Bioinformatic Analysis of Roquin Family Reveals Their Potential Role in Immune System
Abstract
:1. Introduction
2. Results
2.1. Roquin Genes in Metazoan
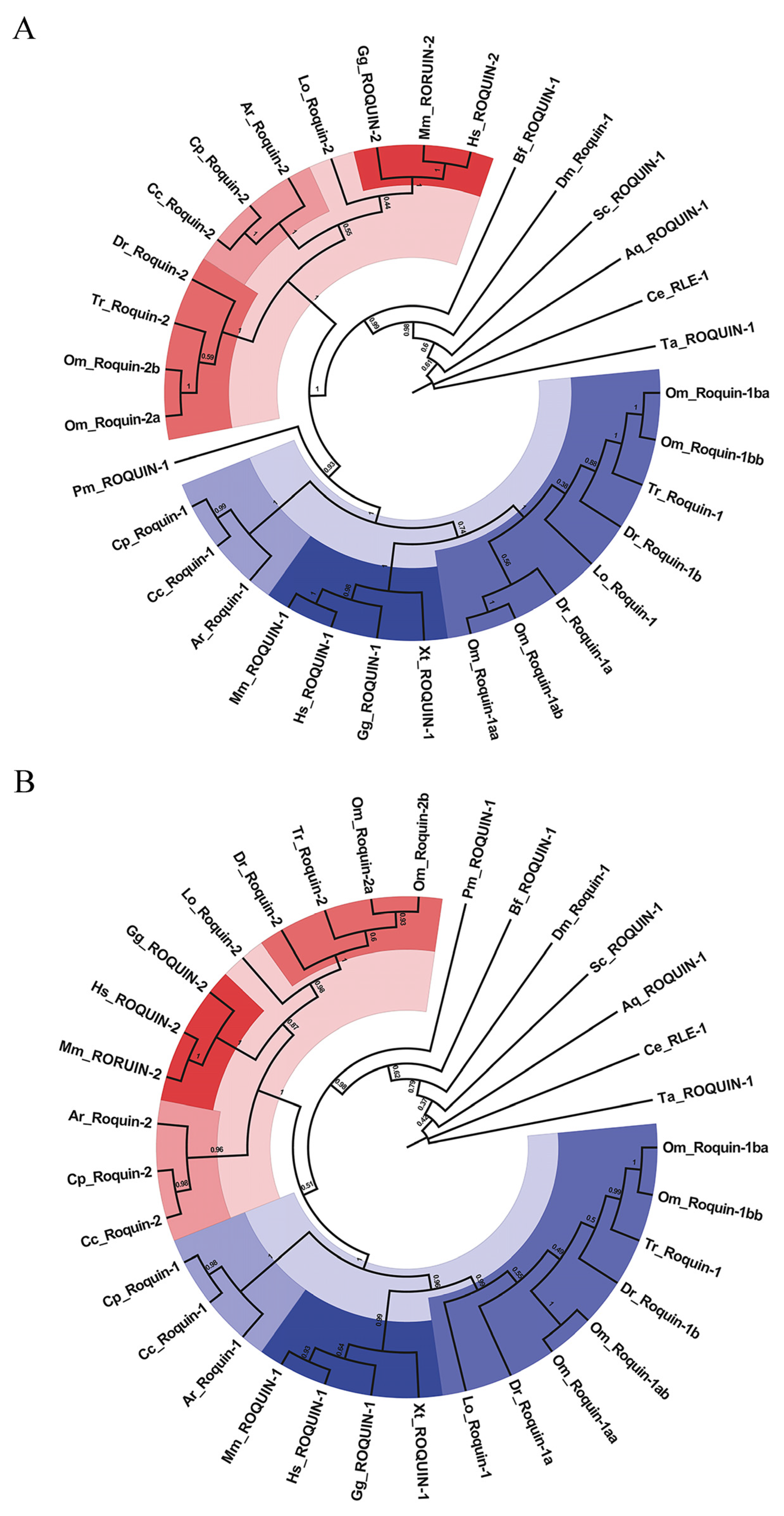
2.2. Evolutionary Relationship of ROQUINS
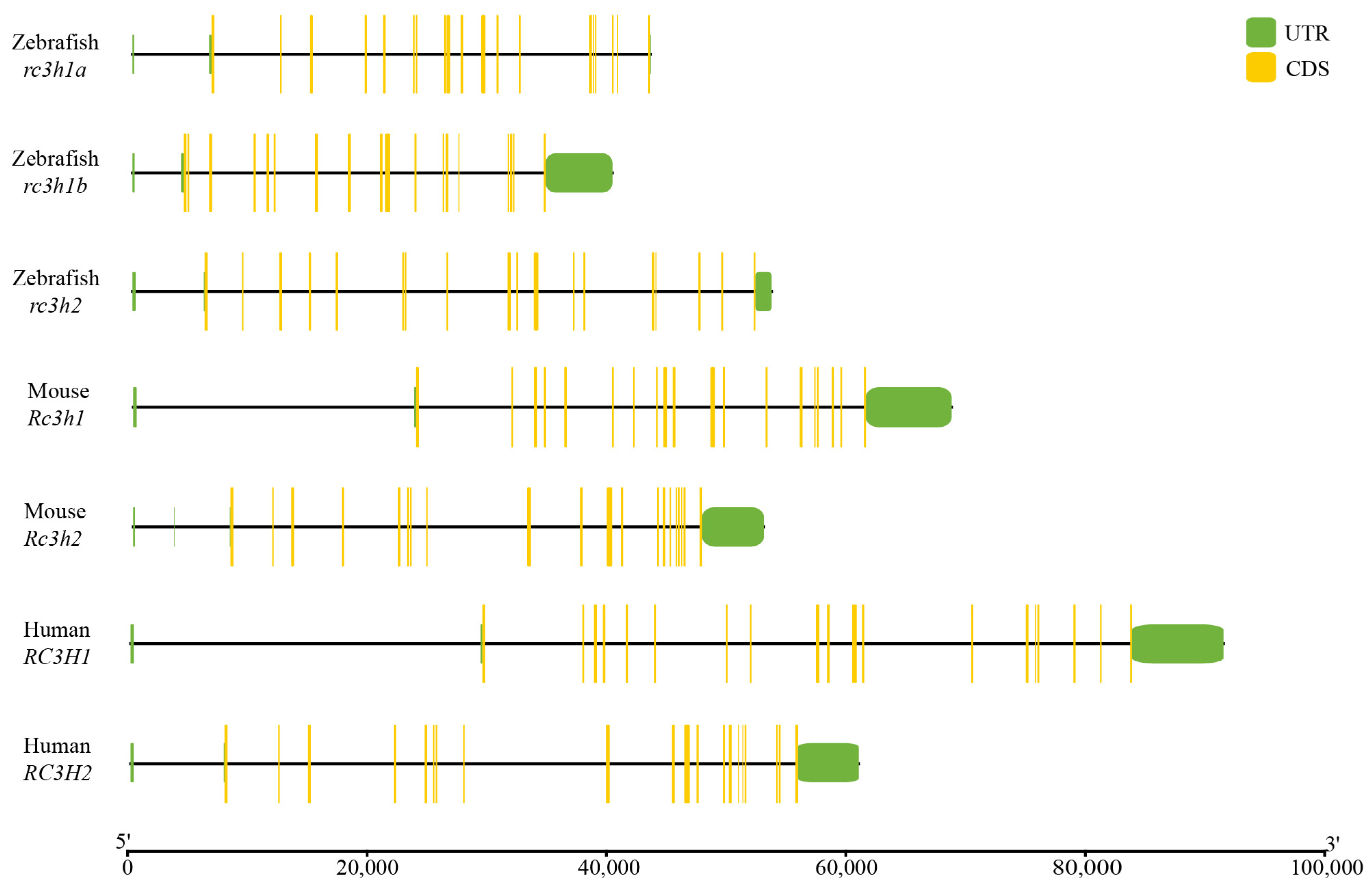
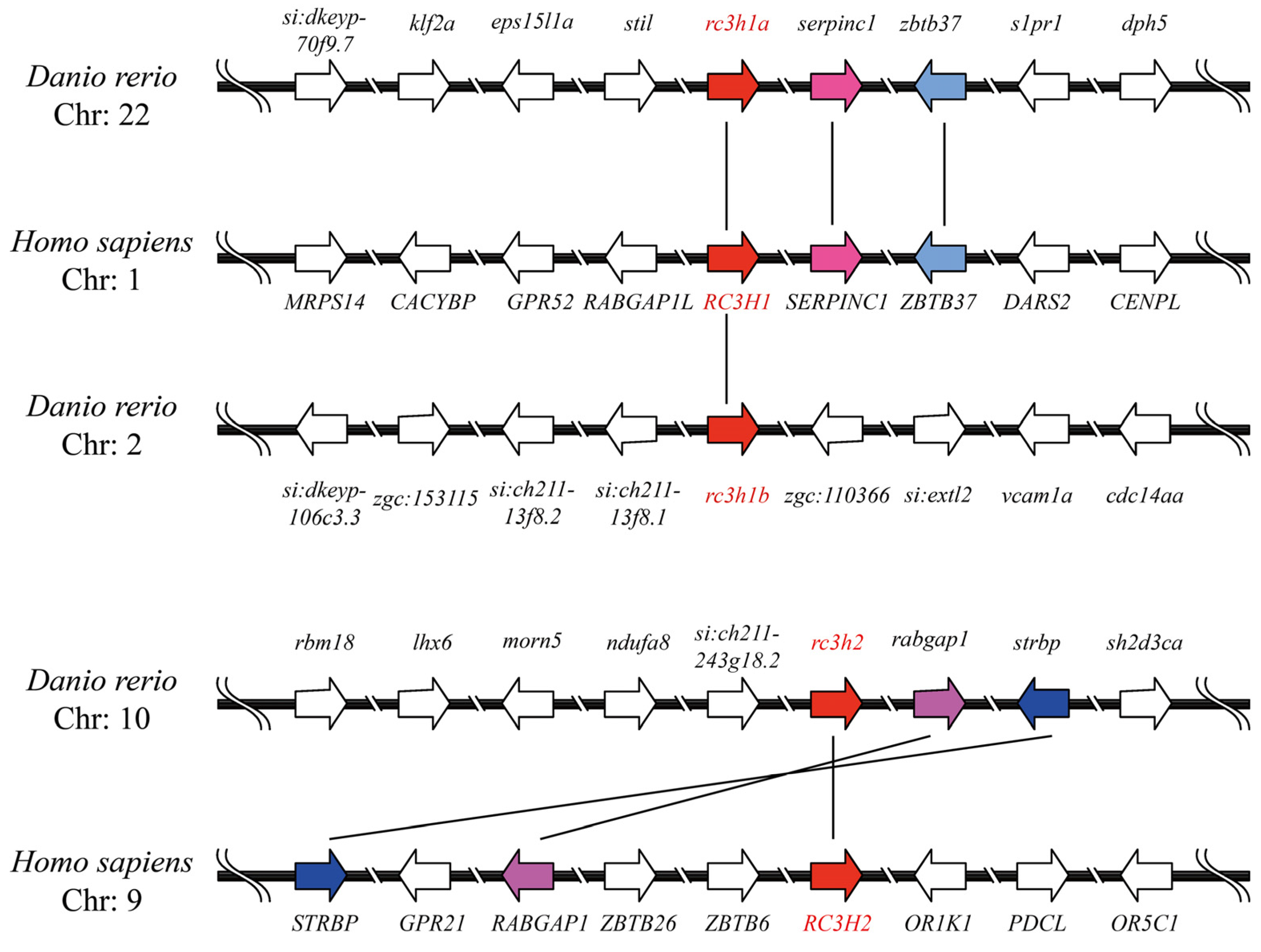
2.3. Genomic Structure and Synteny of Rc3h Genes
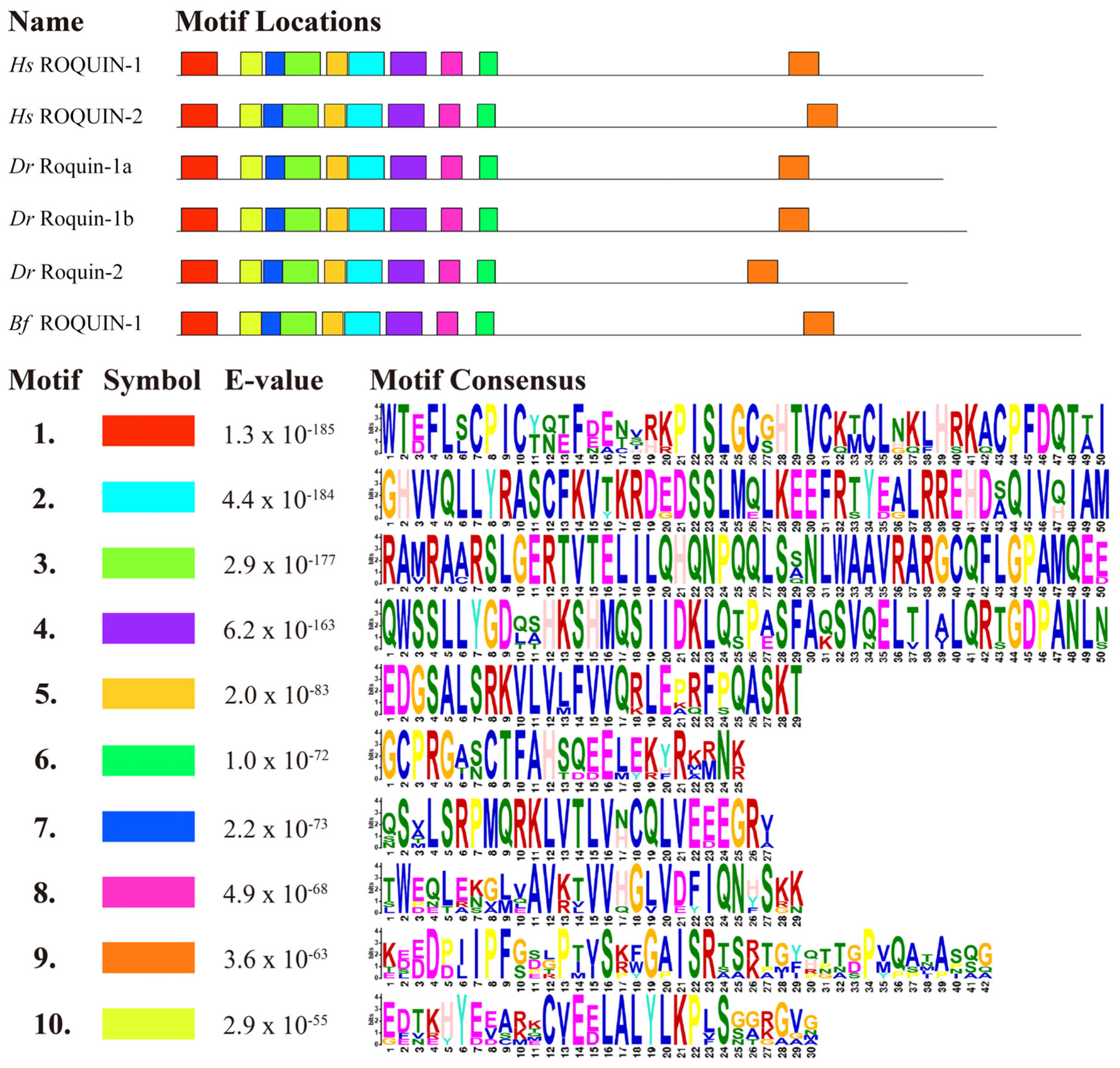
2.4. Domain and Motif Organization of Roquins
2.5. Selective Pressure of rc3h Genes
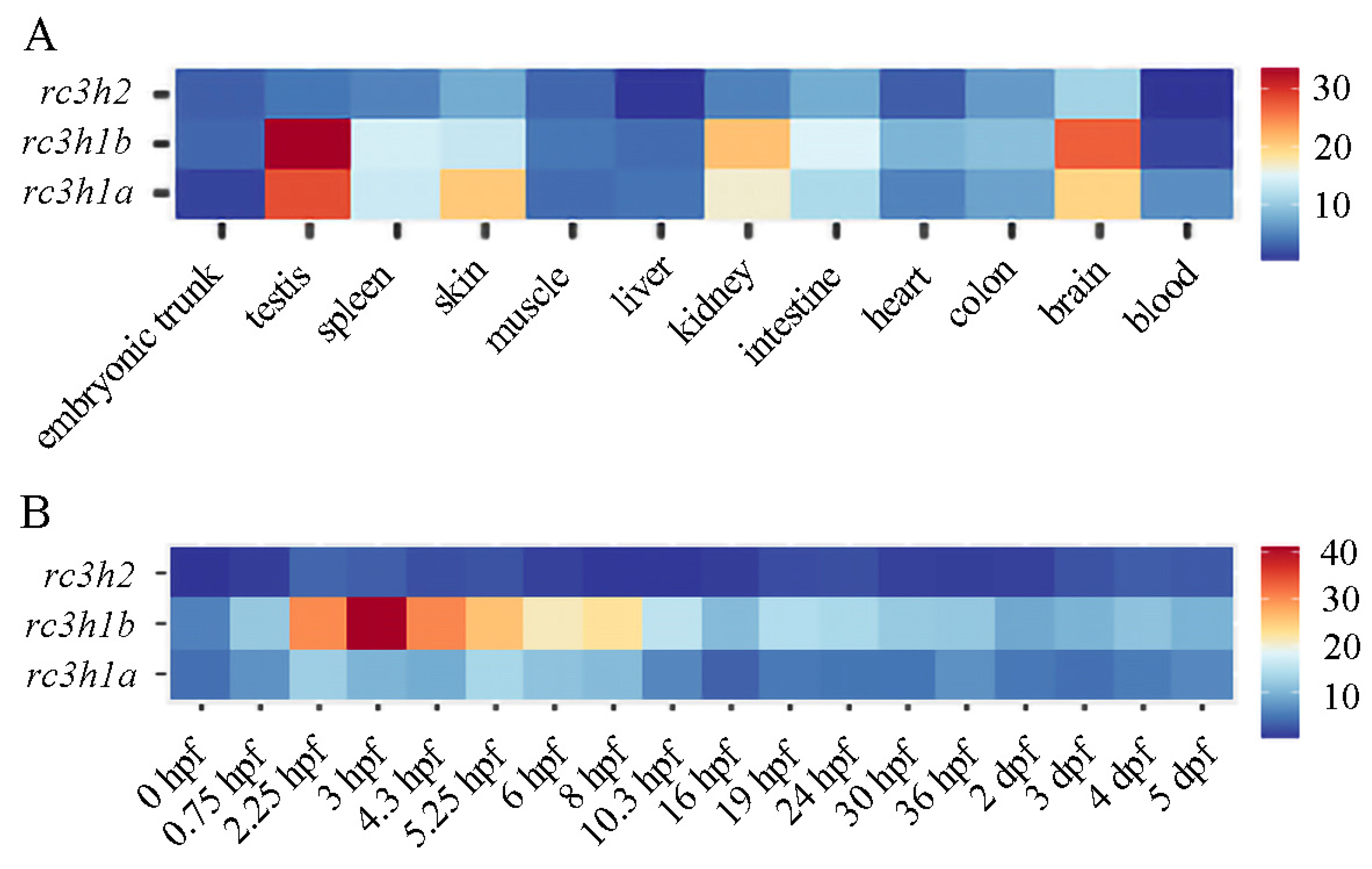
2.6. Expression Profile of rc3h Genes in Zebrafish
2.7. Potential Transcription Factor in the Promoters of rc3h Genes
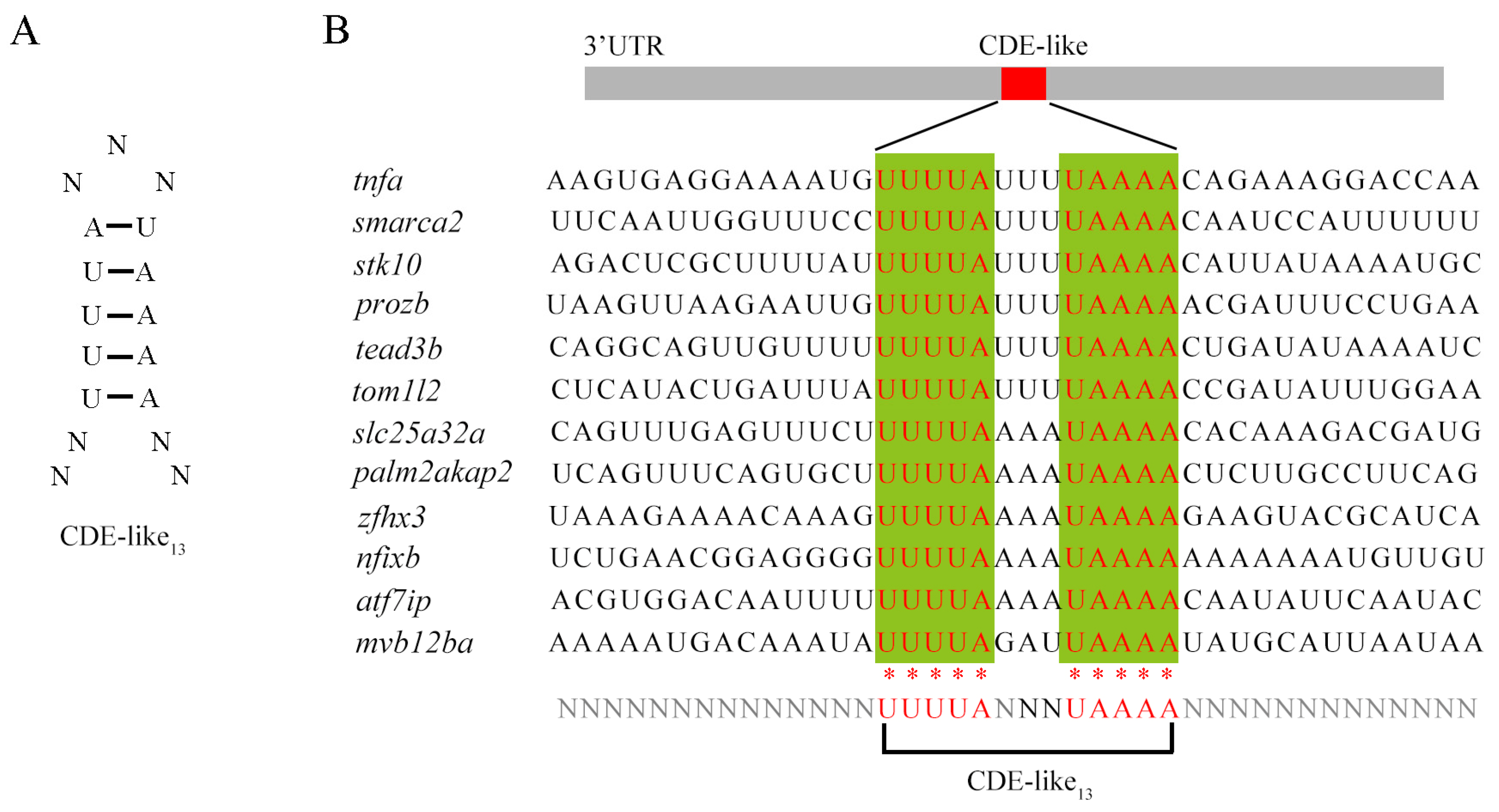
2.8. Interaction Protein Network and Potential Binding Motif of Roquins in Zebrafish
3. Discussion
4. Materials and Methods
4.1. Sequence Retrieval and characterization
4.2. Phylogenetic Analysis
4.3. Genomic Structure, Synteny Analysis and Selective Pressure Analysis
4.4. Expression Analysis of rc3h Genes by Online Data
4.5. Putative Transcription Factors Binding Sites
4.6. Analysis of Protein Interaction Network and Potential Binding Motif of Roquins in Zebrafish
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gan, P.; Wang, Z.; Morales, M.G.; Zhang, Y.; Bassel-Duby, R.; Liu, N.; Olson, E.N. RBPMS is an RNA-binding protein that mediates cardiomyocyte binucleation and cardiovascular development. Dev. Cell 2022, 57, 959–973 e7. [Google Scholar] [CrossRef] [PubMed]
- Zaman, M.M.; Masuda, K.; Nyati, K.K.; Dubey, P.K.; Ripley, B.; Wang, K.; Chalise, J.P.; Higa, M.; Hanieh, H.; Kishimoto, T. Arid5a exacerbates IFN-gamma-mediated septic shock by stabilizing T-bet mRNA. Proc. Natl. Acad. Sci. USA 2016, 113, 11543–11548. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, M.; Saha, S.; Li, J.; Montrose, D.C.; Martinez, L.A. p53 engages the cGAS/STING cytosolic DNA sensing pathway for tumor suppression. Mol. Cell 2023, 83, 266–280.e6. [Google Scholar] [CrossRef]
- Yoshinaga, M.; Takeuchi, O. Regulation of inflammatory diseases via the control of mRNA decay. Inflamm. Regen. 2024, 44, 14. [Google Scholar] [CrossRef]
- Akira, S.; Maeda, K. Control of RNA Stability in Immunity. Annu. Rev. Immunol. 2021, 39, 481–509. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, O. Posttranscriptional control of pro-inflammatory cytokine expression by Regnase-1 and Roquin. Cytokine 2017, 100, 36. [Google Scholar]
- Schaefer, J.S.; Klein, J.R. Roquin-a multifunctional regulator of immune homeostasis. Genes Immun. 2016, 17, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Athanasopoulos, V.; Ramiscal, R.R.; Vinuesa, C.G. ROQUIN signalling pathways in innate and adaptive immunity. Eur. J. Immunol. 2016, 46, 1082–1090. [Google Scholar] [CrossRef] [PubMed]
- Yoshinaga, M.; Takeuchi, O. RNA metabolism governs immune function and response. Adv. Exp. Med. Biol. 2024, 1444, 145–161. [Google Scholar]
- Finan, J.M.; Sutton, T.L.; Dixon, D.A.; Brody, J.R. Targeting the RNA-binding protein HuR in cancer. Cancer Res. 2023, 83, 3507–3516. [Google Scholar] [CrossRef]
- Vinuesa, C.G.; Cook, M.C.; Angelucci, C.; Athanasopoulos, V.; Rui, L.; Hill, K.M.; Yu, D.; Domaschenz, H.; Whittle, B.; Lambe, T.; et al. A RING-type ubiquitin ligase family member required to repress follicular helper T cells and autoimmunity. Nature 2005, 435, 452–458. [Google Scholar] [CrossRef] [PubMed]
- Schlundt, A.; Niessing, D.; Heissmeyer, V.; Sattler, M. RNA recognition by Roquin in posttranscriptional gene regulation. Wiley Interdiscip. Rev. RNA 2016, 7, 455–469. [Google Scholar] [CrossRef] [PubMed]
- Heinemann, U.; Murakawa, Y.; Landthaler, M.; Mayr, F.; Schütz, A. Translational regulation of gene expression by Lin28 and Roquin. Acta Crystallogr. A 2015, 71, S29–S30. [Google Scholar] [CrossRef]
- Glasmacher, E.; Hoefig, K.P.; Vogel, K.U.; Rath, N.; Du, L.; Wolf, C.; Kremmer, E.; Wang, X.; Heissmeyer, V. Roquin binds inducible costimulator mRNA and effectors of mRNA decay to induce microRNA-independent post-transcriptional repression. Nat. Immunol. 2010, 11, 725–733. [Google Scholar] [CrossRef] [PubMed]
- Burkhard, P.; Stetefeld, J.; Strelkov, S.V. Coiled coils: A highly versatile protein folding motif. Trends Cell Biol. 2001, 11, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Athanasopoulos, V.; Barker, A.; Yu, D.; Tan, A.H.; Srivastava, M.; Contreras, N.; Wang, J.; Lam, K.; Brown, S.H.J.; Goodnow, C.C.; et al. The ROQUIN family of proteins localizes to stress granules via the ROQ domain and binds target mRNAs. FEBS J. 2010, 277, 2109–2127. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, J.S.; Montufar-Solis, D.; Klein, J.R. A role for IL-10 in the transcriptional regulation of Roquin-1. Gene 2014, 549, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, J.S.; Montufar-Solis, D.; Vigneswaran, N.; Klein, J.R. Selective upregulation of microRNA expression in peripheral blood leukocytes in IL-10−/− mice precedes expression in the colon. J. Immunol. 2011, 187, 5834–5841. [Google Scholar] [CrossRef] [PubMed]
- Jeltsch, K.M.; Hu, D.; Brenner, S.; Zöller, J.; Heinz, G.A.; Nagel, D.; Vogel, K.U.; Rehage, N.; Warth, S.C.; Edelmann, S.L.; et al. Cleavage of roquin and regnase-1 by the paracaspase MALT1 releases their cooperatively repressed targets to promote T(H)17 differentiation. Nat. Immunol. 2014, 15, 1079–1089. [Google Scholar] [CrossRef]
- Schmidt, H.; Raj, T.; O’Neill, T.J.; Muschaweckh, A.; Giesert, F.; Negraschus, A.; Hoefig, K.P.; Behrens, G.; Esser, L.; Baumann, C.; et al. Unrestrained cleavage of Roquin-1 by MALT1 induces spontaneous T cell activation and the development of autoimmunity. Proc. Natl. Acad. Sci. USA 2023, 120, e2309205120. [Google Scholar] [CrossRef]
- Wimberger, N.; Ober, F.; Avar, G.; Grau, M.; Xu, W.; Lenz, G.; Menden, M.P.; Krappmann, D. Oncogene-induced MALT1 protease activity drives posttranscriptional gene expression in malignant lymphomas. Blood 2023, 142, 1985–2001. [Google Scholar] [CrossRef] [PubMed]
- Janowski, R.; Heinz, G.A.; Schlundt, A.; Wommelsdorf, N.; Brenner, S.; Gruber, A.R.; Blank, M.; Buch, T.; Buhmann, R.; Zavolan, M.; et al. Roquin recognizes a non-canonical hexaloop structure in the 3′-UTR of Ox40. Nat. Commun. 2016, 7, 11032. [Google Scholar] [CrossRef] [PubMed]
- Essig, K.; Kronbeck, N.; Guimaraes, J.C.; Lohs, C.; Schlundt, A.; Hoffmann, A.; Behrens, G.; Brenner, S.; Kowalska, J.; Lopez-Rodriguez, C.; et al. Roquin targets mRNAs in a 3′-UTR-specific manner by different modes of regulation. Nat. Commun. 2018, 9, 3810. [Google Scholar] [CrossRef] [PubMed]
- Tan, D.Z.; Zhou, M.; Kiledjian, M.; Tong, L. The ROQ domain of Roquin recognizes mRNA constitutive-decay element and double-stranded RNA. Nat. Struct. Mol. Biol. 2014, 21, 679–685. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Gao, B.; Lee, S.M.; Bennett, K.; Fang, D. RLE-1, an E3 ubiquitin ligase, regulates C. elegans aging by catalyzing DAF-16 polyubiquitination. Dev. Cell 2007, 12, 235–246. [Google Scholar] [CrossRef] [PubMed]
- Sobańska, D.; Komur, A.A.; Chabowska-Kita, A.; Gumna, J.; Kumari, P.; Pachulska-Wieczorek, K.; Ciosk, R. The silencing of ets-4 mRNA relies on the functional cooperation between REGE-1/Regnase-1 and RLE-1/Roquin-1. Nucleic Acids Res. 2022, 50, 8226–8239. [Google Scholar] [CrossRef]
- Sgromo, A.; Raisch, T.; Bawankar, P.; Bhandari, D.; Chen, Y.; Kuzuoğlu-Öztürk, D.; Weichenrieder, O.; Izaurralde, E. A CAF40-binding motif facilitates recruitment of the CCR4-NOT complex to mRNAs targeted by Drosophila Roquin. Nat. Commun. 2017, 8, 14307. [Google Scholar] [CrossRef] [PubMed]
- Du, B.B.; Liu, L.; Zhu, Y.Y. RNA-binding protein Roquin negatively regulates STING-dependent innate immune response in Drosophila. Yi Chuan 2020, 42, 1201–1210. [Google Scholar]
- Yang, S.; Xu, X.; Zhang, A.; Wang, Y.; Ji, G.; Sun, C.; Li, H. The evolution and immunomodulatory role of Zc3h12 proteins in zebrafish (Danio rerio). Int. J. Biol. Macromol. 2023, 239, 124214. [Google Scholar] [CrossRef]
- Plata, G.; Vitkup, D. Genetic robustness and functional evolution of gene duplicates. Nucleic Acids Res. 2014, 42, 2405–2414. [Google Scholar] [CrossRef]
- Han, M.V.; Demuth, J.P.; McGrath, C.L.; Casola, C.; Hahn, M.W. Adaptive evolution of young gene duplicates in mammals. Genome Res. 2009, 19, 859–867. [Google Scholar] [CrossRef] [PubMed]
- Otto, S.P.; Yong, P. The evolution of gene duplicates. Adv. Genet. 2002, 46, 451–483. [Google Scholar] [PubMed]
- Raes, J.; Van de Peer, Y. Gene duplication, the evolution of novel gene functions, and detecting functional divergence of duplicates in silico. Appl. Bioinform. 2003, 2, 91–101. [Google Scholar]
- Gu, Z.; Steinmetz, L.M.; Gu, X.; Scharfe, C.; Davis, R.W.; Li, W.H. Role of duplicate genes in genetic robustness against null mutations. Nature 2003, 421, 63–66. [Google Scholar] [CrossRef]
- Pratama, A.; Ramiscal, R.R.; Silva, D.G.; Das, S.K.; Athanasopoulos, V.; Fitch, J.; Botelho, N.K.; Chang, P.-P.; Hu, X.; Hogan, J.J.; et al. Roquin-2 shares functions with its paralog Roquin-1 in the repression of mRNAs controlling T follicular helper cells and systemic inflammation. Immunity 2013, 38, 669–680. [Google Scholar] [CrossRef]
- Bertino, S.A.; Craft, J. Roquin paralogs add a new dimension to ICOS regulation. Immunity 2013, 38, 624–626. [Google Scholar] [CrossRef]
- Behrens, G.; Edelmann, S.L.; Raj, T.; Kronbeck, N.; Monecke, T.; Davydova, E.; Wong, E.H.; Kifinger, L.; Giesert, F.; Kirmaier, M.E.; et al. Disrupting Roquin-1 interaction with Regnase-1 induces autoimmunity and enhances antitumor responses. Nat. Immunol. 2021, 22, 1563–1576. [Google Scholar] [CrossRef]
- Leppek, K.; Schott, J.; Reitter, S.; Poetz, F.; Hammond, M.C.; Stoecklin, G. Roquin promotes constitutive mRNA decay via a conserved class of stem-loop recognition motifs. Cell 2013, 153, 869–881. [Google Scholar] [CrossRef] [PubMed]
- Tavernier, S.J.; Athanasopoulos, V.; Verloo, P.; Behrens, G.; Staal, J.; Bogaert, D.J.; Naesens, L.; De Bruyne, M.; Van Gassen, S.; Parthoens, E.; et al. A human immune dysregulation syndrome characterized by severe hyperinflammation with a homozygous nonsense Roquin-1 mutation. Nat. Commun. 2019, 10, 4779. [Google Scholar] [CrossRef]
- Hanet, A.; Räsch, F.; Weber, R.; Ruscica, V.; Fauser, M.; Raisch, T.; Kuzuoğlu-Öztürk, D.; Chang, C.-T.; Bhandari, D.; Igreja, C.; et al. HELZ directly interacts with CCR4-NOT and causes decay of bound mRNAs. Life Sci. Alliance 2019, 2, e201900405. [Google Scholar] [CrossRef]
- Nyati, K.K.; Zaman, M.M.U.; Sharma, P.; Kishimoto, T. Arid5a, an RNA-binding protein in immune regulation: RNA stability, inflammation, and autoimmunity. Trends Immunol. 2020, 41, 255–268. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A "one for all, all for one" bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.T.T.; Vincens, P.; Dufayard, J.F.; Roest Crollius, H.; Louis, A. Genomicus in 2022: Comparative tools for thousands of genomes and reconstructed ancestors. Nucleic Acids Res. 2022, 50, D1025–D1031. [Google Scholar] [CrossRef] [PubMed]
- White, R.J.; Collins, J.E.; Sealy, I.M.; Wali, N.; Dooley, C.M.; Digby, Z.; Stemple, D.L.; Murphy, D.N.; Billis, K.; Hourlier, T.; et al. A high-resolution mRNA expression time course of embryonic development in zebrafish. eLife 2017, 6, e30860. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Luan, Y.; Liu, T.; Lee, H.J.; Fang, L.; Wang, Y.; Wang, X.; Zhang, B.; Jin, Q.; Ang, K.C.; et al. A map of cis-regulatory elements and 3D genome structures in zebrafish. Nature 2020, 588, 337–343. [Google Scholar] [CrossRef]
- Farré, D.; Roset, R.; Huerta, M.; Adsuara, J.E.; Roselló, L.; Albà, M.M.; Messeguer, X. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. 2003, 31, 3651–3653. [Google Scholar] [CrossRef]



| Class | Species | Rc3h1 | Rc3h2 | Total |
|---|---|---|---|---|
| Mammalia | Human | 1 | 1 | 2 |
| Mouse | 1 | 1 | 2 | |
| Aves | Chicken | 1 | 1 | 2 |
| Amphibia | Frog | 1 | 0 | 1 |
| Osteichthyes | Spotted gar | 1 | 1 | 2 |
| Rainbow trout | 4 (1aa, 1ab, 1ba, 1bb) | 2 (2a, 2b) | 6 | |
| Torafugu | 1 | 1 | 2 | |
| Zebrafish | 2 (1a, 1b) | 1 | 3 | |
| Chondrichthyes 1 | Shark | 1 | 1 | 2 |
| Ray | 1 | 1 | 2 | |
| Cyclostomata | Lamprey | 1 | 1 | |
| Leptocardii | Amphioxi | 1 | 1 | |
| Ascidiacea | Ascidian | 1 | 1 | |
| Insecta | Fly | 1 | 1 | |
| Phasmidia | Nematode | 1 | 1 | |
| Demospongiae | Sponge | 1 | 1 | |
| Trichoplacoidea | Trichoplax | 1 | 1 | |
| Gene | Chromosome | No. Exon | CDS (bp) | Amino Acids (aa) | Molecular Weight (Da) | pI |
|---|---|---|---|---|---|---|
| rc3h1a | 22 | 20 | 3237 | 1078 | 120125.74 | 7.87 |
| rc3h1b | 2 | 19 | 3336 | 1111 | 122613.65 | 7.41 |
| rc3h2 | 10 | 19 | 3087 | 1028 | 113962.46 | 6.56 |
| Seq_1 | Seq_2 | Ka | Ks | Ka/Ks |
|---|---|---|---|---|
| Bf-rc3h1 | Dr-rc3h1a | 0.494146491 | 1.804421487 | 0.27385314 |
| Bf-rc3h1 | Dr-rc3h1b | 0.445397577 | 2.009173201 | 0.221682022 |
| Bf-rc3h1 | Dr-rc3h2 | 0.492221298 | 2.375139538 | 0.207238897 |
| Dr-rc3h1a | Dr-rc3h1b | 0.221189798 | 2.031204763 | 0.108895864 |
| Dr-rc3h1a | Dr-rc3h2 | 0.428709628 | 2.433626247 | 0.176160834 |
| Dr-rc3h1b | Dr-rc3h2 | 0.416147921 | 1.995249095 | 0.208569407 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Yang, S.; Zhang, X.; Zhang, Y.; Zhang, Y.; Li, H. Bioinformatic Analysis of Roquin Family Reveals Their Potential Role in Immune System. Int. J. Mol. Sci. 2024, 25, 5859. https://doi.org/10.3390/ijms25115859
Li X, Yang S, Zhang X, Zhang Y, Zhang Y, Li H. Bioinformatic Analysis of Roquin Family Reveals Their Potential Role in Immune System. International Journal of Molecular Sciences. 2024; 25(11):5859. https://doi.org/10.3390/ijms25115859
Chicago/Turabian StyleLi, Xianpeng, Shuaiqi Yang, Xiangmin Zhang, Yi Zhang, Yu Zhang, and Hongyan Li. 2024. "Bioinformatic Analysis of Roquin Family Reveals Their Potential Role in Immune System" International Journal of Molecular Sciences 25, no. 11: 5859. https://doi.org/10.3390/ijms25115859
APA StyleLi, X., Yang, S., Zhang, X., Zhang, Y., Zhang, Y., & Li, H. (2024). Bioinformatic Analysis of Roquin Family Reveals Their Potential Role in Immune System. International Journal of Molecular Sciences, 25(11), 5859. https://doi.org/10.3390/ijms25115859






