A.; et al. Spontaneous immortalization of chicken fibroblasts generates stable, high-yield cell lines for serum-free production of cultured meat. Nat. Food 2023, 4, 35–50. [Google Scholar] [CrossRef] [PubMed]
- Pazos, P.; Boveri, M.; Gennari, A.; Casado, J.; Fernandez, F.; Prieto, P. Culturing cells without serum: Lessons learnt using molecules of plant origin. ALTEX 2004, 21, 67–72. [Google Scholar] [PubMed]
- Cox, R.P.; Gesner, B.M. Studies on the effects of simple sugars on mammalian cells in culture and characterization of the inhibition of 3T3 fibroblasts by L-fucose. Cancer Res. 1968, 28, 1162–1172. [Google Scholar]
- Ukeda, H.; Kawana, D.; Maeda, S.; Sawamura, M. Spectrophotometric Assay for Superoxide Dismutase Based on the Reduction of Highly Water-soluble Tetrazolium Salts by Xanthine-Xanthine Oxidase. Biosci. Biotechnol. Biochem. 1999, 63, 485–488. [Google Scholar] [CrossRef]

| Gene_ID | Transcript_ID | Gene_Symbol | Description | PK15_F1H_5_20_40/PK15_F1H0.fc | PK15_F1H_5_20_40/PK15_F1H0.logCPM | PK15_F1H_5_20_40/PK15_F1H0.raw.pval | PK15_F1H_5_20_40/PK15_F1H0.bh.pval |
|---|---|---|---|---|---|---|---|
| 100049691 | NM_001098597 | OSTN | osteocrin | 142.744964 | 6.229014 | 4.9843 × 10−33 | 7.5307 × 10−29 |
| 100520218 | XM_003132298,XM_013981851,XM_013981853,XM_013981854,XM_013981855,XM_021070779,XM_021070780,XM_021070781 | ADAMTS9 | ADAM metallopeptidase with thrombospondin type 1 motif 9, transcript variant X1 | −104.994789 | 1.934609 | 1.2212 × 10−28 | 9.2253 × 10−25 |
| 100152394 | NM_001244640,NM_001244641 | RTN1 | reticulon 1 | 20.525415 | 5.414605 | 3.2805 × 10−20 | 1.6522 × 10−16 |
| 100513560 | XM_013979738,XM_013979739,XM_013979740,XM_013979741,XM_021063401,XM_021063402,XM_021063403 | ETV1 | ETS variant 1, transcript variant X7 | 16.908247 | 6.160216 | 1.4798 × 10−19 | 5.5895 × 10−16 |
| 100737483 | XM_021091686 | LOC100737483 | keratin, type II cytoskeletal 6A | 45.172367 | 5.278455 | 3.7805 × 10−18 | 1.1424 × 10−14 |
| 403103 | NM_214412,XM_021098317 | CYP1A1 | cytochrome P450 1A1 | 21.822831 | 4.754509 | 1.0019 × 10−17 | 2.523 × 10−14 |
| 110260665 | XM_021091687 | LOC110260665 | keratin, type II cytoskeletal 6A-like | 32.843664 | 6.004388 | 4.9299 × 10−17 | 1.0641 × 10−13 |
| 100624115 | XM_021100590 | ADGRL3 | adhesion G protein-coupled receptor L3 | −6.342352 | 5.136530 | 1.3642 × 10−16 | 2.5765 × 10−13 |
| 106507259 | XR_002343949 | LOC106507259 | keratin, type II cytoskeletal 6A-like | 41.605454 | 4.962795 | 1.7087 × 10−16 | 2.8685 × 10−13 |
| 102164968 | XM_013984758,XM_021091688,XM_021091689 | KRT6A | keratin 6A, transcript variant X2 | 44.892932 | 8.266441 | 4.7843 × 10−16 | 7.2286 × 10−13 |
| Term_id | Term_Name | Adjusted_p-Value | Term_ Size | Query_Size | Intersec-tion_Size | Effective_Domain_Size | Intersection_Size_UP | Intersection_Size_ DOWN |
|---|---|---|---|---|---|---|---|---|
| GO *:0005737 | cytoplasm | 1.15186 × 10−10 | 9056 | 1107 | 628 | 19,669 | 288 | 340 |
| GO:0031323 | regulation of the cellular metabolic process | 5.11208 × 10−8 | 4452 | 1060 | 339 | 19,136 | 111 | 228 |
| GO:0048522 | positive regulation of the cellular process | 5.11208 × 10−8 | 4306 | 1060 | 330 | 19,136 | 123 | 207 |
| GO:0048518 | positive regulation of the biological process | 5.19773 × 10−8 | 4924 | 1060 | 367 | 19,136 | 144 | 223 |
| GO:0051171 | regulation of nitrogen compound metabolic process | 9.9293 × 10−7 | 4665 | 1060 | 344 | 19,136 | 120 | 224 |
| GO:0080090 | regulation of the primary metabolic process | 1.10779 × 10−6 | 4793 | 1060 | 351 | 19,136 | 122 | 229 |
| GO:0009891 | positive regulation of the biosynthetic process | 1.53459 × 10−6 | 1613 | 1060 | 146 | 19,136 | 43 | 103 |
| GO:0031325 | positive regulation of the cellular metabolic process | 2.19895 × 10−6 | 2373 | 1060 | 196 | 19,136 | 63 | 133 |
| GO:0010557 | positive regulation of macromolecule biosynthetic process | 2.19895 × 10−6 | 1520 | 1060 | 138 | 19,136 | 39 | 99 |
| GO:0031328 | positive regulation of cellular biosynthetic process | 2.19895 × 10−6 | 1580 | 1060 | 142 | 19,136 | 41 | 101 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seo, Y.A.; Cha, M.J.; Park, S.; Lee, S.; Lim, Y.J.; Son, D.W.; Lee, E.J.; Kim, P.; Chang, S. Development of a Normal Porcine Cell Line Growing in a Heme-Supplemented, Serum-Free Condition for Cultured Meat. Int. J. Mol. Sci. 2024, 25, 5824. https://doi.org/10.3390/ijms25115824
Seo YA, Cha MJ, Park S, Lee S, Lim YJ, Son DW, Lee EJ, Kim P, Chang S. Development of a Normal Porcine Cell Line Growing in a Heme-Supplemented, Serum-Free Condition for Cultured Meat. International Journal of Molecular Sciences. 2024; 25(11):5824. https://doi.org/10.3390/ijms25115824
Chicago/Turabian StyleSeo, Yeon Ah, Min Jeong Cha, Sehyeon Park, Seungki Lee, Ye Jin Lim, Dong Woo Son, Eun Ji Lee, Pil Kim, and Suhwan Chang. 2024. "Development of a Normal Porcine Cell Line Growing in a Heme-Supplemented, Serum-Free Condition for Cultured Meat" International Journal of Molecular Sciences 25, no. 11: 5824. https://doi.org/10.3390/ijms25115824
APA StyleSeo, Y. A., Cha, M. J., Park, S., Lee, S., Lim, Y. J., Son, D. W., Lee, E. J., Kim, P., & Chang, S. (2024). Development of a Normal Porcine Cell Line Growing in a Heme-Supplemented, Serum-Free Condition for Cultured Meat. International Journal of Molecular Sciences, 25(11), 5824. https://doi.org/10.3390/ijms25115824







