miR-107 Targets NSG1 to Regulate Hypopharyngeal Squamous Cell Carcinoma Progression through ERK Pathway
Abstract
1. Introduction
2. Results
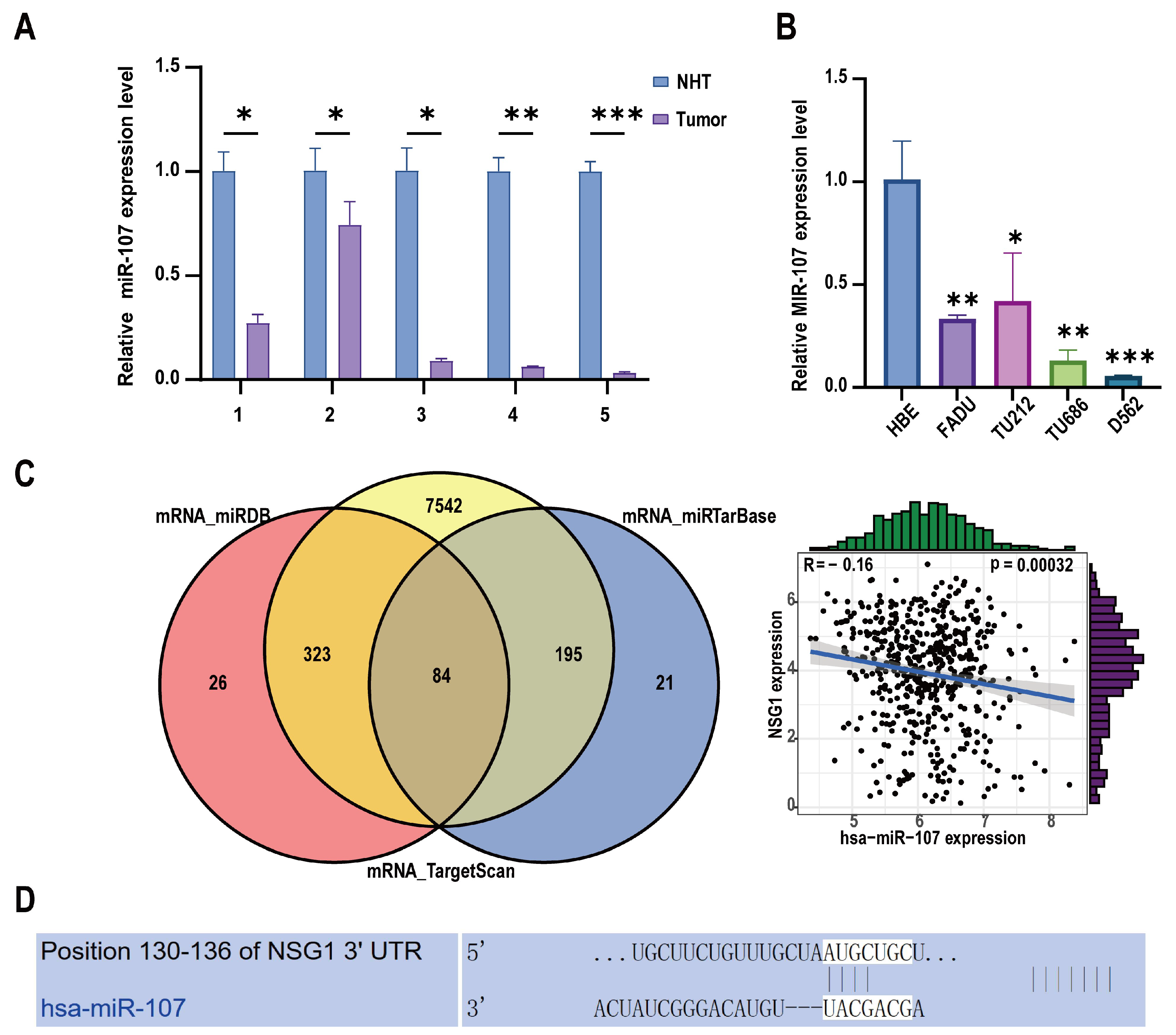
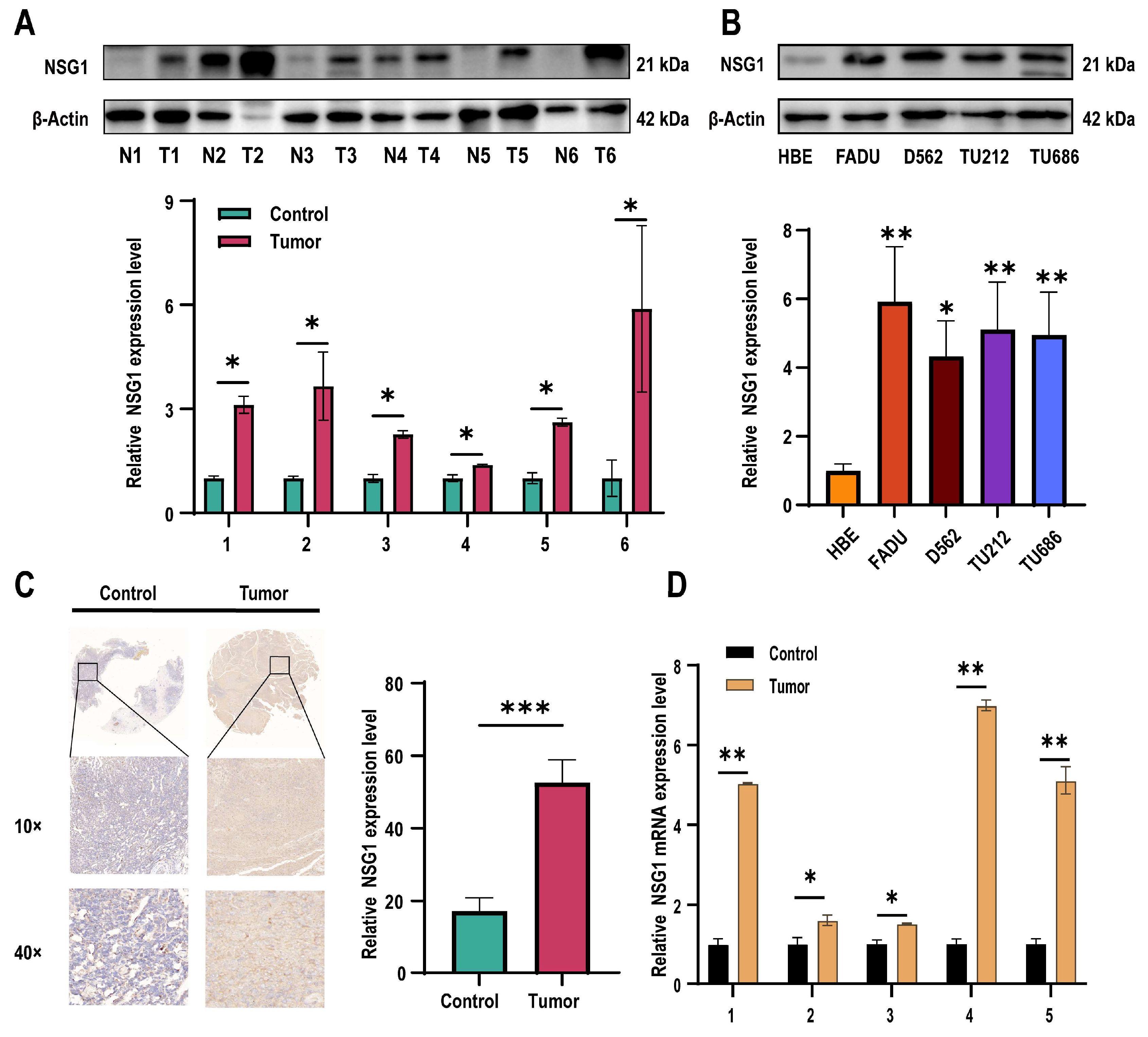
2.1. Decreased miR-107 Expression and Upregulation of NSG1 in HSCC Tissue and Cells
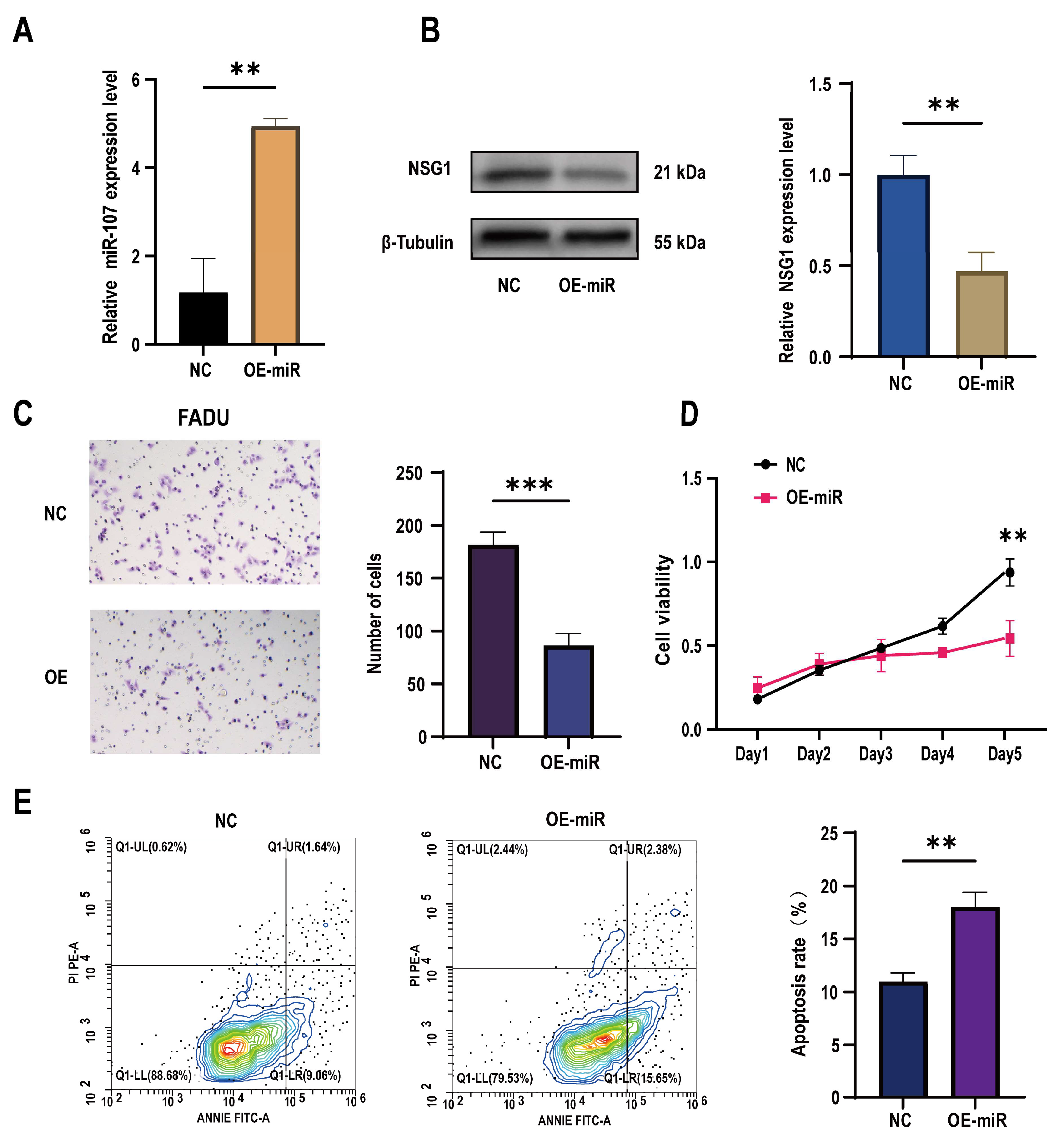
2.2. Overexpression of miR-107 Targeted the Downregulation of NSG1 and Promoted the Apoptosis of FaDu Cells, Which Inhibited Their Proliferation and Invasion
2.3. Knocking down NSG1 Inhibited the Proliferation, Migration, and Invasion Ability of the HSCC Cells
2.4. Overexpression of NSG1 Promoted the Proliferation, Migration, and Invasion of Hypopharyngeal Cancer Cells
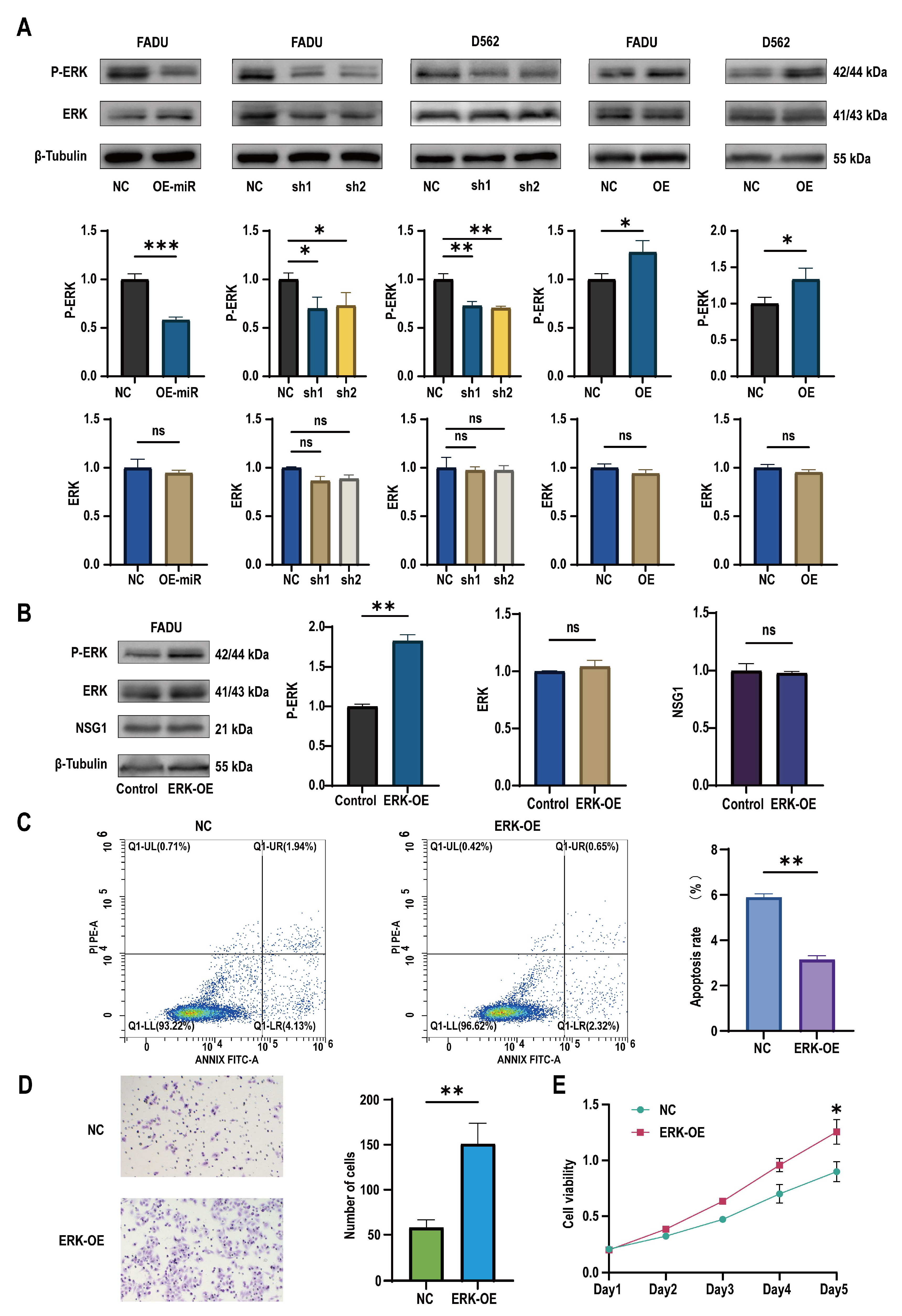
2.5. miR-107 Targeted NSG1 to Affect the Invasion and Cell Viability of HSCC Cells by Regulating the ERK Pathways
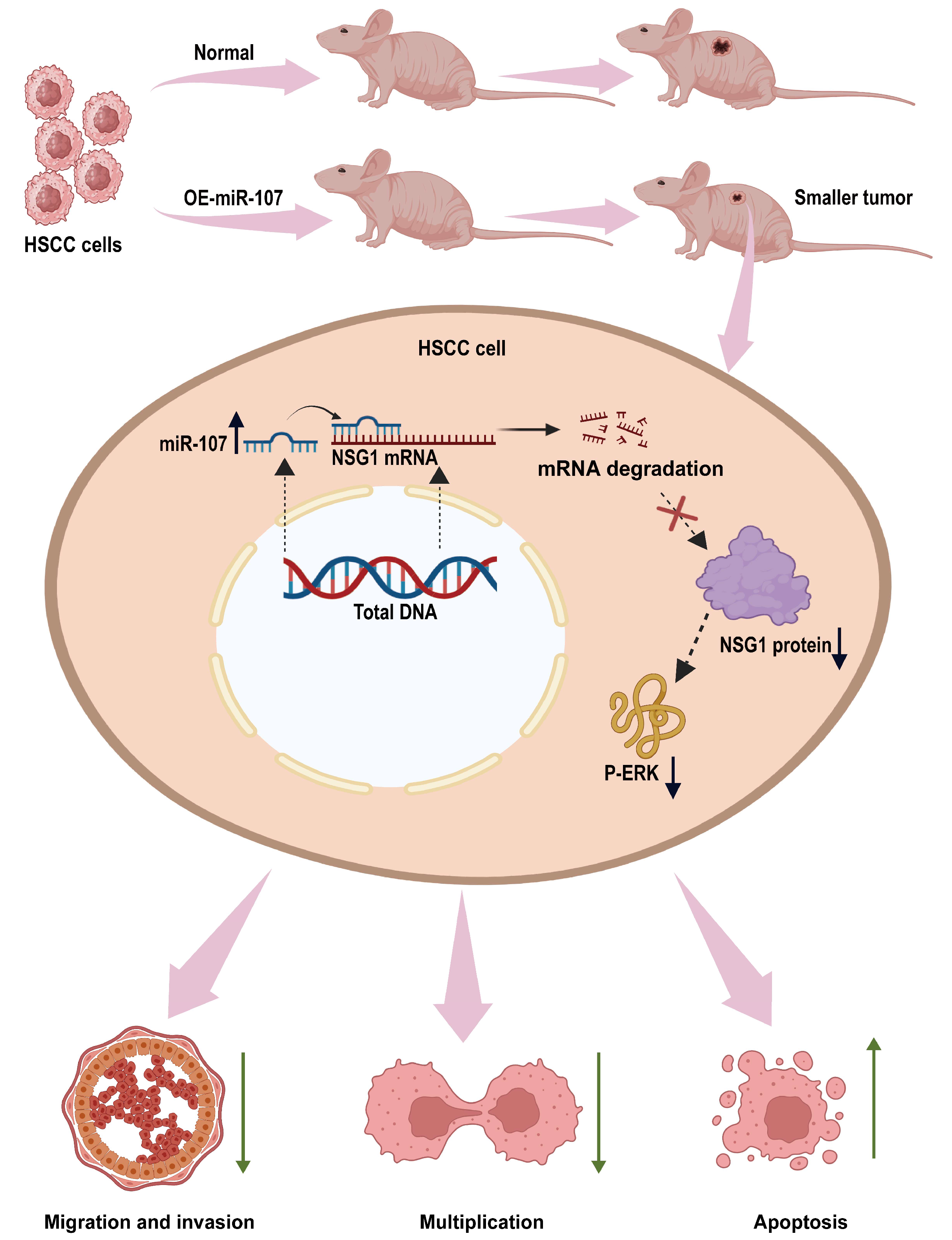
2.6. Overexpression of miR-107 Weakened the Tumorigenic Ability of HSCC Cells In Vivo
3. Discussion
4. Materials and Methods
4.1. Bioinformatics Analysis
4.2. Patients and Specimens
4.3. Cell Culture
4.4. PCR Assay
4.5. Western Blot
4.6. Apoptosis Detection by Flow Cytometry
4.7. Cell Counting Kit 8
4.8. Cell Transfection
4.9. Wound-Healing Assay
4.10. Colony Formation
4.11. Transwell Invasion Assay
4.12. Animal Study
4.13. Hematoxylin and Eosin Staining
4.14. Immunohistochemistry
4.15. Data Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Garneau, J.C.; Bakst, R.L.; Miles, B.A. Hypopharyngeal cancer: A state of the art review. Oral Oncol. 2018, 86, 244–250. [Google Scholar] [CrossRef]
- Chan, J.Y.W.; Wei, W.I. Current management strategy of hypopharyngeal carcinoma. Auris Nasus Larynx 2013, 40, 2–6. [Google Scholar] [CrossRef]
- Bozec, A.; Poissonnet, G.; Dassonville, O.; Culié, D. Current Therapeutic Strategies for Patients with Hypopharyngeal Carcinoma: Oncologic and Functional Outcomes. J. Clin. Med. 2023, 12, 1237. [Google Scholar] [CrossRef]
- Liu, J.; Pandya, P.; Afshar, S. Therapeutic Advances in Oncology. Int. J. Mol. Sci. 2021, 22, 2008. [Google Scholar] [CrossRef]
- Fan, Y.; Li, H.; Yu, Z.; Dong, W.; Cui, X.; Ma, J.; Li, S. Long non-coding RNA FGD5-AS1 promotes non-small cell lung cancer cell proliferation through sponging hsa-miR-107 to up-regulate FGFRL1. Biosci. Rep. 2020, 40, BSR20193309. [Google Scholar] [CrossRef]
- Nelson, P.T.; Wang, W.X.; Mao, G.; Wilfred, B.R.; Xie, K.; Jennings, M.H.; Gao, Z.; Wang, X. Specific sequence determinants of miR-15/107 microRNA gene group targets. Nucleic Acids Res. 2011, 39, 8163–8172. [Google Scholar] [CrossRef]
- Chava, S.; Reynolds, C.P.; Pathania, A.S.; Gorantla, S.; Poluektova, L.Y.; Coulter, D.W.; Gupta, S.C.; Pandey, M.K.; Challagundla, K.B. miR-15a-5p, miR-15b-5p, and miR-16-5p inhibit tumor progression by directly targeting MYCN in neuroblastoma. Mol. Oncol. 2020, 14, 180–196. [Google Scholar] [CrossRef]
- Xue, G.; Yan, H.-L.; Zhang, Y.; Hao, L.-Q.; Zhu, X.-T.; Mei, Q.; Sun, S.-H. c-Myc-mediated repression of miR-15-16 in hypoxia is induced by increased HIF-2α and promotes tumor angiogenesis and metastasis by upregulating FGF2. Oncogene 2015, 34, 1393–1406. [Google Scholar] [CrossRef]
- Moncini, S.; Lunghi, M.; Valmadre, A.; Grasso, M.; Del Vescovo, V.; Riva, P.; Denti, M.A.; Venturin, M. The miR-15/107 Family of microRNA Genes Regulates CDK5R1/p35 with Implications for Alzheimer’s Disease Pathogenesis. Mol. Neurobiol. 2017, 54, 4329–4342. [Google Scholar] [CrossRef]
- Tian, X.; Wu, Y.; Yang, Y.; Wang, J.; Niu, M.; Gao, S.; Qin, T.; Bao, D. Long noncoding RNA LINC00662 promotes M2 macrophage polarization and hepatocellular carcinoma progression via activating Wnt/β-catenin signaling. Mol. Oncol. 2020, 14, 462–483. [Google Scholar] [CrossRef]
- Wang, S.; Ma, G.; Zhu, H.; Lv, C.; Chu, H.; Tong, N.; Wu, D.; Qiang, F.; Gong, W.; Zhao, Q.; et al. miR-107 regulates tumor progression by targeting NF1 in gastric cancer. Sci. Rep. 2016, 6, 36531. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Han, Y.; Hu, S.; Cai, Y.; Yang, J.; Ren, S.; Zhao, Y.; Lu, T.; Zhou, X.; Wang, X. Circulating Exosomal MiR-107 Restrains Tumorigenesis in Diffuse Large B-Cell Lymphoma by Targeting 14-3-3η. Front. Cell Dev. Biol. 2021, 9, 667800. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.; Sun, C.; Sun, S.; Li, A.; Hu, Q.; Liu, Y.; Zhang, Y. Phosphatidylinositol (3,5)-bisphosphate machinery regulates neurite thickness through neuron-specific endosomal protein NSG1/NEEP21. J. Biol. Chem. 2023, 299, 102775. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Lin, X.; Zhang, L.; Yu, L.; Wu, Q.; Zhang, S.; Xue, F.; Huang, Y. Development of a panel of serum IgG and IgA autoantibodies for early diagnosis of colon cancer. Int. J. Med. Sci. 2020, 17, 2744–2750. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Zhang, L.; Tu, M.; Yin, X.; Cai, L.; Zhang, S.; Yu, L.; Pan, X.; Huang, Y. Development of a panel of autoantibody against NSG1 with CEA, CYFRA21-1, and SCC-Ag for the diagnosis of esophageal squamous cell carcinoma. Clin. Chim. Acta 2021, 520, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Tu, M.; Zhang, Y.; Zhuang, W.; Cai, L.; Zhang, L.; Yu, L.; Zhang, Z.; Huang, Y. Aberrant NSG1 Expression Promotes Esophageal Squamous Cell Carcinoma Cell EMT by the Activation of ERK Signaling Pathway. Dig. Dis. Sci. 2023, 68, 1847–1857. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Ding, Y.; He, Y.; Yu, Z.; Zhou, Y.; Gong, A.; Xu, M. LncRNA UCA1 promotes pancreatic cancer cell migration by regulating mitochondrial dynamics via the MAPK pathway. Arch. Biochem. Biophys. 2023, 748, 109783. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.-C.; Chen, T.-Y.; Liao, L.-T.; Chen, T.; Li, Q.-L.; Hu, J.-W.; Zhou, P.-H.; Zhang, Y.-Q. NETO2 promotes esophageal cancer progression by inducing proliferation and metastasis via PI3K/AKT and ERK pathway. Int. J. Biol. Sci. 2021, 17, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Kim, W.-J.; Kang, H.-G.; Jang, J.-H.; Choi, I.J.; Chun, K.-H.; Kim, S.-J. Upregulation of LAMB1 via ERK/c-Jun Axis Promotes Gastric Cancer Growth and Motility. Int. J. Mol. Sci. 2021, 22, 626. [Google Scholar] [CrossRef]
- Shang, R.; Lee, S.; Senavirathne, G.; Lai, E.C. microRNAs in action: Biogenesis, function and regulation. Nat. Rev. Genet. 2023, 24, 816–833. [Google Scholar] [CrossRef]
- Hussen, B.M.; Hidayat, H.J.; Salihi, A.; Sabir, D.K.; Taheri, M.; Ghafouri-Fard, S. MicroRNA: A signature for cancer progression. Biomed. Pharmacother. 2021, 138, 111528. [Google Scholar] [CrossRef] [PubMed]
- Ding, R.; Hong, W.; Huang, L.; Shao, J.; Yu, W.; Xu, X. Examination of the effects of microRNA-145-5p and phosphoserine aminotransferase 1 in colon cancer. Bioengineered 2022, 13, 12794–12806. [Google Scholar] [CrossRef] [PubMed]
- Nan, Y.; Guo, L.; Lu, Y.; Guo, G.; Hong, R.; Zhao, L.; Wang, L.; Ren, B.; Yu, K.; Zhong, Y.; et al. miR-451 suppresses EMT and metastasis in glioma cells. Cell Cycle 2021, 20, 1270–1278. [Google Scholar] [CrossRef] [PubMed]
- Yan, T.; Wang, X.; Wei, G.; Li, H.; Hao, L.; Liu, Y.; Yu, X.; Zhu, W.; Liu, P.; Zhu, Y.; et al. Exosomal miR-10b-5p mediates cell communication of gastric cancer cells and fibroblasts and facilitates cell proliferation. J. Cancer 2021, 12, 2140–2150. [Google Scholar] [CrossRef] [PubMed]
- Finnerty, J.R.; Wang, W.X.; Hébert, S.S.; Wilfred, B.R.; Mao, G.; Nelson, P.T. The miR-15/107 group of microRNA genes: Evolutionary biology, cellular functions, and roles in human diseases. J. Mol. Biol. 2010, 402, 491–509. [Google Scholar] [CrossRef] [PubMed]
- Quann, K.; Jing, Y.; Rigoutsos, I. Post-transcriptional regulation of BRCA1 through its coding sequence by the miR-15/107 group of miRNAs. Front. Genet. 2015, 6, 242. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Huang, H.; Wang, L.; Liu, Y.; Wang, M.; Zhao, S.; Lu, G.; Kang, X. Cancer-associated fibroblasts secreted miR-103a-3p suppresses apoptosis and promotes cisplatin resistance in non-small cell lung cancer. Aging 2021, 13, 14456–14468. [Google Scholar] [CrossRef]
- Bu, L.; Tian, Y.; Wen, H.; Jia, W.; Yang, S. miR-195-5p exerts tumor-suppressive functions in human lung cancer cells through targeting TrxR2. Acta Biochim. Biophys. Sin. 2021, 53, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Turco, C.; Donzelli, S.; Fontemaggi, G. miR-15/107 microRNA Gene Group: Characteristics and Functional Implications in Cancer. Front. Cell Dev. Biol. 2020, 8, 427. [Google Scholar] [CrossRef]
- Chen, H.A.; Li, C.C.; Lin, Y.J.; Wang, T.F.; Chen, M.C.; Su, Y.H.; Yeh, Y.L.; Padma, V.V.; Liao, P.H.; Huang, C.Y. Hsa-miR-107 regulates chemosensitivity and inhibits tumor growth in hepatocellular carcinoma cells. Aging 2021, 13, 12046–12057. [Google Scholar] [CrossRef]
- Loosen, S.H.; Castoldi, M.; Jördens, M.S.; Roy, S.; Vucur, M.; Kandler, J.; Hammerich, L.; Mohr, R.; Tacke, F.; Ulmer, T.F.; et al. Serum levels of circulating microRNA-107 are elevated in patients with early-stage HCC. PLoS ONE 2021, 16, e0247917. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Wang, Y.; Jiang, C.; Wang, Q.; Mi, J.; Zhang, Y.; Zuo, L.; Geng, Z.; Song, X.; Ge, S.; et al. miR-107 regulates the effect of MCM7 on the proliferation and apoptosis of colorectal cancer via the PAK2 pathway. Biochem. Pharmacol. 2021, 190, 114610. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Yang, Z.; Huang, F.; Yin, L.; Yan, G.; Gong, G. microRNA-107 inhibits gastric cancer cell proliferation and metastasis by targeting PI3K/AKT pathway. Microb. Pathog. 2018, 121, 110–114. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Kobeissi, A.; Dong, Y.; Kaplan, N.; Yang, W.; He, C.; Zeng, K.; Peng, H. MicroRNAs-103/107 Regulate Autophagy in the Epidermis. J. Investig. Dermatol. 2018, 138, 1481–1490. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Lan, T.; Liao, H.; Feng, X.; Chen, X.; Liao, W.; Hou, G.; Xu, L.; Feng, Q.; Xie, K.; et al. CircNFIB inhibits tumor growth and metastasis through suppressing MEK1/ERK signaling in intrahepatic cholangiocarcinoma. Mol. Cancer 2022, 21, 18. [Google Scholar] [CrossRef] [PubMed]
- Ishola, A.A.; Chien, C.-S.; Yang, Y.-P.; Chien, Y.; Yarmishyn, A.A.; Tsai, P.-H.; Chen, J.C.-Y.; Hsu, P.-K.; Luo, Y.-H.; Chen, Y.-M.; et al. Oncogenic circRNA C190 Promotes Non–Small Cell Lung Cancer via Modulation of the EGFR/ERK Pathway. Cancer Res. 2022, 82, 75–89. [Google Scholar] [CrossRef]
- Wang, Q.; Liao, C.; Tan, Z.; Li, X.; Guan, X.; Li, H.; Tian, Z.; Liu, J.; An, J. FUT6 inhibits the proliferation, migration, invasion, and EGF-induced EMT of head and neck squamous cell carcinoma (HNSCC) by regulating EGFR/ERK/STAT signaling pathway. Cancer Gene Ther. 2023, 30, 182–191. [Google Scholar] [CrossRef] [PubMed]
- Granda-Díaz, R.; Manterola, L.; Hermida-Prado, F.; Rodríguez, R.; Santos, L.; García-De-La-Fuente, V.; Fernández, M.T.; Corte-Torres, M.D.; Rodrigo, J.P.; Álvarez-Teijeiro, S.; et al. Targeting oncogenic functions of miR-301a in head and neck squamous cell carcinoma by PI3K/PTEN and MEK/ERK pathways. Biomed. Pharmacother. 2023, 161, 114512. [Google Scholar] [CrossRef] [PubMed]
- Dar, G.M.; Agarwal, S.; Kumar, A.; Sharma, A.K.; Verma, R.; Sattar, R.S.A.; Ahmad, E.; Ali, A.; Mahajan, B.; Saluja, S.S.; et al. A non-invasive miRNA-based approach in early diagnosis and therapeutics of oral cancer. Crit. Rev. Oncol. Hematol. 2022, 180, 103850. [Google Scholar] [CrossRef]
- He, B.; Zhao, Z.; Cai, Q.; Zhang, Y.; Zhang, P.; Shi, S.; Xie, H.; Peng, X.; Yin, W.; Tao, Y.; et al. miRNA-based biomarkers, therapies, and resistance in Cancer. Int. J. Biol. Sci. 2020, 16, 2628–2647. [Google Scholar] [CrossRef]
- Kasinski, A.L.; Slack, F.J. miRNA-34 prevents cancer initiation and progression in a therapeutically resistant K-ras and p53-induced mouse model of lung adenocarcinoma. Cancer Res. 2012, 72, 5576–5587. [Google Scholar] [CrossRef] [PubMed]
- Tassone, P.; Di Martino, M.T.; Arbitrio, M.; Fiorillo, L.; Staropoli, N.; Ciliberto, D.; Cordua, A.; Scionti, F.; Bertucci, B.; Salvino, A.; et al. Safety and activity of the first-in-class locked nucleic acid (LNA) miR-221 selective inhibitor in refractory advanced cancer patients: A first-in-human, phase 1, open-label, dose-escalation study. J. Hematol. Oncol. 2023, 16, 68. [Google Scholar] [CrossRef] [PubMed]
- Kara, G.; Calin, G.A.; Ozpolat, B. RNAi-based therapeutics and tumor targeted delivery in cancer. Adv. Drug Deliv. Rev. 2022, 182, 114113. [Google Scholar] [CrossRef] [PubMed]



| miR-107-RT | GTCGTATCGACTGCAGGGTCCGAGGTATTCGCAGTCGATACGACTGATAG | |
|---|---|---|
| Forward | Reverse | |
| miR-107 | GCAGCAGCATTGTACAGGG | ACTGCAGGGTCCGAGGTATT |
| NSG1 | GTTGGGGAACAATTTCGCAG | GGTGACACCCTCCGTGATG |
| β-Actin | AAGTGTGACGTTGACATCCG | GATCCACATCTGCTGGAAGG |
| U6 | TGCCCCACATAATGCTACC | TATGTCCGTCTGTGGAAACC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, Y.; He, Z.; Han, B.; Lin, Z.; Zhou, P.; Li, S.; Huang, S.; Chen, X. miR-107 Targets NSG1 to Regulate Hypopharyngeal Squamous Cell Carcinoma Progression through ERK Pathway. Int. J. Mol. Sci. 2024, 25, 5961. https://doi.org/10.3390/ijms25115961
Hu Y, He Z, Han B, Lin Z, Zhou P, Li S, Huang S, Chen X. miR-107 Targets NSG1 to Regulate Hypopharyngeal Squamous Cell Carcinoma Progression through ERK Pathway. International Journal of Molecular Sciences. 2024; 25(11):5961. https://doi.org/10.3390/ijms25115961
Chicago/Turabian StyleHu, Yifan, Zhizhen He, Baoai Han, Zehua Lin, Peng Zhou, Shuang Li, Shuo Huang, and Xiong Chen. 2024. "miR-107 Targets NSG1 to Regulate Hypopharyngeal Squamous Cell Carcinoma Progression through ERK Pathway" International Journal of Molecular Sciences 25, no. 11: 5961. https://doi.org/10.3390/ijms25115961
APA StyleHu, Y., He, Z., Han, B., Lin, Z., Zhou, P., Li, S., Huang, S., & Chen, X. (2024). miR-107 Targets NSG1 to Regulate Hypopharyngeal Squamous Cell Carcinoma Progression through ERK Pathway. International Journal of Molecular Sciences, 25(11), 5961. https://doi.org/10.3390/ijms25115961





