A Humanized CB1R Yeast Biosensor Enables Facile Screening of Cannabinoid Compounds
Abstract
:1. Introduction
2. Results
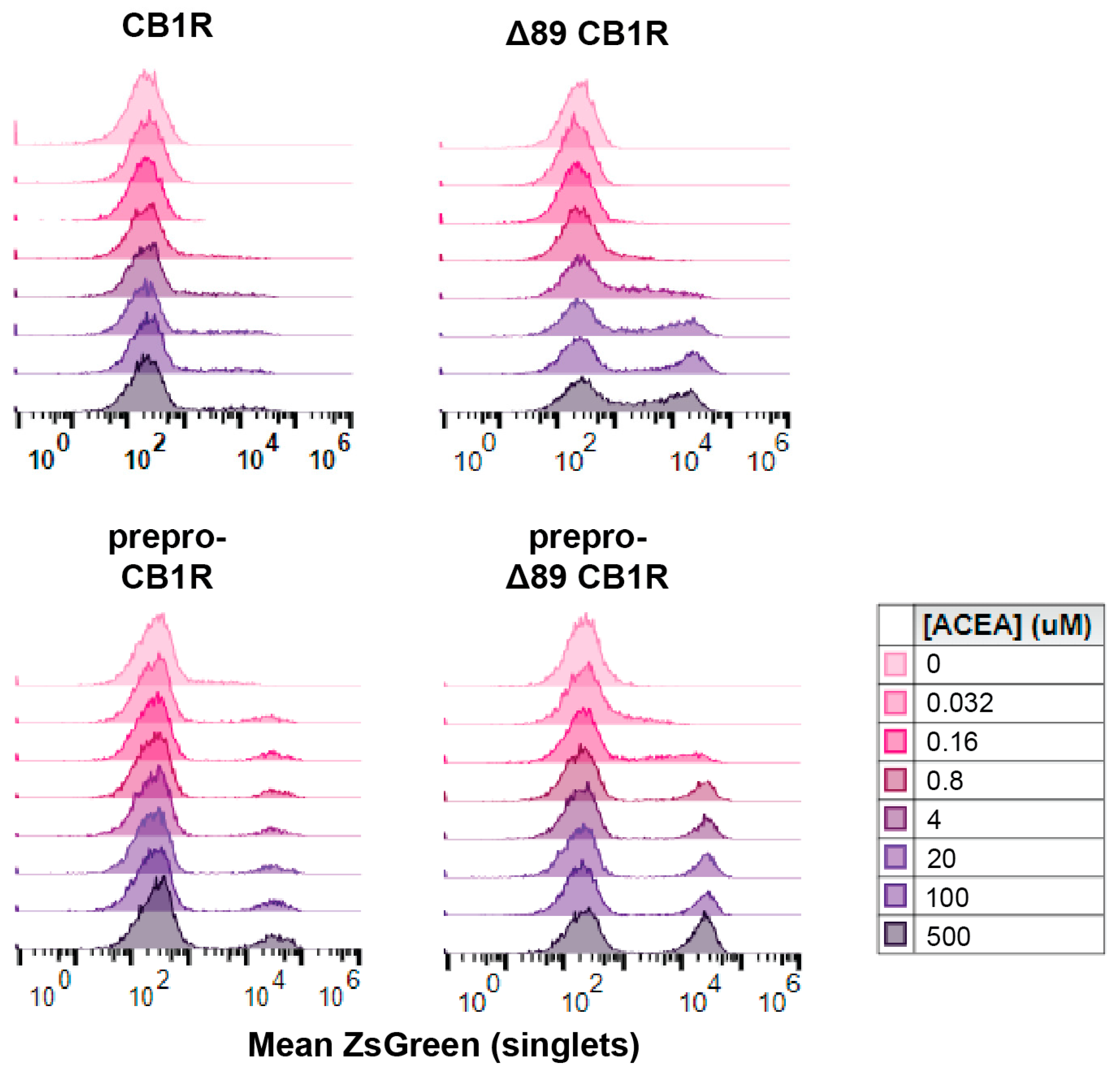
2.1. Engineering Cannabinoid Receptor Function in Yeast
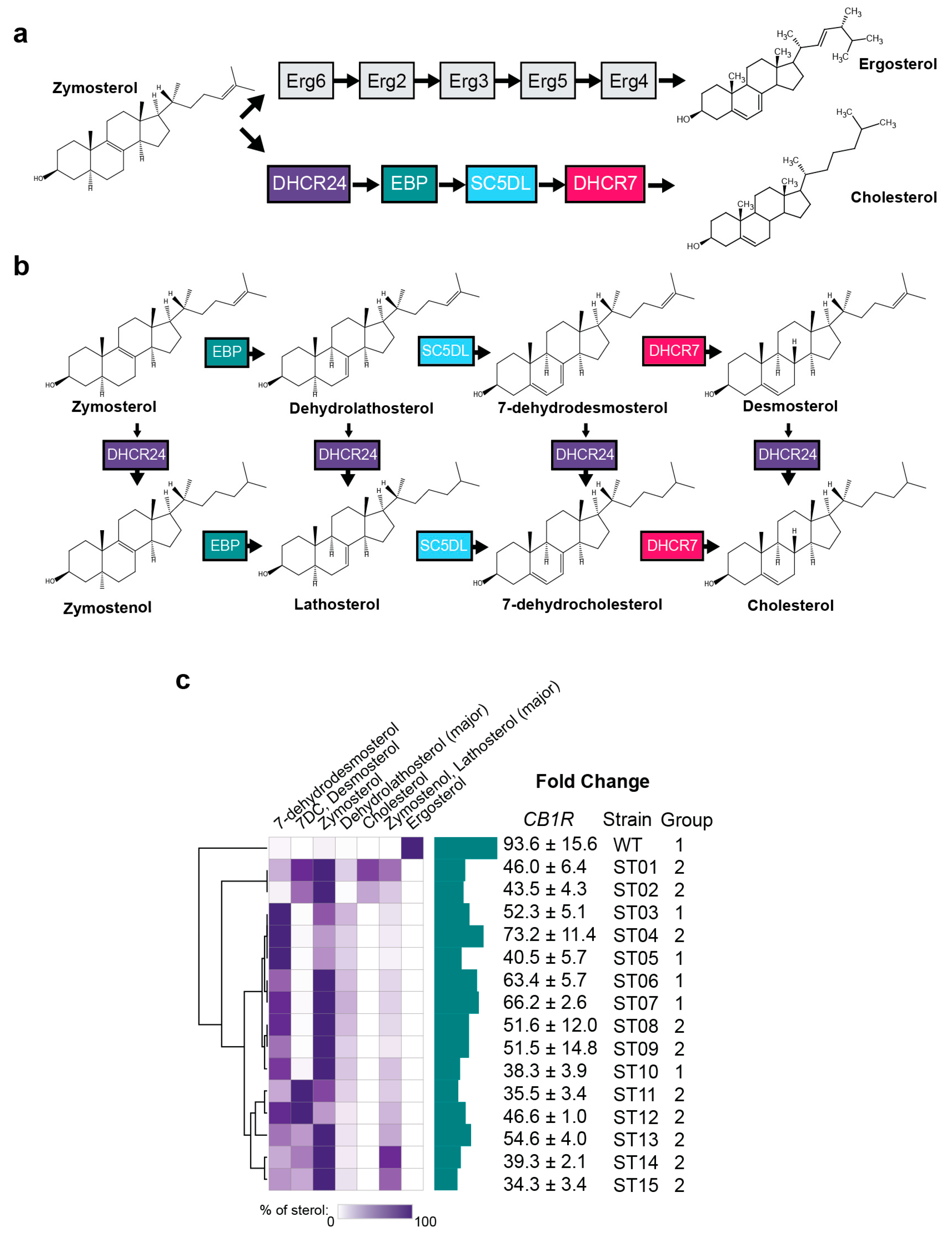
2.2. Yeast Sterol Composition Modulates Cannabinoid Receptor Activity
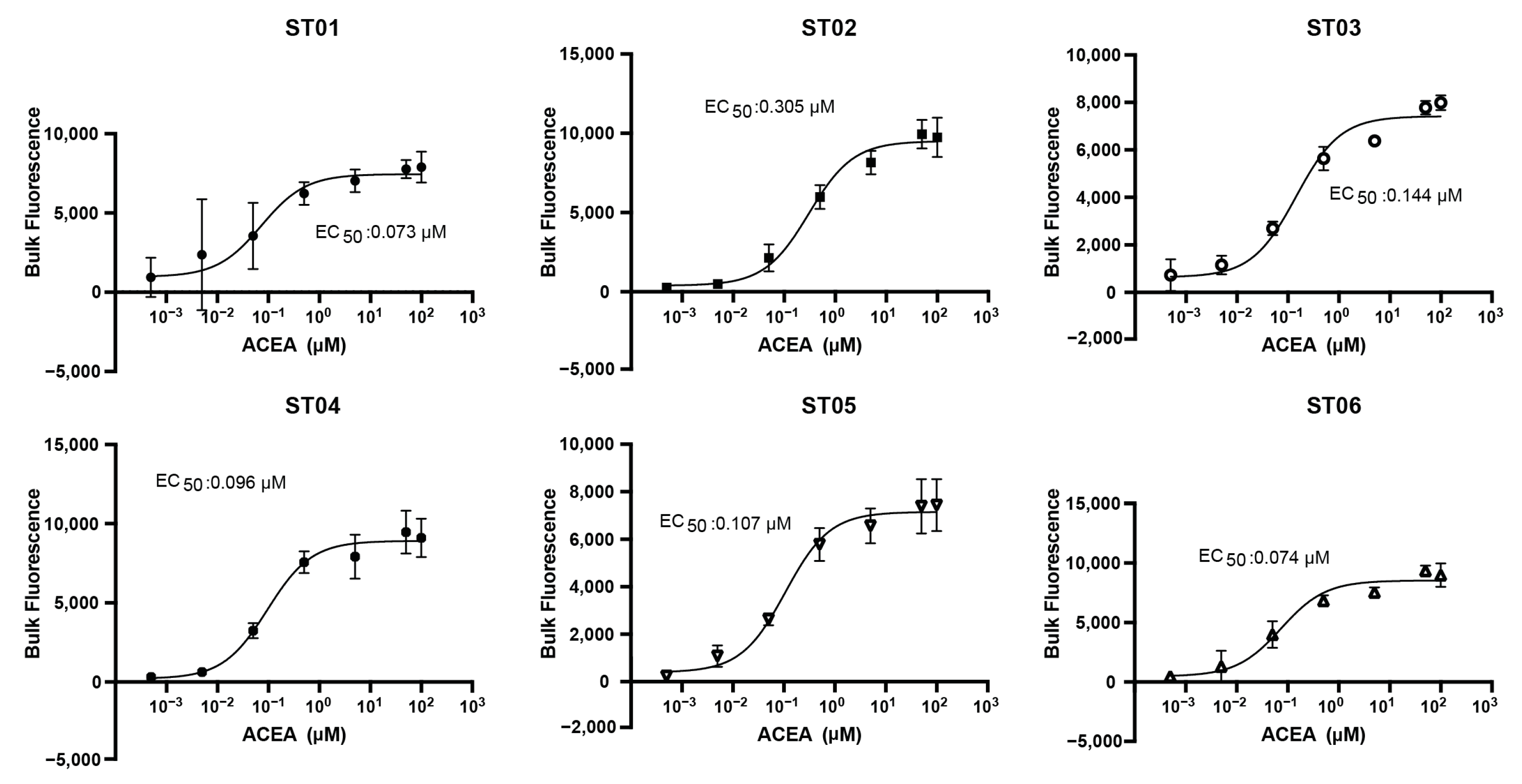
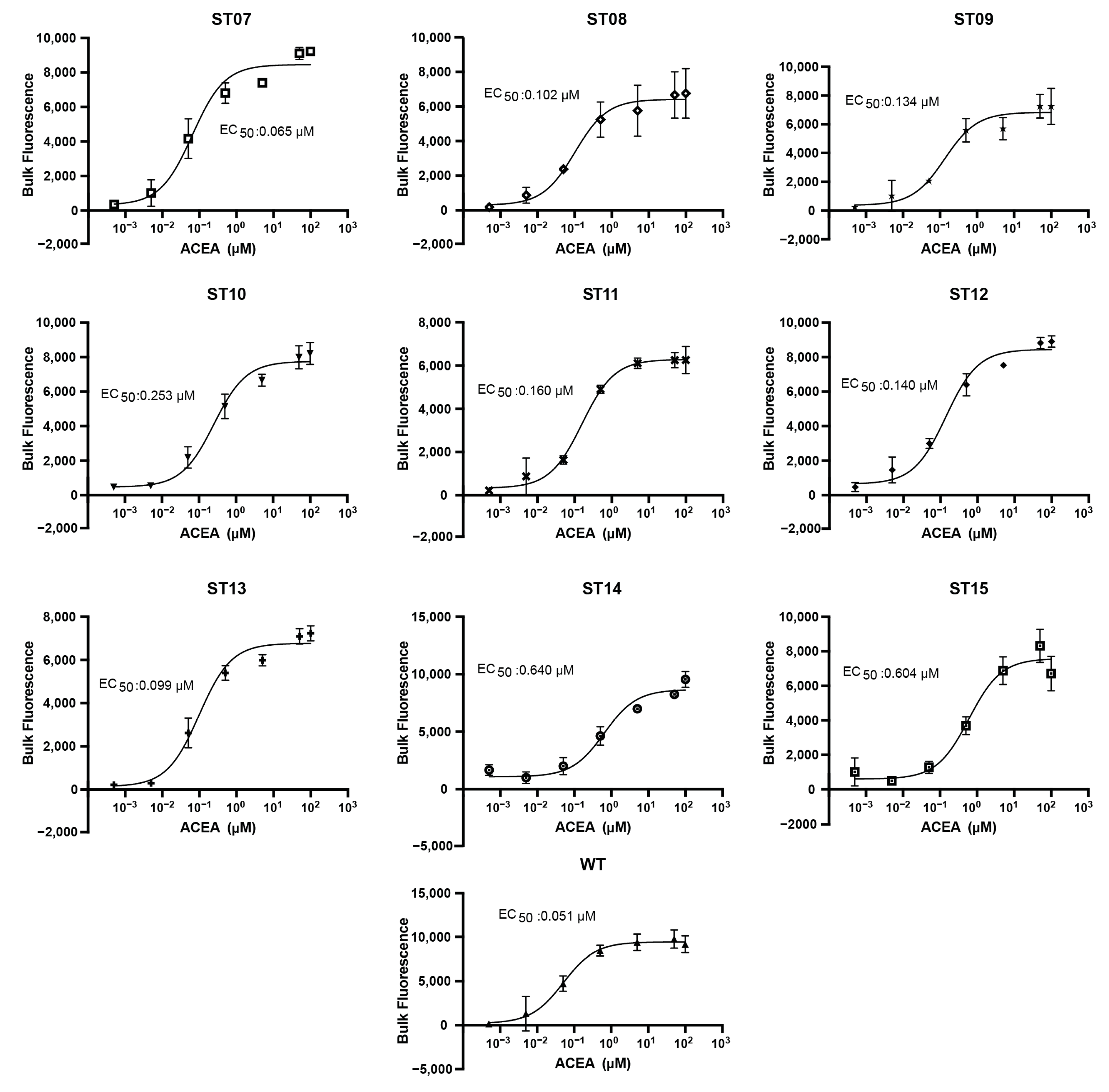
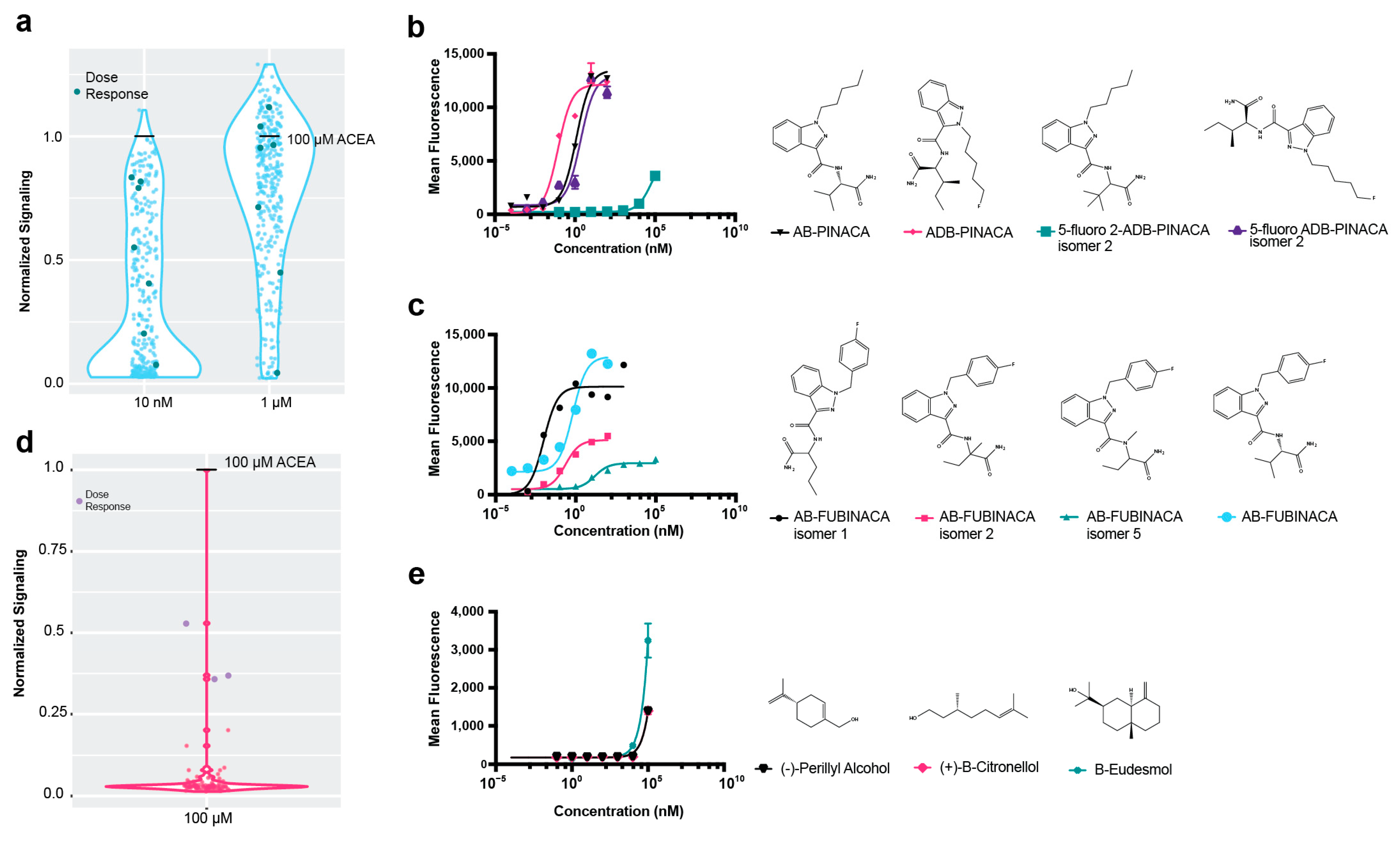
2.3. High-Throughput Drug Screening of Terpenes with Dual Cannabinoid Receptor Biosensor Strains
2.4. High-Throughput Drug Screening of Terpenes with Dual Cannabinoid Receptor Biosensor Strains
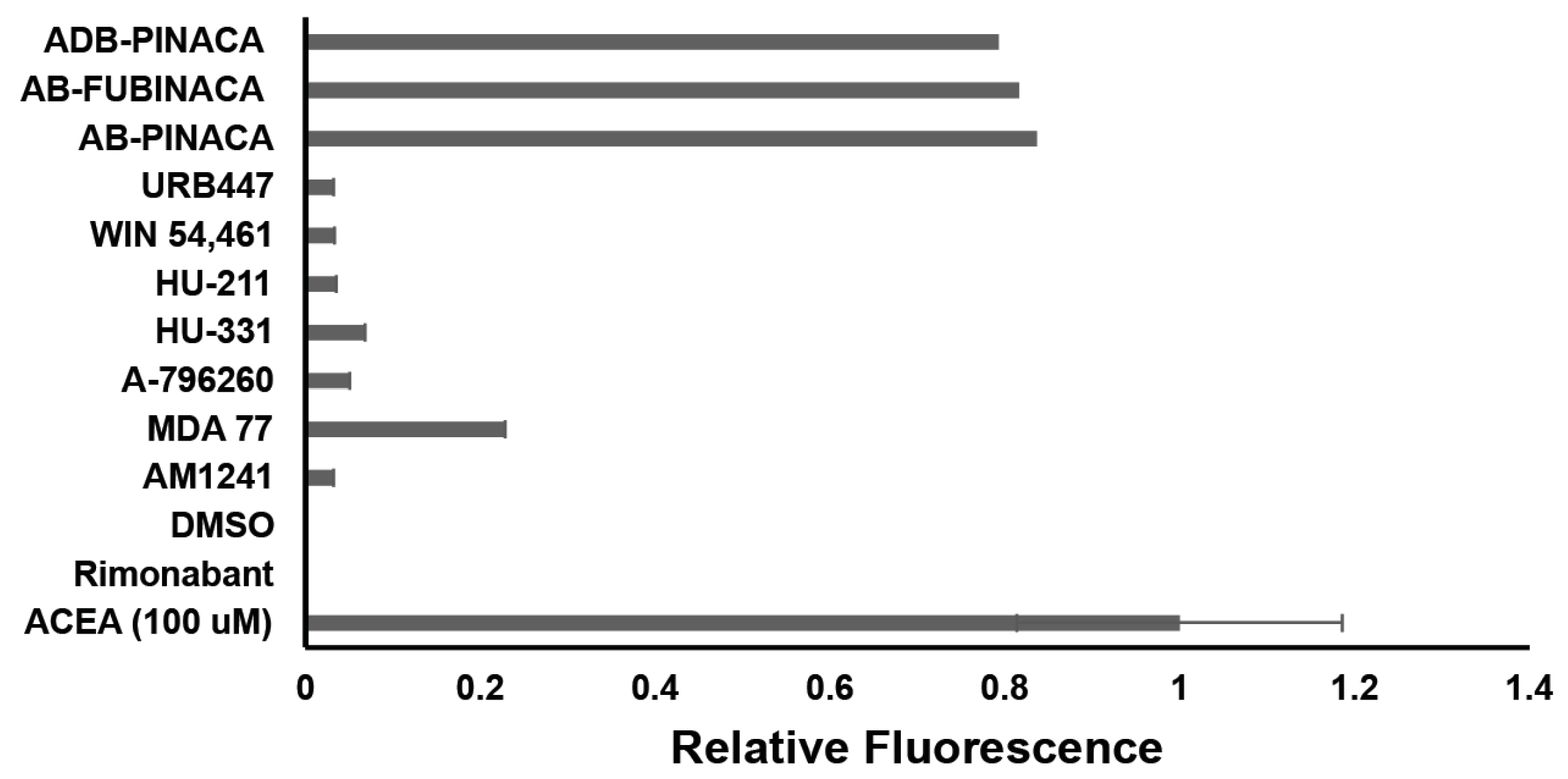
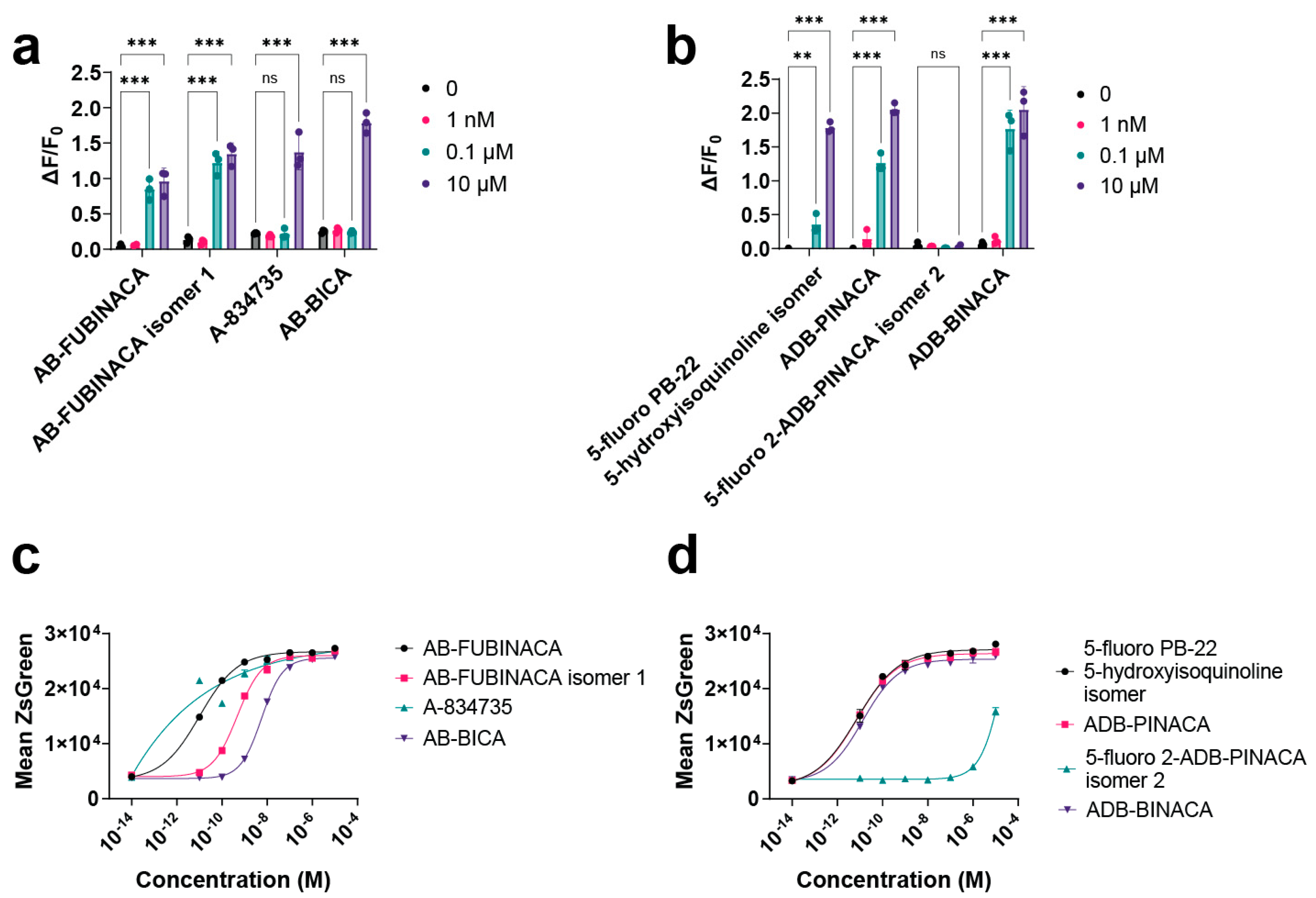
2.5. Mammalian Cell Assays Validate Hits from Yeast Screening
3. Discussion
4. Materials and Methods
4.1. Molecular Biology
4.2. Yeast Transformations
4.3. Yeast Functional Assays
4.4. Mammalian Functional Assays
5. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Hauser, A.S.; Attwood, M.M.; Rask-Andersen, M.; Schiöth, H.B.; Gloriam, D.E. Trends in GPCR Drug Discovery: New Agents, Targets and Indications. Nat. Rev. Drug Discov. 2017, 16, 829–842. [Google Scholar] [CrossRef]
- Versele, M.; Lemaire, K.; Thevelein, J.M. Sex and Sugar in Yeast: Two Distinct GPCR Systems. EMBO Rep. 2001, 2, 574–579. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.; Dyos, S.; Whiteway, M.; White, J.; Watson, M.; Marzioch, M.; Clare, J.; Cousens, D.; Paddon, C.; Plumpton, C.; et al. Functional Coupling of Mammalian Receptors to the Yeast Mating Pathway Using Novel Yeast/Mammalian G Protein Alpha-Subunit Chimeras. Yeast 2000, 16, 11–22. [Google Scholar] [CrossRef]
- Irving, A.J.; McDonald, N.A.; Harkany, T. CB1 Cannabinoid Receptors: Molecular Biology, Second Messenger Coupling and Polarized Trafficking in Neurons. In Cannabinoids and the Brain; Köfalvi, A., Ed.; Springer: Boston, MA, USA, 2008; pp. 59–73. ISBN 978-0-387-74349-3. [Google Scholar]
- Zou, S.; Kumar, U. Cannabinoid Receptors and the Endocannabinoid System: Signaling and Function in the Central Nervous System. Int. J. Mol. Sci. 2018, 19, 833. [Google Scholar] [CrossRef]
- Lu, H.-C.; Mackie, K. An Introduction to the Endogenous Cannabinoid System. Biol. Psychiatry 2016, 79, 516–525. [Google Scholar] [CrossRef] [PubMed]
- Ranganathan, M.; Cortes, J.; Radhakrishnan, R.; Thurnauer, H.; Planeta, B.; Skosnik, P.; Gao, H.; Labaree, D.; Neumeister, A.; Pittman, B.; et al. Reduced Brain Cannabinoid Receptor Availability in Schizophrenia. Biol. Psychiatry 2016, 79, 997–1005. [Google Scholar] [CrossRef]
- Tai, S.; Fantegrossi, W.E. Synthetic Cannabinoids: Pharmacology, Behavioral Effects, and Abuse Potential. Curr. Addict. Rep. 2014, 1, 129–136. [Google Scholar] [CrossRef]
- Adams, A.J.; Banister, S.D.; Irizarry, L.; Trecki, J.; Schwartz, M.; Gerona, R. “Zombie” Outbreak Caused by the Synthetic Cannabinoid AMB-FUBINACA in New York. N. Engl. J. Med. 2017, 376, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Lobato-Freitas, C.; Brito-da-Costa, A.M.; Dinis-Oliveira, R.J.; Carmo, H.; Carvalho, F.; Silva, J.P.; Dias-da-Silva, D. Overview of Synthetic Cannabinoids ADB-FUBINACA and AMB-FUBINACA: Clinical, Analytical, and Forensic Implications. Pharmaceuticals 2021, 14, 186. [Google Scholar] [CrossRef]
- Tokarczyk, B.; Jurczyk, A.; Krupińska, J.; Adamowicz, P. Fatal Intoxication with New Synthetic Cannabinoids 5F-MDMB-PICA and 4F-MDMB-BINACA—Parent Compounds and Metabolite Identification in Blood, Urine and Cerebrospinal Fluid. Forensic Sci. Med. Pathol. 2022, 18, 393–402. [Google Scholar] [CrossRef]
- Hua, T.; Li, X.; Wu, L.; Iliopoulos-Tsoutsouvas, C.; Wang, Y.; Wu, M.; Shen, L.; Brust, C.A.; Nikas, S.P.; Song, F.; et al. Activation and Signaling Mechanism Revealed by Cannabinoid Receptor-Gi Complex Structures. Cell 2020, 180, 655–665.e18. [Google Scholar] [CrossRef] [PubMed]
- Miettinen, K.; Leelahakorn, N.; Almeida, A.; Zhao, Y.; Hansen, L.R.; Nikolajsen, I.E.; Andersen, J.B.; Givskov, M.; Staerk, D.; Bak, S.; et al. A GPCR-Based Yeast Biosensor for Biomedical, Biotechnological, and Point-of-Use Cannabinoid Determination. Nat. Commun. 2022, 13, 3664. [Google Scholar] [CrossRef]
- Shaw, W.M.; Zhang, Y.; Lu, X.; Khalil, A.S.; Ladds, G.; Luo, X.; Ellis, T. Screening Microbially Produced Δ9-Tetrahydrocannabinol Using a Yeast Biosensor Workflow. Nat. Commun. 2022, 13, 5509. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Wong, W.; IJzerman, A.P. Human G Protein-Coupled Receptor Studies in Saccharomyces Cerevisiae. Biochem. Pharmacol. 2016, 114, 103–115. [Google Scholar] [CrossRef]
- Lengger, B.; Jensen, M.K. Engineering G Protein-Coupled Receptor Signalling in Yeast for Biotechnological and Medical Purposes. FEMS Yeast Res. 2019, 20, foz087. [Google Scholar] [CrossRef]
- Mukherjee, K.; Bhattacharyya, S.; Peralta-Yahya, P. GPCR-Based Chemical Biosensors for Medium-Chain Fatty Acids. ACS Synth. Biol. 2015, 4, 1261–1269. [Google Scholar] [CrossRef] [PubMed]
- Prather, P.L.; Martin, N.A.; Breivogel, C.S.; Childers, S.R. Activation of Cannabinoid Receptors in Rat Brain by WIN 55212-2 Produces Coupling to Multiple G Protein α-Subunits with Different Potencies. Mol. Pharmacol. 2000, 57, 1000–1010. [Google Scholar]
- Dohlman, H.G.; Song, J.; Ma, D.; Courchesne, W.E.; Thorner, J. Sst2, a Negative Regulator of Pheromone Signaling in the Yeast Saccharomyces Cerevisiae: Expression, Localization, and Genetic Interaction and Physical Association with Gpa1 (the G-Protein Alpha Subunit). Mol. Cell. Biol. 1996, 16, 5194–5209. [Google Scholar] [CrossRef]
- Nakamura, Y.; Ishii, J.; Kondo, A. Bright Fluorescence Monitoring System Utilizing Zoanthus Sp. Green Fluorescent Protein (ZsGreen) for Human G-Protein-Coupled Receptor Signaling in Microbial Yeast Cells. PLoS ONE 2013, 8, e82237. [Google Scholar] [CrossRef]
- Lee, M.E.; DeLoache, W.C.; Cervantes, B.; Dueber, J.E. A Highly Characterized Yeast Toolkit for Modular, Multipart Assembly. ACS Synth. Biol. 2015, 4, 975–986. [Google Scholar] [CrossRef]
- Membrane Assembly of the Cannabinoid Receptor 1: Impact of a Long N-Terminal Tail | Molecular Pharmacology. Available online: https://molpharm.aspetjournals.org/content/64/3/570 (accessed on 1 August 2022).
- Hebert-Chatelain, E.; Desprez, T.; Serrat, R.; Bellocchio, L.; Soria-Gomez, E.; Busquets-Garcia, A.; Pagano Zottola, A.C.; Delamarre, A.; Cannich, A.; Vincent, P.; et al. A Cannabinoid Link between Mitochondria and Memory. Nature 2016, 539, 555–559. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.; Wilson, R.; O’Brien, S.; Tufarelli, C.; Anderson, S.I.; O’Sullivan, S.E. The Effects of the Endocannabinoids Anandamide and 2-Arachidonoylglycerol on Human Osteoblast Proliferation and Differentiation. PLoS ONE 2015, 10, e0136546. [Google Scholar] [CrossRef] [PubMed]
- Gimpl, G. Interaction of G Protein Coupled Receptors and Cholesterol. Chem. Phys. Lipids 2016, 199, 61–73. [Google Scholar] [CrossRef]
- Sarkar, P.; Chattopadhyay, A. Cholesterol in GPCR Structures: Prevalence and Relevance. J. Membr. Biol. 2022, 255, 99–106. [Google Scholar] [CrossRef]
- Vallée, M.; Vitiello, S.; Bellocchio, L.; Hébert-Chatelain, E.; Monlezun, S.; Martin-Garcia, E.; Kasanetz, F.; Baillie, G.L.; Panin, F.; Cathala, A.; et al. Pregnenolone Can Protect the Brain from Cannabis Intoxication. Science 2014, 343, 94–98. [Google Scholar] [CrossRef]
- Bari, M.; Paradisi, A.; Pasquariello, N.; Maccarrone, M. Cholesterol-Dependent Modulation of Type 1 Cannabinoid Receptors in Nerve Cells. J. Neurosci. Res. 2005, 81, 275–283. [Google Scholar] [CrossRef] [PubMed]
- Oddi, S.; Dainese, E.; Fezza, F.; Lanuti, M.; Barcaroli, D.; De Laurenzi, V.; Centonze, D.; Maccarrone, M. Functional Characterization of Putative Cholesterol Binding Sequence (CRAC) in Human Type-1 Cannabinoid Receptor. J. Neurochem. 2011, 116, 858–865. [Google Scholar] [CrossRef]
- Bean, B.D.M.; Mulvihill, C.J.; Garge, R.K.; Boutz, D.R.; Rousseau, O.; Floyd, B.M.; Cheney, W.; Gardner, E.C.; Ellington, A.D.; Marcotte, E.M.; et al. Functional Expression of Opioid Receptors and Other Human GPCRs in Yeast Engineered to Produce Human Sterols. Nat. Commun. 2022, 13, 2882. [Google Scholar] [CrossRef]
- LoVerme, J.; Duranti, A.; Tontini, A.; Spadoni, G.; Mor, M.; Rivara, S.; Stella, N.; Xu, C.; Tarzia, G.; Piomelli, D. Synthesis and Characterization of a Peripherally Restricted CB1 Cannabinoid Antagonist, URB447, That Reduces Feeding and Body-Weight Gain in Mice. Bioorg. Med. Chem. Lett. 2009, 19, 639–643. [Google Scholar] [CrossRef]
- Eissenstat, M.A.; Bell, M.R.; D’Ambra, T.E.; Alexander, E.J.; Daum, S.J.; Ackerman, J.H.; Gruett, M.D.; Kumar, V.; Estep, K.G.; Olefirowicz, E.M. Aminoalkylindoles: Structure-Activity Relationships of Novel Cannabinoid Mimetics. J. Med. Chem. 1995, 38, 3094–3105. [Google Scholar] [CrossRef]
- Jüttler, E.; Potrovita, I.; Tarabin, V.; Prinz, S.; Dong-Si, T.; Fink, G.; Schwaninger, M. The Cannabinoid Dexanabinol Is an Inhibitor of the Nuclear Factor-Kappa B (NF-κB). Neuropharmacology 2004, 47, 580–592. [Google Scholar] [CrossRef] [PubMed]
- Mechoulam, R.; Ben-Zvi, Z.; Gaoni, Y. Hashish—XIII: On the Nature of the Beam Test. Tetrahedron 1968, 24, 5615–5624. [Google Scholar] [CrossRef] [PubMed]
- Russo, E.B. Taming THC: Potential Cannabis Synergy and Phytocannabinoid-Terpenoid Entourage Effects. Br. J. Pharmacol. 2011, 163, 1344–1364. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, R.S.; McKay, J. Industrial Hemp Cannabis Cultivars and Seeds with Stable Cannabinoid Profiles. U.S. Patent 10,499,584, 10 December 2019. [Google Scholar]
- Fischedick, J.; Elzinga, S. Cannabinoids and Terpenes as Chemotaxonomic Markers in Cannabis. Nat. Prod. Chem. Res. 2015, 3, 181. [Google Scholar] [CrossRef]
- Luo, X.; Reiter, M.A.; d’Espaux, L.; Wong, J.; Denby, C.M.; Lechner, A.; Zhang, Y.; Grzybowski, A.T.; Harth, S.; Lin, W.; et al. Complete Biosynthesis of Cannabinoids and Their Unnatural Analogues in Yeast. Nature 2019, 567, 123–126. [Google Scholar] [CrossRef]
- Ehrenworth, A.M.; Claiborne, T.; Peralta-Yahya, P. Medium-Throughput Screen of Microbially Produced Serotonin via a G-Protein-Coupled Receptor-Based Sensor. Available online: https://pubs.acs.org/doi/pdf/10.1021/acs.biochem.7b00605 (accessed on 1 August 2022).

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mulvihill, C.J.; Lutgens, J.D.; Gollihar, J.D.; Bachanová, P.; Tramont, C.; Marcotte, E.M.; Ellington, A.D.; Gardner, E.C. A Humanized CB1R Yeast Biosensor Enables Facile Screening of Cannabinoid Compounds. Int. J. Mol. Sci. 2024, 25, 6060. https://doi.org/10.3390/ijms25116060
Mulvihill CJ, Lutgens JD, Gollihar JD, Bachanová P, Tramont C, Marcotte EM, Ellington AD, Gardner EC. A Humanized CB1R Yeast Biosensor Enables Facile Screening of Cannabinoid Compounds. International Journal of Molecular Sciences. 2024; 25(11):6060. https://doi.org/10.3390/ijms25116060
Chicago/Turabian StyleMulvihill, Colleen J., Joshua D. Lutgens, Jimmy D. Gollihar, Petra Bachanová, Caitlin Tramont, Edward M. Marcotte, Andrew D. Ellington, and Elizabeth C. Gardner. 2024. "A Humanized CB1R Yeast Biosensor Enables Facile Screening of Cannabinoid Compounds" International Journal of Molecular Sciences 25, no. 11: 6060. https://doi.org/10.3390/ijms25116060




