Alpha Carbonic Anhydrase from Nitratiruptor tergarcus Engineered for Increased Activity and Thermostability
Abstract
1. Introduction
2. Results and Discussion
2.1. Mutation Selection Process Yields 13 Single Mutants from Seven Selected Residues
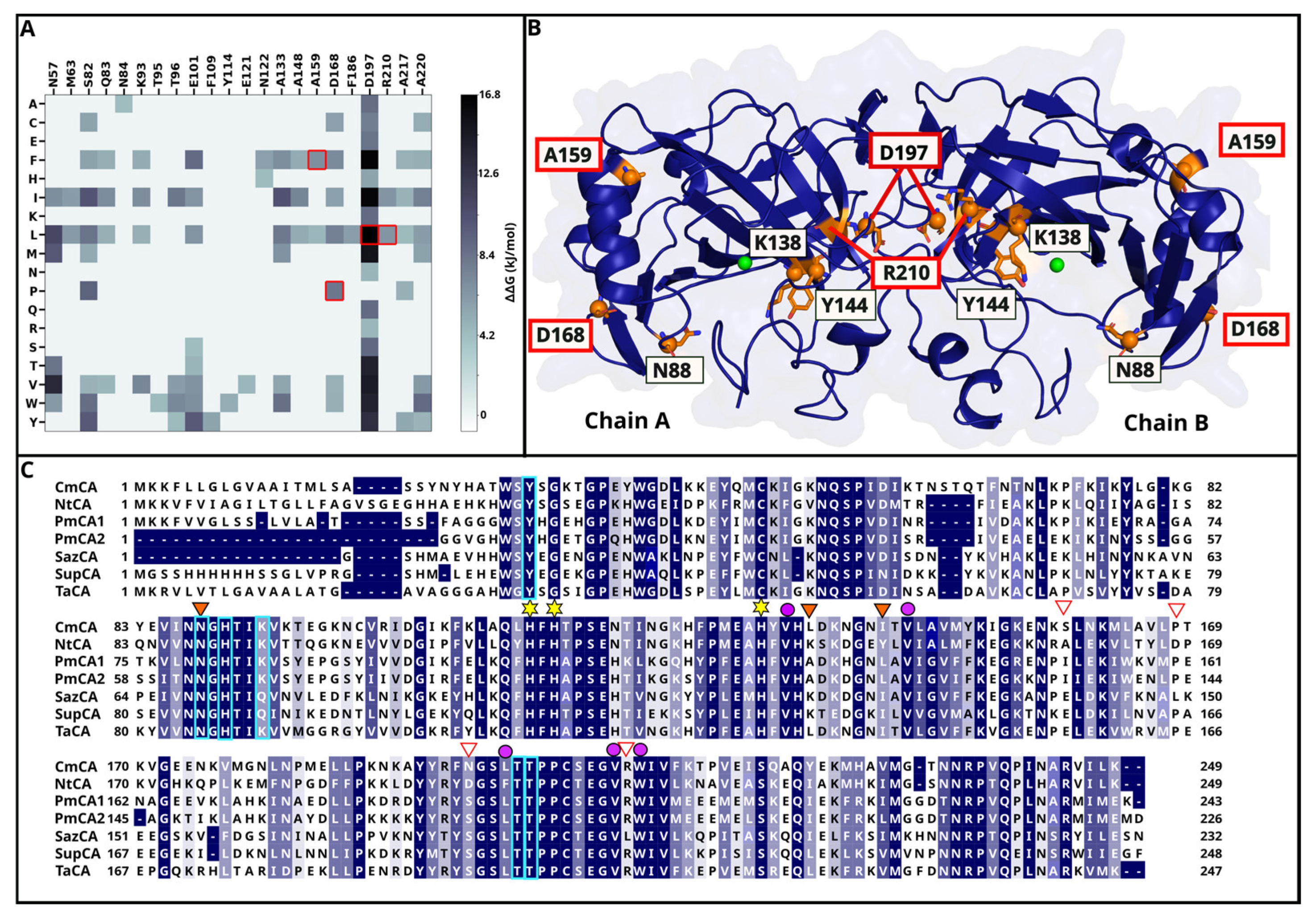
2.1.1. DEEPDDG Analysis Yields Two Surface and Two Interface Residues as Mutation Targets
2.1.2. Three Catalytic Pocket Residues Are Selected Resulting in Five Different Single Mutants
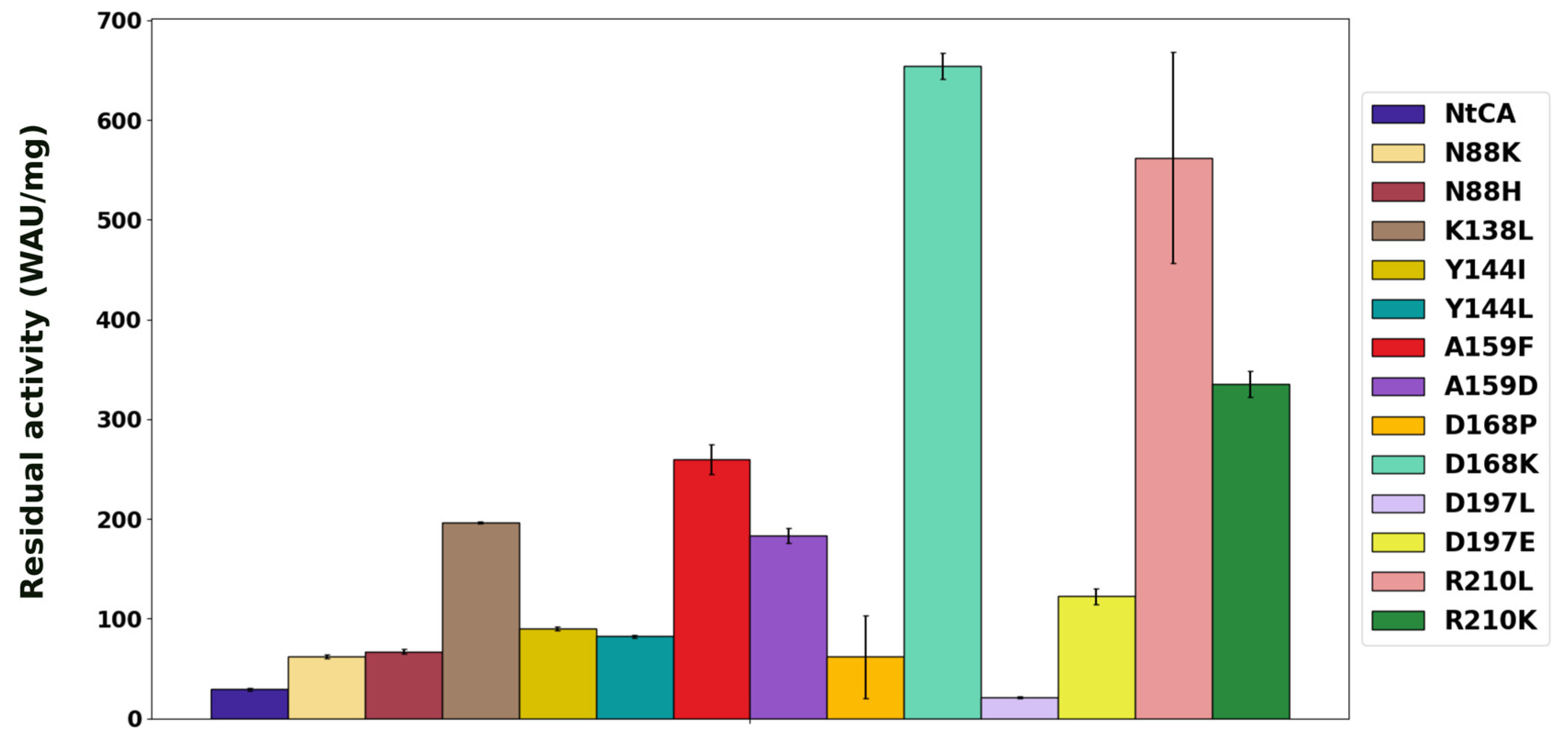
2.2. Characterization of Single Mutants Reveal the Majority to Exhibit Improved Catalytic Efficiency
2.3. Trajectory Analyses of MD Simulations of Mutants Relate Changes in Residue Interactions and Behaviour upon Mutation to Thermostability Profiles
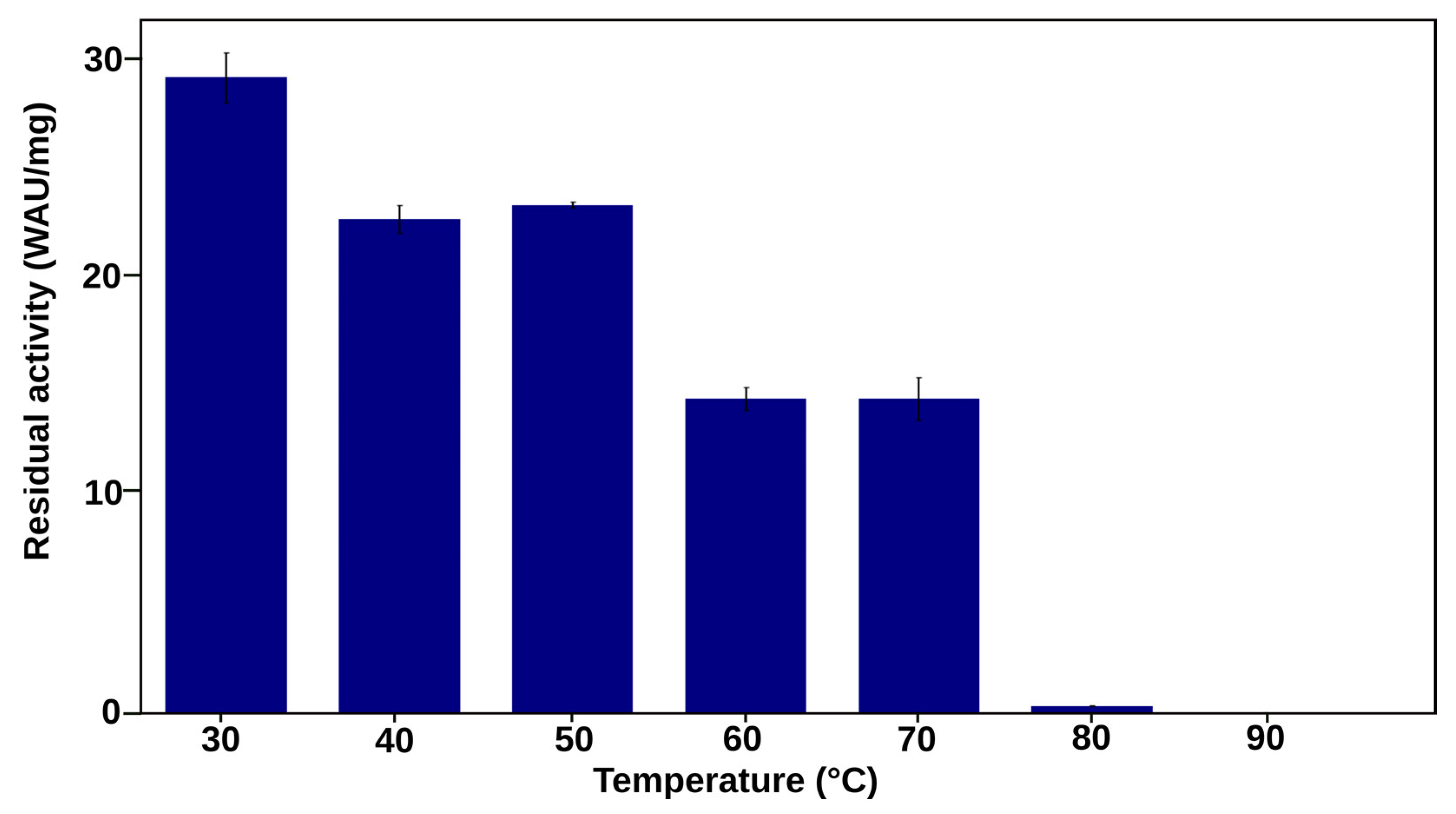
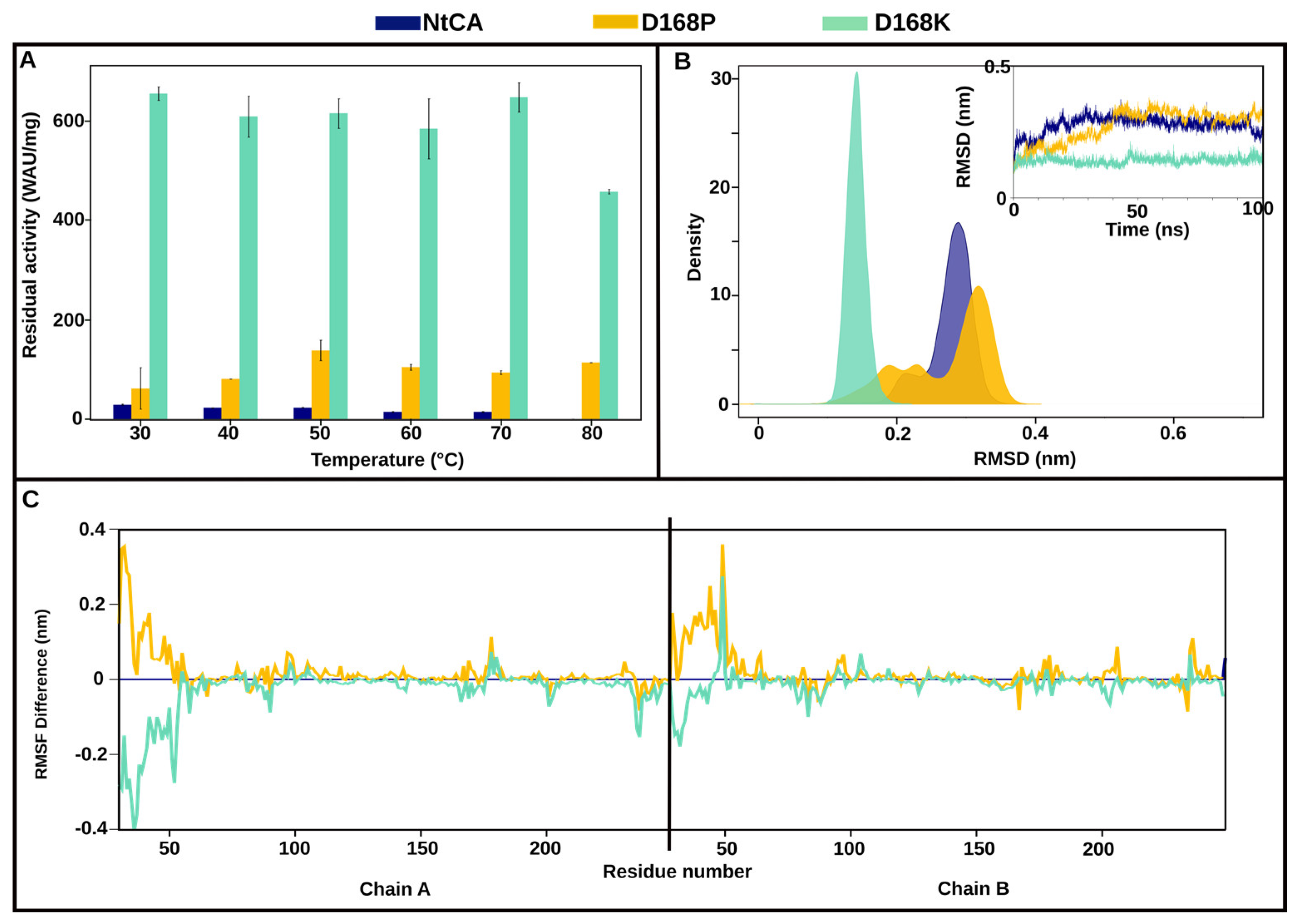
2.3.1. The Most Efficient Mutant, D168K, Maintains High Activity up to 80 °C
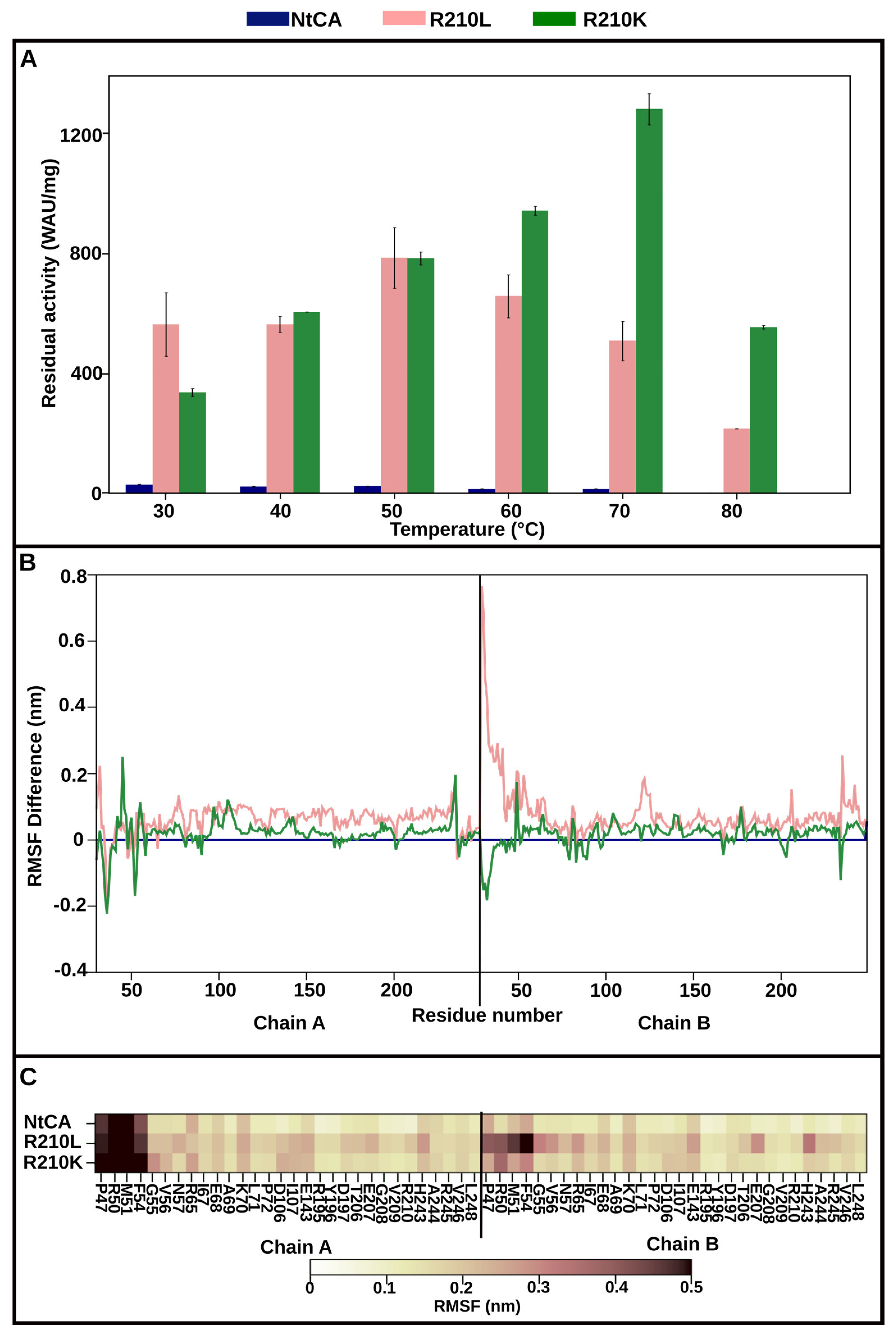
2.3.2. Mutant R210K Is Activated by Increasing Temperatures up to 70 °C and Exhibits Highest Activity at 80 °C
2.3.3. A159F Displays Exceptional Thermostability up to 70 °C Supported by Stable Structure RMSD and Reduced Residue Fluctuations
2.4. Combining Mutations Results in One Mutant Retaining Activity after Incubation at 90 °C
3. Materials and Methods
3.1. Sequence Retrieval, Analyses and Mutation Identification
3.2. Bacterial Strains, Plasmids and Mutagenesis
3.3. Protein Expression, Protein Purification and SDS-PAGE
3.4. CO2 Hydration Assay
3.5. Thermostability Assays
3.6. Molecular Dynamics Simulations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Silverman, D.N.; Lindskog, S. The catalytic mechanism of carbonic anhydrase: Implications of a rate-limiting protolysis of water. Acc. Chem. Res. 1988, 21, 30–36. [Google Scholar] [CrossRef]
- Tripp, B.C.; Smith, K.; Ferry, J.G. Carbonic anhydrase: New insights for an ancient enzyme. J. Biol. Chem. 2001, 276, 48615–48618. [Google Scholar] [CrossRef] [PubMed]
- Coleman, J.E. Mechanism of action of carbonic anhydrase substrate, sulfonamide, and anion binding. J. Biol. Chem. 1967, 242, 5212–5219. [Google Scholar] [CrossRef] [PubMed]
- Di Fiore, A.; Alterio, V.; Monti, S.M.; De Simone, G.; D’Ambrosio, K. Thermostable carbonic anhydrases in biotechnological applications. Int. J. Mol. Sci. 2015, 16, 15456–15480. [Google Scholar] [CrossRef] [PubMed]
- DiMario, R.J.; Machingura, M.C.; Waldrop, G.L.; Moroney, J.V. The many types of carbonic anhydrases in photosynthetic organisms. Plant Sci. 2018, 268, 11–17. [Google Scholar] [CrossRef]
- Kikutani, S.; Nakajima, K.; Nagasato, C.; Tsuji, Y.; Miyatake, A.; Matsuda, Y. Thylakoid luminal θ-carbonic anhydrase critical for growth and photosynthesis in the marine diatom Phaeodactylum tricornutum. Proc. Natl. Acad. Sci. USA 2016, 113, 9828–9833. [Google Scholar] [CrossRef] [PubMed]
- Jensen, E.L.; Clement, R.; Kosta, A.; Maberly, S.C.; Gontero, B. A new widespread subclass of carbonic anhydrase in marine phytoplankton. ISME J. 2019, 13, 2094–2106. [Google Scholar] [CrossRef] [PubMed]
- Lindskog, S. Structure and mechanism of carbonic anhydrase. Pharmacol. Ther. 1997, 74, 1–20. [Google Scholar] [CrossRef]
- Capasso, C.; Supuran, C.T. An overview of the alpha-, beta-and gamma-carbonic anhydrases from Bacteria: Can bacterial carbonic anhydrases shed new light on evolution of bacteria? J. Enzym. Inhib. Med. Chem. 2015, 30, 325–332. [Google Scholar] [CrossRef]
- Supuran, C.T. Structure and function of carbonic anhydrases. Biochem. J. 2016, 473, 2023–2032. [Google Scholar] [CrossRef]
- Boone, C.D.; Pinard, M.; McKenna, R.; Silverman, D. Catalytic mechanism of α−class carbonic anhydrases: CO2 hydration and proton transfer. In Carbonic Anhydrase: Mechanism, Regulation, Links to Disease, and Industrial Applications; Frost, S., McKenna, R., Eds.; Springer: Dordrecht, The Netherlands, 2014; Volume 75, pp. 31–52. [Google Scholar]
- Ferry, J.G. The γ class of carbonic anhydrases. Biochim. Biophys. Acta Proteins Proteom. 2010, 1804, 374–381. [Google Scholar] [CrossRef] [PubMed]
- Lane, T.W.; Morel, F.M. A biological function for cadmium in marine diatoms. Proc. Natl. Acad. Sci. USA 2000, 97, 4627–4631. [Google Scholar] [CrossRef] [PubMed]
- Tripp, B.C.; Bell, C.B.; Cruz, F.; Krebs, C.; Ferry, J.G. A role for iron in an ancient carbonic anhydrase. J. Biol. Chem. 2004, 279, 6683–6687. [Google Scholar] [CrossRef] [PubMed]
- Alterio, V.; Langella, E.; De Simone, G.; Monti, S.M. Cadmium-containing carbonic anhydrase CDCA1 in marine diatom Thalassiosira weissflogii. Mar. Drugs 2015, 13, 1688–1697. [Google Scholar] [CrossRef]
- Xu, Y.; Feng, L.; Jeffrey, P.D.; Shi, Y.; Morel, F.M. Structure and metal exchange in the cadmium carbonic anhydrase of marine diatoms. Nature 2008, 452, 56–61. [Google Scholar] [CrossRef] [PubMed]
- Liljas, A.; Kannan, K.K.; Bergsten, P.C.; Waara, I.; Fridborg, K.; Strandberg, B.; Petef, M. Crystal structure of human carbonic anhydrase C. Nat. New Biol. 1972, 235, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Cox, E.H.; McLendon, G.L.; Morel, F.M.; Lane, T.W.; Prince, R.C.; Pickering, I.J.; George, G.N. The active site structure of Thalassiosira weissflogii carbonic anhydrase 1. Biochemistry 2000, 39, 12128–12130. [Google Scholar] [CrossRef]
- De Simone, G.; Di Fiore, A.; Capasso, C.; Supuran, C.T. The zinc coordination pattern in the η-carbonic anhydrase from Plasmodium falciparum is different from all other carbonic anhydrase genetic families. Bioorg. Med. Chem. Lett. 2015, 25, 1385–1389. [Google Scholar] [CrossRef]
- Strop, P.; Smith, K.S.; Iverson, T.M.; Ferry, J.G.; Rees, D.C. Crystal structure of the “cab”-type β class carbonic anhydrase from the archaeon Methanobacterium thermoautotrophicum. J. Biol. Chem. 2001, 276, 10299–10305. [Google Scholar] [CrossRef]
- Smith, K.S.; Ferry, J.G. Prokaryotic carbonic anhydrases. FEMS Microbiol. Rev. 2000, 24, 335–366. [Google Scholar] [CrossRef]
- Savile, C.K.; Lalonde, J.J. Biotechnology for the acceleration of carbon dioxide capture and sequestration. Curr. Opin. Biotechnol. 2011, 22, 818–823. [Google Scholar] [CrossRef] [PubMed]
- De Simone, G.; Monti, S.M.; Alterio, V.; Buonanno, M.; De Luca, V.; Rossi, M.; Carginale, V.; Supuran, C.T.; Capasso, C.; Di Fiore, A. Crystal structure of the most catalytically effective carbonic anhydrase enzyme known, SazCA from the thermophilic bacterium Sulfurihydrogenibium azorense. Bioorganic Med. Chem. Lett. 2015, 25, 2002–2006. [Google Scholar] [CrossRef] [PubMed]
- Aguiar, P.; Beveridge, T.; Reysenbach, A.-L. Sulfurihydrogenibium azorense, sp. nov., a thermophilic hydrogen-oxidizing microaerophile from terrestrial hot springs in the Azores. Int. J. Syst. Evol. Microbiol. 2004, 54, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Vetriani, C.; Speck, M.D.; Ellor, S.V.; Lutz, R.A.; Starovoytov, V. Thermovibrio ammonificans sp. nov., a thermophilic, chemolithotrophic, nitrate-ammonifying bacterium from deep-sea hydrothermal vents. Int. J. Syst. Evol. Microbiol. 2004, 54, 175–181. [Google Scholar] [CrossRef] [PubMed]
- James, P.; Isupov, M.N.; Sayer, C.; Saneei, V.; Berg, S.; Lioliou, M.; Kotlar, H.K.; Littlechild, J.A. The structure of a tetrameric α-carbonic anhydrase from Thermovibrio ammonificans reveals a core formed around intermolecular disulfides that contribute to its thermostability. Acta Crystallogr. Sect. D Biol. Crystallogr. 2014, 70, 2607–2618. [Google Scholar] [CrossRef] [PubMed]
- Götz, D.; Banta, A.; Beveridge, T.J.; Rushdi, A.I.; Simoneit, B.R.T.; Reysenbach, A.L. Persephonella marina gen. nov., sp. nov. and Persephonella guaymasensis sp. nov., two novel, thermophilic, hydrogen-oxidizing microaerophiles from deep-sea hydrothermal vents. Int. J. Syst. Evol. Microbiol. 2002, 52, 1349–1359. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Sung, J.; Yeon, J.; Choi, S.H.; Jin, M.S. Crystal Structure of a Highly Thermostable α-Carbonic Anhydrase from Persephonella marina EX-H1. Mol. Cells 2019, 42, 460. [Google Scholar] [CrossRef] [PubMed]
- Fredslund, F.; Borchert, M.S.; Poulsen, J.C.N.; Mortensen, S.B.; Perner, M.; Streit, W.R.; Leggio, L.L. Structure of a hyperthermostable carbonic anhydrase identified from an active hydrothermal vent chimney. Enzym. Microb. Technol. 2018, 114, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Parra-Cruz, R.; Jäger, C.M.; Lau, P.L.; Gomes, R.L.; Pordea, A. Rational design of thermostable carbonic anhydrase mutants using molecular dynamics simulations. J. Phys. Chem. B 2018, 122, 8526–8536. [Google Scholar] [CrossRef]
- Parra-Cruz, R.; Lau, P.L.; Loh, H.-S.; Pordea, A. Engineering of Thermovibrio ammonificans carbonic anhydrase mutants with increased thermostability. J. CO2 Util. 2020, 37, 1–8. [Google Scholar] [CrossRef]
- Warden, A.C.; Williams, M.; Peat, T.S.; Seabrook, S.A.; Newman, J.; Dojchinov, G.; Haritos, V.S. Rational engineering of a mesohalophilic carbonic anhydrase to an extreme halotolerant biocatalyst. Nat. Commun. 2015, 6, 10278. [Google Scholar] [CrossRef]
- Vogler, M.; Karan, R.; Renn, D.; Vancea, A.; Vielberg, M.-T.; Grotzinger, S.W.; DasSarma, P.; DasSarma, S.; Eppinger, J.; Groll, M.; et al. Crystal structure and active site engineering of a halophilic γ-carbonic anhydrase. Front. Microbiol. 2020, 11, 742. [Google Scholar] [CrossRef]
- Pires, D.E.; Ascher, D.B.; Blundell, T.L. DUET: A server for predicting effects of mutations on protein stability using an integrated computational approach. Nucleic Acids Res. 2014, 42, W314–W319. [Google Scholar] [CrossRef]
- Rodrigues, C.H.; Pires, D.E.; Ascher, D.B. DynaMut: Predicting the impact of mutations on protein conformation, flexibility and stability. Nucleic Acids Res. 2018, 46, W350–W355. [Google Scholar] [CrossRef]
- Quan, L.; Lv, Q.; Zhang, Y. STRUM: Structure-based prediction of protein stability changes upon single-point mutation. Bioinformatics 2016, 32, 2936–2946. [Google Scholar] [CrossRef]
- Parthiban, V.; Gromiha, M.M.; Schomburg, D. CUPSAT: Prediction of protein stability upon point mutations. Nucleic Acids Res. 2006, 34, W239–W242. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Wang, J.; He, L.; Qi, Y.; Zhang, J.Z. DeepDDG: Predicting the stability change of protein point mutations using neural networks. J. Chem. Inf. Model. 2019, 59, 1508–1514. [Google Scholar] [CrossRef] [PubMed]
- De Luca, V.; Del Prete, S.; Carginale, V.; Vullo, D.; Supuran, C.T.; Capasso, C. A failed tentative to design a super carbonic anhydrase having the biochemical properties of the most thermostable CA (SspCA) and the fastest (SazCA) enzymes. J. Enzym. Inhib. Med. Chem. 2015, 30, 989–994. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, S.; Takai, K.; Inagaki, F.; Horikoshi, K.; Sako, Y. Nitratiruptor tergarcus gen. nov., sp. nov. and Nitratifractor salsuginis gen. nov., sp. nov., nitrate-reducing chemolithoautotrophs of the ε-Proteobacteria isolated from a deep-sea hydrothermal system in the Mid-Okinawa Trough. Int. J. Syst. Evol. Microbiol. 2005, 55, 925–933. [Google Scholar] [CrossRef]
- Manyumwa, C.V.; Emameh, R.Z.; Tastan Bishop, Ö. Alpha-Carbonic Anhydrases from Hydrothermal Vent Sources as Potential Carbon Dioxide Sequestration Agents: In Silico Sequence, Structure and Dynamics Analyses. Int. J. Mol. Sci. 2020, 21, 8066. [Google Scholar] [CrossRef]
- Nair, S.K.; Calderone, T.; Christianson, D.W.; Fierke, C. Altering the mouth of a hydrophobic pocket. Structure and kinetics of human carbonic anhydrase II mutants at residue Val-121. J. Biol. Chem. 1991, 266, 17320–17325. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.P.-Y.; Liang, F.-C.; Lee, C.-T.; Zerella, R.; Chan, S.I. Contributions of a surface hydrophobic cluster to the folding and structural stability of ubiquitin. J. Chin. Chem. Soc. 2008, 55, 772–781. [Google Scholar] [CrossRef]
- Sayers, E.W.; Barrett, T.; Benson, D.A.; Bolton, E.; Bryant, S.H.; Canese, K.; Chetvernin, V.; Church, D.M.; DiCuccio, M.; Federhen, S.; et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2010, 39, D38–D51. [Google Scholar] [CrossRef] [PubMed]
- Kiefer, F.; Arnold, K.; Künzli, M.; Bordoli, L.; Schwede, T. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 2009, 37, D387–D392. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; De Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Eisenberg, D.; Lüthy, R.; Bowie, J.U. [20] VERIFY3D: Assessment of protein models with three-dimensional profiles. In Methods in Enzymology; Academic Press: Cambridge, MA, USA, 1997; Volume 277, pp. 396–404. [Google Scholar] [CrossRef]
- Notredame, C.; Higgins, D.G.; Heringa, J. T-coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000, 302, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Di Tommaso, P.; Moretti, S.; Xenarios, I.; Orobitg, M.; Montanyola, A.; Chang, J.-M.; Taly, J.-F.; Notredame, C. T-Coffee: A web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 2011, 39, W13–W17. [Google Scholar] [CrossRef] [PubMed]
- De Luca, V.; Vullo, D.; Scozzafava, A.; Carginale, V.; Rossi, M.; Supuran, C.T.; Capasso, C. An α-carbonic anhydrase from the thermophilic bacterium Sulphurihydrogenibium azorense is the fastest enzyme known for the CO2 hydration reaction. Bioorganic Med. Chem. 2013, 21, 1465–1469. [Google Scholar] [CrossRef]
- Capasso, C.; De Luca, V.; Carginale, V.; Cannio, R.; Rossi, M. Biochemical properties of a novel and highly thermostable bacterial α-carbonic anhydrase from Sulfurihydrogenibium yellowstonense YO3AOP1. J. Enzym. Inhib. Med. Chem. 2012, 27, 892–897. [Google Scholar] [CrossRef]
- Di Fiore, A.; Capasso, C.; De Luca, V.; Monti, S.M.; Carginale, V.; Supuran, C.T.; Scozzafava, A.; Pedone, C.; Rossi, M.; De Simone, G. X-ray structure of the first extremo-α-carbonic anhydrase’, a dimeric enzyme from the thermophilic bacterium Sulfurihydrogenibium yellowstonense YO3AOP1. Acta Crystallogr. Sect. D Biol. Crystallogr. 2013, 69, 1150–1159. [Google Scholar] [CrossRef] [PubMed]
- Kanth, B.K.; Jun, S.-Y.; Kumari, S.; Pack, S.P. Highly thermostable carbonic anhydrase from Persephonella marina EX-H1: Its expression and characterization for CO2-sequestration applications. Process Biochem. 2014, 49, 2114–2121. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The protein data bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Waite, D.W.; Vanwonterghem, I.; Rinke, C.; Parks, D.H.; Zhang, Y.; Takai, K.; Sievert, S.M.; Simon, J.; Campbell, B.J.; Hanson, T.E.; et al. Comparative genomic analysis of the class Epsilonproteobacteria and proposed reclassification to Epsilonbacteraeota (phyl. nov.). Front. Microbiol. 2017, 8, 682. [Google Scholar] [CrossRef]
- Borchert, M.; Saunders, P. Heat−Stable Carbonic Anhydrases and Their Use. US Patent 8,945,826, 2015. [Google Scholar]
- Gibson, D.G.; Young, L.; Chuang, R.-Y.; Venter, J.C.; Hutchison, C.A., III; Smith, H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 2009, 6, 343–345. [Google Scholar] [CrossRef]
- Wilbur, K.M.; Anderson, N.G. Electrometric and colorimetric determination of carbonic anhydrase. J. Biol. Chem. 1948, 176, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Sanyanga, T.A.; Nizami, B.; Tastan Bishop, Ö. Mechanism of Action of Non-Synonymous Single Nucleotide Variations Associated with α-Carbonic Anhydrase II Deficiency. Molecules 2019, 24, 3987. [Google Scholar] [CrossRef]
- Case, D.A.; Aktulga, H.M.; Belfon, K.; Ben-Shalom, I.Y.; Berryman, J.T.; Brozell, S.R.; Cerutti, D.S.; Cheatham, T.E., III; Cisneros, G.A.; Cruzeiro, V.W.D.; et al. AmberTools22; University of California: San Francisco, CA, USA, 2022. [Google Scholar]
- Case, D.; Cerutti, D.S.; Cheatham, T.E., III; Darden, T.A.; Duke, R.E.; Giese, T.J.; Gohlke, H.; Goetz, A.W.; Greene, D.; Homeyer, N.; et al. AMBER 2017; University of California: San Francisco, CA, USA, 2017. [Google Scholar]
- Gordon, J.C.; Myers, J.B.; Folta, T.; Shoja, V.; Heath, L.S.; Onufriev, A. H++: A server for estimating pKas and adding missing hydrogens to macromolecules. Nucleic Acids Res. 2005, 33, W368–W371. [Google Scholar] [CrossRef] [PubMed]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef]
- Roe, D.R.; Cheatham, T.E., III. PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef] [PubMed]


| Mutation Site | Residue Position | Mutant Residue | Rationale |
|---|---|---|---|
| N88 | Active site | K H | Catalytic pocket engineering |
| K138 | Close to active site | L | Catalytic pocket engineering |
| Y144 | Close to active site | I L | Catalytic pocket engineering |
| A159 | Surface residue | F D | DEEPDDG/Surface engineering |
| D168 | Surface residue | P K | DEEPDDG/Surface engineering |
| D197 | Interface residue | L E | DEEPDDG/Interface engineering |
| R210 | Interface residue | L K | DEEPDDG/Interface/Active site engineering |
| Mutations | Residue Functions | Mutant Name |
|---|---|---|
| A159F, R210L | Surface and interface | A159F_R210L |
| A159F, R210K | Surface and interface | A159F_R210K |
| N88K, R210L | Active site and interface | N88K_R210L |
| N88K, R210K | Active site and interface | N88K_R210K |
| D168K, R210L | Surface and interface | D168K_R210L |
| D168K, R210K | Surface and interface | D168K_R210K |
| A159F, D168P, D197L, R210L | 2 Surface and 2 interface | Q1 |
| A159F, D168P, D197E, R210L | 2 Surface and 2 interface | Q2 |
| A159F, D168P, D197L, R210K | 2 Surface and 2 interface | Q3 |
| A159F, D168P, D197E, R210K | 2 Surface and 2 interface | Q4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manyumwa, C.V.; Zhang, C.; Jers, C.; Mijakovic, I. Alpha Carbonic Anhydrase from Nitratiruptor tergarcus Engineered for Increased Activity and Thermostability. Int. J. Mol. Sci. 2024, 25, 5853. https://doi.org/10.3390/ijms25115853
Manyumwa CV, Zhang C, Jers C, Mijakovic I. Alpha Carbonic Anhydrase from Nitratiruptor tergarcus Engineered for Increased Activity and Thermostability. International Journal of Molecular Sciences. 2024; 25(11):5853. https://doi.org/10.3390/ijms25115853
Chicago/Turabian StyleManyumwa, Colleen Varaidzo, Chenxi Zhang, Carsten Jers, and Ivan Mijakovic. 2024. "Alpha Carbonic Anhydrase from Nitratiruptor tergarcus Engineered for Increased Activity and Thermostability" International Journal of Molecular Sciences 25, no. 11: 5853. https://doi.org/10.3390/ijms25115853
APA StyleManyumwa, C. V., Zhang, C., Jers, C., & Mijakovic, I. (2024). Alpha Carbonic Anhydrase from Nitratiruptor tergarcus Engineered for Increased Activity and Thermostability. International Journal of Molecular Sciences, 25(11), 5853. https://doi.org/10.3390/ijms25115853








