Targeted Antisense Oligonucleotide-Mediated Skipping of Murine Postn Exon 17 Partially Addresses Fibrosis in D2.mdx Mice
Abstract
:1. Introduction
2. Results
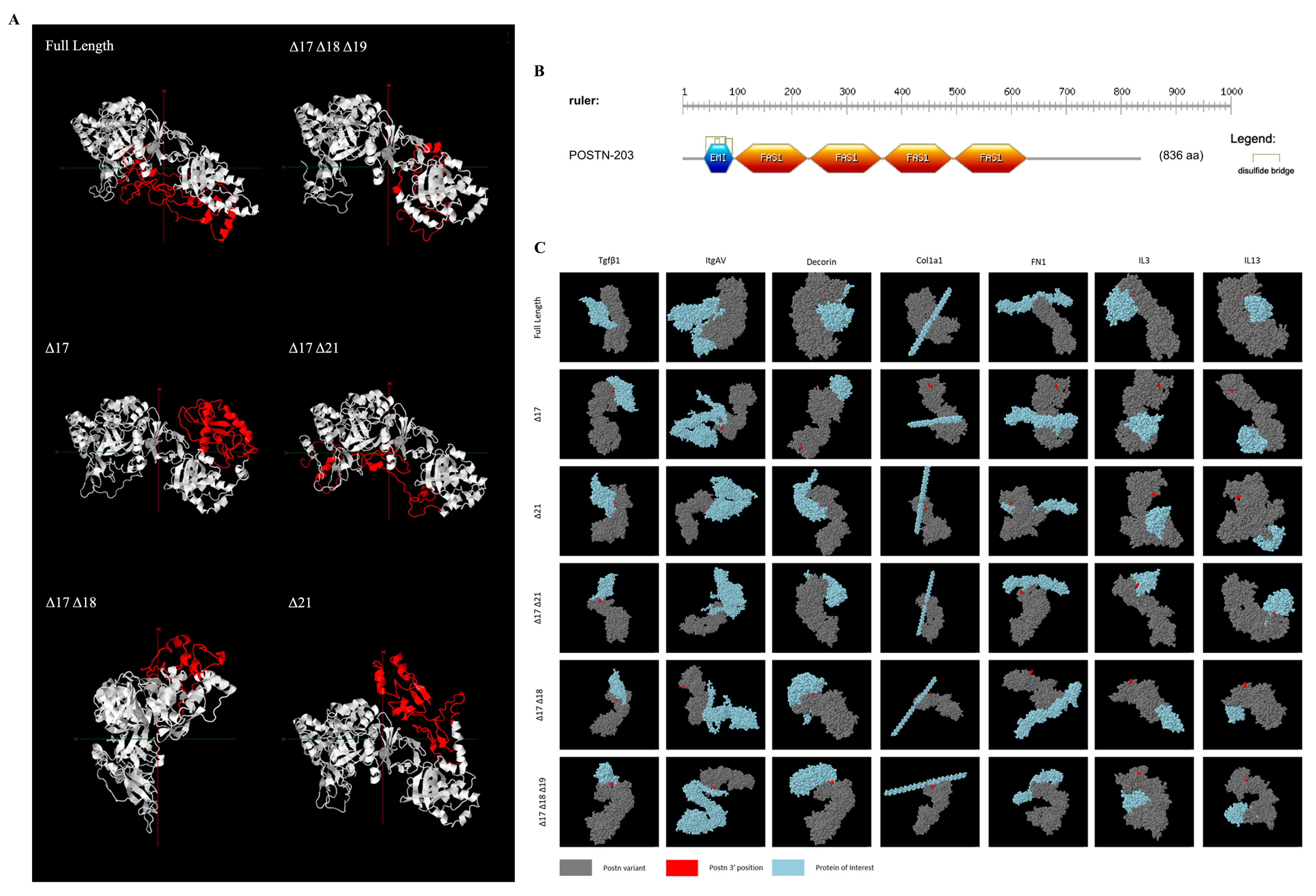
2.1. Bioinformatic Analysis of Periostin Variants Reveals Structural and Docking Disparities
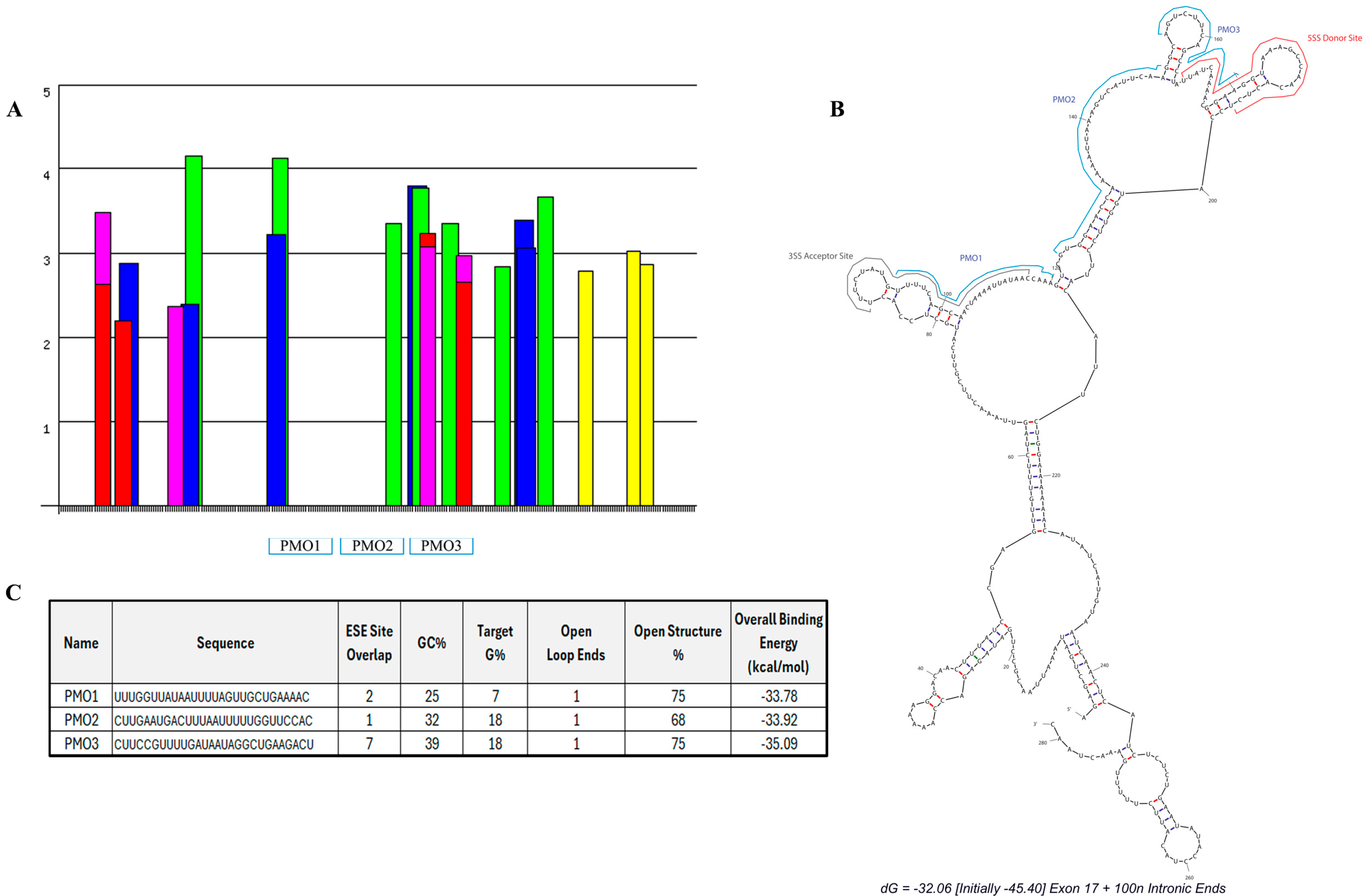
2.2. Design of Antisense Oligonucleotides for the Targeted Skipping of Postn Exon 17
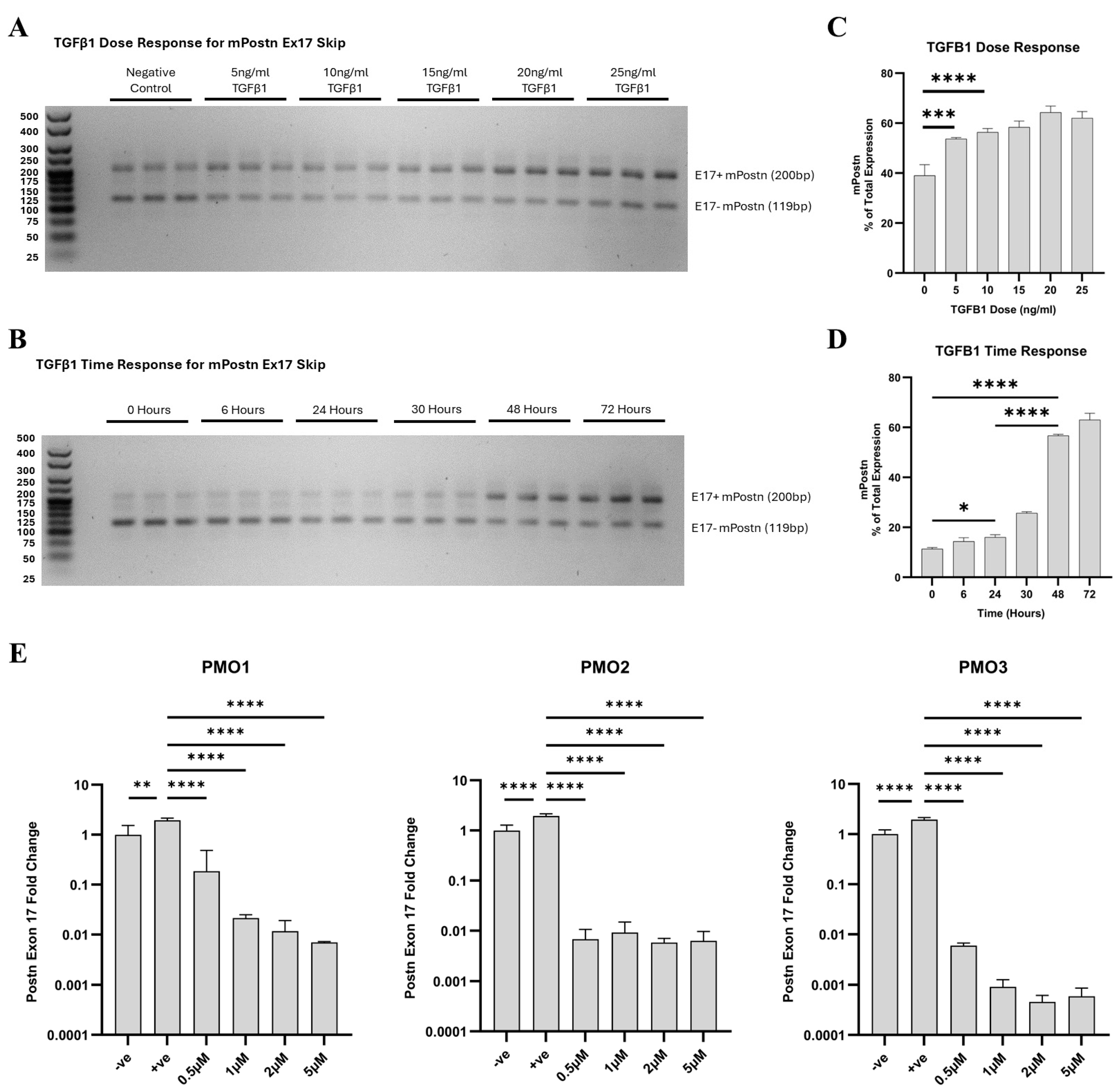
2.3. TGF-β1-Induced Expression of e17+ mPostn Is Mitigated by Targeted mPostn Exon 17 Skipping In Vitro
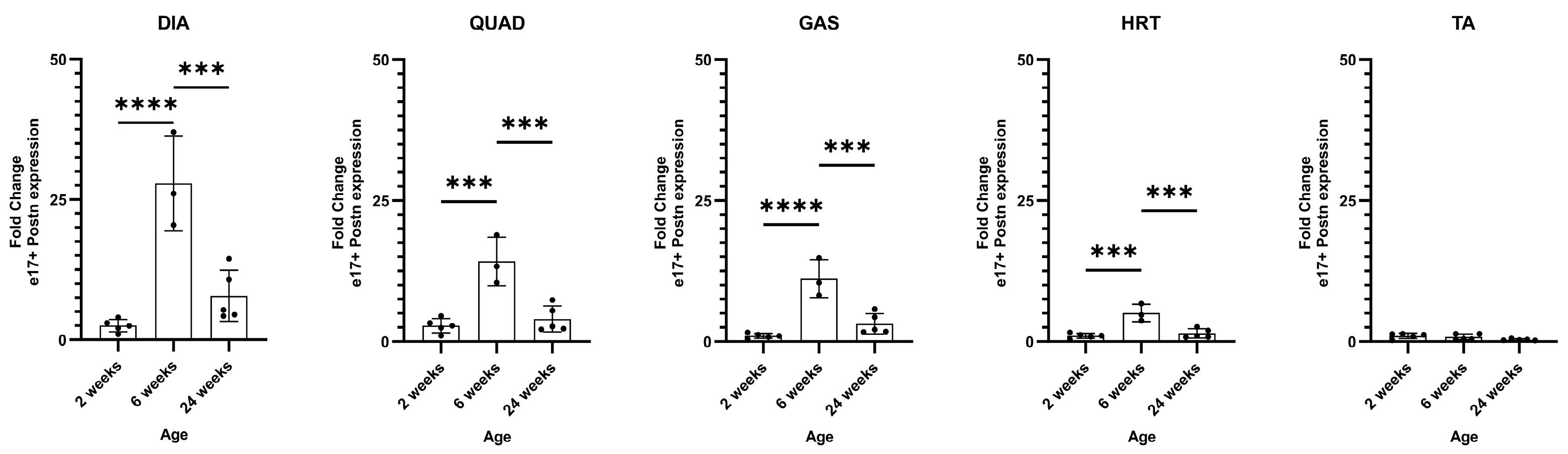
2.4. Intraperitoneal vivoPMO3 Delivery Significantly Decreases mPostn Exon 17 Expression and Improves Grip Strength Muscle Function in the D2.mdx Mouse Model
3. Discussion
4. Materials and Methods
4.1. Structural Analysis of Periostin Protein Products
4.2. Design of the Antisense Sequences to Skip Postn Exon 17
4.3. Cell Culture Experiments in Mh Mouse Fibroblast Cell Line
4.4. Animals and In Vivo Experimental Design
4.5. Fatigue Resistance Analysis
4.6. Forelimb Grip Strength Analysis
4.7. Sample Collection and Processing
4.8. RNA Extraction from Tissues, cDNA Synthesis, and Quantitative Polymerase Chain Reaction
4.9. Protein Extraction and Western Blot
4.10. Hydroxyproline Quantification
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hoffman, E.P.; Brown, R.H.; Kunkel, L.M. Dystrophin: The protein product of the Duchenne muscular dystrophy locus. Cell 1987, 51, 919–928. [Google Scholar] [CrossRef] [PubMed]
- Mah, J.K.; Korngut, L.; Dykeman, J.; Day, L.; Pringsheim, T.; Jette, N. A systematic review and meta-analysis on the epidemiology of Duchenne and Becker muscular dystrophy. Neuromuscul. Disord. NMD 2014, 24, 482–491. [Google Scholar] [CrossRef] [PubMed]
- Bez Batti Angulski, A.; Hosny, N.; Cohen, H.; Martin, A.A.; Hahn, D.; Bauer, J.; Metzger, J.M. Duchenne muscular dystrophy: Disease mechanism and therapeutic strategies. Front. Physiol. 2023, 14, 1183101. [Google Scholar] [CrossRef] [PubMed]
- Bernasconi, P.; Torchiana, E.; Confalonieri, P.; Brugnoni, R.; Barresi, R.; Mora, M.; Cornelio, F.; Morandi, L.; Mantegazza, R. Expression of transforming growth factor-beta 1 in dystrophic patient muscles correlates with fibrosis. Pathogenetic role of a fibrogenic cytoki. J. Clin. Investig. 1995, 96, 1137–1144. [Google Scholar] [CrossRef] [PubMed]
- Hartel, J.V.; Granchelli, J.A.; Hudecki, M.S.; Pollina, C.M.; Gosselin, L.E. Impact of prednisone on TGF-beta1 and collagen in diaphragm muscle from mdx mice. Muscle Nerve 2001, 24, 428–432. [Google Scholar] [CrossRef]
- Hwang, J.H.; Yang, S.H.; Kim, Y.C.; Kim, J.H.; An, J.N.; Moon, K.C.; Oh, Y.K.; Park, J.Y.; Kim, D.K.; Kim, Y.S.; et al. Experimental Inhibition of Periostin Attenuates Kidney Fibrosis. Am. J. Nephrol. 2017, 46, 501–517. [Google Scholar] [CrossRef]
- Butcher, J.T.; Norris, R.A.; Hoffman, S.; Mjaatvedt, C.H.; Markwald, R.R. Periostin promotes atrioventricular mesenchyme matrix invasion and remodeling mediated by integrin signaling through Rho/PI 3-kinase. Dev. Biol. 2007, 302, 256–266. [Google Scholar] [CrossRef]
- Sidhu, S.S.; Yuan, S.; Innes, A.L.; Kerr, S.; Woodruff, P.G.; Hou, L.; Muller, S.J.; Fahy, J.V. Roles of epithelial cell-derived periostin in TGF-beta activation, collagen production, and collagen gel elasticity in asthma. Proc. Natl. Acad. Sci. USA 2010, 107, 14170–14175. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Fan, D.; Wang, C.; Wang, J.-Y.; Cui, X.-B.; Wu, D.; Zhou, Y.; Wu, L.-L. Angiotensin II increases periostin expression via Ras/p38 MAPK/CREB and ERK1/2/TGF-β1 pathways in cardiac fibroblasts. Cardiovasc. Res. 2011, 91, 80–89. [Google Scholar] [CrossRef]
- Maruhashi, T.; Kii, I.; Saito, M.; Kudo, A. Interaction between periostin and BMP-1 promotes proteolytic activation of lysyl oxidase. J. Biol. Chem. 2010, 285, 13294–13303. [Google Scholar] [CrossRef]
- Goetsch, S.C.; Hawke, T.J.; Gallardo, T.D.; Richardson, J.A.; Garry, D.J. Transcriptional profiling and regulation of the extracellular matrix during muscle regeneration. Physiol. Genom. 2003, 14, 261–271. [Google Scholar] [CrossRef] [PubMed]
- Özdemir, C.; Akpulat, U.; Sharafi, P.; Yıldız, Y.; Onbaşılar, İ.; Kocaefe, Ç. Periostin is temporally expressed as an extracellular matrix component in skeletal muscle regeneration and differentiation. Gene 2014, 553, 130–139. [Google Scholar] [CrossRef] [PubMed]
- Lorts, A.; Schwanekamp, J.A.; Baudino, T.A.; McNally, E.M.; Molkentin, J.D. Deletion of periostin reduces muscular dystrophy and fibrosis in mice by modulating the transforming growth factor-β pathway. Proc. Natl. Acad. Sci. USA 2012, 109, 10978–10983. [Google Scholar] [CrossRef] [PubMed]
- Marotta, M.; Ruiz-Roig, C.; Sarria, Y.; Peiro, J.L.; Nuñez, F.; Ceron, J.; Munell, F.; Roig-Quilis, M. Muscle genome-wide expression profiling during disease evolution in mdx mice. Physiol. Genom. 2009, 37, 119–132. [Google Scholar] [CrossRef] [PubMed]
- Holland, A.; Dowling, P.; Meleady, P.; Henry, M.; Zweyer, M.; Mundegar, R.R.; Swandulla, D.; Ohlendieck, K. Label-free mass spectrometric analysis of the mdx-4cv diaphragm identifies the matricellular protein periostin as a potential factor involved in dystrophinopathy-related fibrosis. Proteomics 2015, 15, 2318–2331. [Google Scholar] [CrossRef] [PubMed]
- Trundle, J.; Cernisova, V.; Boulinguiez, A.; Lu-Nguyen, N.; Malerba, A.; Popplewell, L. Expression of the Pro-Fibrotic Marker Periostin in a Mouse Model of Duchenne Muscular Dystrophy. Biomedicines 2024, 12, 216. [Google Scholar] [CrossRef] [PubMed]
- Horiuchi, K.; Amizuka, N.; Takeshita, S.; Takamatsu, H.; Katsuura, M.; Ozawa, H.; Toyama, Y.; Bonewald, L.F.; Kudo, A. Identification and Characterization of a Novel Protein, Periostin, with Restricted Expression to Periosteum and Periodontal Ligament and Increased Expression by Transforming Growth Factor β. J. Bone Miner. Res. 1999, 14, 1239–1249. [Google Scholar] [CrossRef]
- Taniyama, Y.; Katsuragi, N.; Sanada, F.; Azuma, J.; Iekushi, K.; Koibuchi, N.; Okayama, K.; Ikeda-Iwabu, Y.; Muratsu, J.; Otsu, R.; et al. Selective Blockade of Periostin Exon 17 Preserves Cardiac Performance in Acute Myocardial Infarction. Hypertension 2016, 67, 356–361. [Google Scholar] [CrossRef] [PubMed]
- Cernisova, V.; Lu-Nguyen, N.; Trundle, J.; Herath, S.; Malerba, A.; Popplewell, L. Microdystrophin Gene Addition Significantly Improves Muscle Functionality and Diaphragm Muscle Histopathology in a Fibrotic Mouse Model of Duchenne Muscular Dystrophy. Int. J. Mol. Sci. 2023, 24, 8174. [Google Scholar] [CrossRef]
- van Putten, M.; Putker, K.; Overzier, M.; Adamzek, W.A.; Pasteuning-Vuhman, S.; Plomp, J.J.; Aartsma-Rus, A. Natural disease history of the D2-mdx mouse model for Duchenne muscular dystrophy. FASEB J. 2019, 33, 8110–8124. [Google Scholar] [CrossRef]
- Hammers, D.W.; Hart, C.C.; Matheny, M.K.; Wright, L.A.; Armellini, M.; Barton, E.R.; Sweeney, H.L. The D2.mdx mouse as a preclinical model of the skeletal muscle pathology associated with Duchenne muscular dystrophy. Sci. Rep. 2020, 10, 14070. [Google Scholar] [CrossRef] [PubMed]
- Yates, A.D.; Achuthan, P.; Akanni, W.; Allen, J.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Azov, A.G.; Bennett, R.; et al. Ensembl 2020. Nucleic Acids Res. 2020, 48, D682–D688. [Google Scholar] [CrossRef] [PubMed]
- Cai, L.; Brophy, R.H.; Tycksen, E.D.; Duan, X.; Nunley, R.M.; Rai, M.F. Distinct expression pattern of periostin splice variants in chondrocytes and ligament progenitor cells. FASEB J. 2019, 33, 8386–8405. [Google Scholar] [CrossRef]
- Takeshita, S.; Fau, K.R.; Tezuka, K.; Fau, T.K.; Amann, E.; Amann, E. Osteoblast-specific factor 2: Cloning of a putative bone adhesion protein with homology with the insect protein fasciclin I. Biochem. J. 1993, 294, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [PubMed]
- Siriett, V.; Salerno, M.S.; Berry, C.; Nicholas, G.; Bower, R.; Kambadur, R.; Sharma, M. Antagonism of myostatin enhances muscle regeneration during sarcopenia. Mol. Ther. 2007, 15, 1463–1470. [Google Scholar] [CrossRef] [PubMed]
- de Castro, E.; Sigrist, C.J.A.; Gattiker, A.; Bulliard, V.; Langendijk-Genevaux, P.S.; Gasteiger, E.; Bairoch, A.; Hulo, N. ScanProsite: Detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 2006, 34, 362. [Google Scholar] [CrossRef] [PubMed]
- Kozakov, D.; Hall, D.R.; Xia, B.; Porter, K.A.; Padhorny, D.; Yueh, C.; Beglov, D.; Vajda, S. The ClusPro web server for protein-protein docking. Nat. Protoc. 2017, 12, 255–278. [Google Scholar] [CrossRef]
- Cartegni, L.; Wang, J.; Zhu, Z.; Zhang, M.Q.; Krainer, A.R. ESEfinder: A web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003, 31, 3568–3571. [Google Scholar] [CrossRef]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef]
- Cadoni, E.; De Paepe, L.; Manicardi, A.; Madder, A. Beyond small molecules: Targeting G-quadruplex structures with oligonucleotides and their analogues. Nucleic Acids Res. 2021, 49, 6638–6659. [Google Scholar] [CrossRef] [PubMed]
- Rusbjerg-Weberskov, C.E.; Johansen, M.L.; Nowak, J.S.; Otzen, D.E.; Pedersen, J.S.; Enghild, J.J.; Nielsen, N.S. Periostin C-Terminal Is Intrinsically Disordered and Interacts with 143 Proteins in an In Vitro Epidermal Model of Atopic Dermatitis. Biochemistry 2023, 62, 2803–2815. [Google Scholar] [CrossRef] [PubMed]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2021, 50, D439–D444. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Jin, R.; Norris, R.A.; Zhang, L.; Yu, S.; Wu, F.; Markwald, R.R.; Nanda, A.; Conway, S.J.; Smyth, S.S.; et al. Periostin mediates vascular smooth muscle cell migration through the integrins ανβ3 and ανβ5 and focal adhesion kinase (FAK) pathway. Atherosclerosis 2010, 208, 358–365. [Google Scholar] [CrossRef] [PubMed]
- Na Nan, D.; Klincumhom, N.; Trachoo, V.; Everts, V.; Ferreira, J.N.; Osathanon, T.; Pavasant, P. Periostin-integrin interaction regulates force-induced TGF-β1 and α-SMA expression by hPDLSCs. Oral Dis. 2023, 30, 2570–2579. [Google Scholar] [CrossRef] [PubMed]
- González-González, L.; Alonso, J. Periostin: A Matricellular Protein With Multiple Functions in Cancer Development and Progression. Front. Oncol. 2018, 8, 225. [Google Scholar] [CrossRef] [PubMed]
- Hoersch, S.; Andrade-Navarro, M.A. Periostin shows increased evolutionary plasticity in its alternatively spliced region. BMC Evol. Biol. 2010, 10, 30. [Google Scholar] [CrossRef] [PubMed]
- Parimon, T.; Yao, C.; Habiel, D.M.; Ge, L.; Bora, S.A.; Brauer, R.; Evans, C.M.; Xie, T.; Alonso-Valenteen, F.; Medina-Kauwe, L.K.; et al. Syndecan-1 promotes lung fibrosis by regulating epithelial reprogramming through extracellular vesicles. JCI Insight 2019, 5, e129359. [Google Scholar] [CrossRef] [PubMed]
- Tanino, Y.; Wang, X.; Nikaido, T.; Misa, K.; Sato, Y.; Togawa, R.; Kawamata, T.; Kikuchi, M.; Frevert, C.W.; Tanino, M.; et al. Syndecan-4 Inhibits the Development of Pulmonary Fibrosis by Attenuating TGF-β Signaling. Int. J. Mol. Sci. 2019, 20, 4989. [Google Scholar] [CrossRef]
- Kudo, A.; Kii, I. Periostin function in communication with extracellular matrices. J. Cell Commun. Signal 2017, 12, 301–308. [Google Scholar] [CrossRef]
- Ito, N.; Miyagoe-Suzuki, Y.; Takeda, S.i.; Kudo, A. Periostin Is Required for the Maintenance of Muscle Fibers during Muscle Regeneration. Int. J. Mol. Sci. 2021, 22, 3627. [Google Scholar] [CrossRef] [PubMed]
- Hakim, C.H.; Wasala, N.B.; Pan, X.; Kodippili, K.; Yue, Y.; Zhang, K.; Yao, G.; Haffner, B.; Duan, S.X.; Ramos, J.; et al. A Five-Repeat Micro-Dystrophin Gene Ameliorated Dystrophic Phenotype in the Severe DBA/2J-mdx Model of Duchenne Muscular Dystrophy. Mol. Ther.-Methods Clin. Dev. 2017, 6, 216–230. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, D.P.; Dangott, L.J.; Schmitt, E.E.; Vellers, H.L.; Lightfoot, J.T. Differential skeletal muscle proteome of high- and low-active mice. J. Appl. Physiol. 2014, 116, 1057–1067. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, D.P.; Dangott, L.J.; Lightfoot, J.T. Lessons learned from vivo-morpholinos: How to avoid vivo-morpholino toxicity. BioTechniques 2014, 56, 251–256. [Google Scholar] [CrossRef] [PubMed]
- GENE-TOOLS. Available online: https://www.gene-tools.com/vivomorpholinos (accessed on 18 March 2024).
- Forand, A.; Muchir, A.; Mougenot, N.; Sevoz-Couche, C.; Peccate, C.; Lemaitre, M.; Izabelle, C.; Wood, M.; Lorain, S.; Piétri-Rouxel, F. Combined Treatment with Peptide-Conjugated Phosphorodiamidate Morpholino Oligomer-PPMO and AAV-U7 Rescues the Severe DMD Phenotype in Mice. Molecular Therapy. Methods Clin. Dev. 2020, 17, 695–708. [Google Scholar] [CrossRef] [PubMed]
- Peccate, C.; Mollard, A.; Le Hir, M.; Julien, L.; McClorey, G.; Jarmin, S.; Le Heron, A.; Dickson, G.; Benkhelifa-Ziyyat, S.; Piétri-Rouxel, F.; et al. Antisense pre-treatment increases gene therapy efficacy in dystrophic muscles. Hum. Mol. Genet. 2016, 25, 3555–3563. [Google Scholar] [CrossRef] [PubMed]
- Lu-Nguyen, N.; Malerba, A.; Popplewell, L.; Schnell, F.; Hanson, G.; Dickson, G. Systemic Antisense Therapeutics for Dystrophin and Myostatin Exon Splice Modulation Improve Muscle Pathology of Adult mdx Mice. Mol. Therapy. Nucleic Acids 2017, 6, 15–28. [Google Scholar] [CrossRef]
- Beach, M.A.; Nayanathara, U.; Gao, Y.; Zhang, C.; Xiong, Y.; Wang, Y.; Such, G.K. Polymeric Nanoparticles for Drug Delivery. Chem. Rev. 2024, 124, 5505–5616. [Google Scholar] [CrossRef]
- Kaspute, G.; Arunagiri, B.D.; Alexander, R.; Ramanavicius, A.; Samukaite-Bubniene, U. Development of Essential Oil Delivery Systems by ‘Click Chemistry’ Methods: Possible Ways to Manage Duchenne Muscular Dystrophy. Materials 2023, 16, 6537. [Google Scholar] [CrossRef]
- Huang, Y.; Ning, X.; Ahrari, S.; Cai, Q.; Rajora, N.; Saxena, R.; Yu, M.; Zheng, J. Physiological principles underlying the kidney targeting of renal nanomedicines. Nat. Rev. Nephrol. 2024, 20, 354–370. [Google Scholar] [CrossRef]
- Sunkyung, K.; Yong, Z.; Paul, A.R.; Deborah, J.F.; Daniel, W.S.; Rodolfo, V.; Dennis, M.; Brent, J.B. Associations between timing of corticosteroid treatment initiation and clinical outcomes in Duchenne muscular dystrophy. Neuromuscul. Disord. 2017, 27, 730–737. [Google Scholar] [CrossRef] [PubMed]
- Woodman, K.G.; Coles, C.A.; Lamandé, S.R.; White, J.D. Nutraceuticals and Their Potential to Treat Duchenne Muscular Dystrophy: Separating the Credible from the Conjecture. Nutrients 2016, 8, 713. [Google Scholar] [CrossRef] [PubMed]
- RCSB Protein Data Bank. Available online: https://www.rcsb.org/ (accessed on 18 March 2024).
- Stothard, P. The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. BioTechniques 2000, 28, 1102–1104. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Chan, C.Y.; Lawrence, C.E. Sfold web server for statistical folding and rational design of nucleic acids. Nucleic Acids Res. 2004, 32, W135–W141. [Google Scholar] [CrossRef] [PubMed]
- Gruber, A.R.; Lorenz, R.; Bernhart, S.H.; Neuböck, R.; Hofacker, I.L. The Vienna RNA websuite. Nucleic Acids Res. 2008, 36 (Suppl. 2), W70–W74. [Google Scholar] [CrossRef]
- Morcos, P.A.; Li, Y.; Jiang, S. Vivo-Morpholinos: A non-peptide transporter delivers Morpholinos into a wide array of mouse tissues. Biotechniques 2008, 45, 613–623. [Google Scholar] [CrossRef]
- Lazar, I.; Lazar, I. Gel Analyzer 19.1. 2020. Available online: http://www.gelanalyzer.com (accessed on 18 March 2024).

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trundle, J.; Lu-Nguyen, N.; Malerba, A.; Popplewell, L. Targeted Antisense Oligonucleotide-Mediated Skipping of Murine Postn Exon 17 Partially Addresses Fibrosis in D2.mdx Mice. Int. J. Mol. Sci. 2024, 25, 6113. https://doi.org/10.3390/ijms25116113
Trundle J, Lu-Nguyen N, Malerba A, Popplewell L. Targeted Antisense Oligonucleotide-Mediated Skipping of Murine Postn Exon 17 Partially Addresses Fibrosis in D2.mdx Mice. International Journal of Molecular Sciences. 2024; 25(11):6113. https://doi.org/10.3390/ijms25116113
Chicago/Turabian StyleTrundle, Jessica, Ngoc Lu-Nguyen, Alberto Malerba, and Linda Popplewell. 2024. "Targeted Antisense Oligonucleotide-Mediated Skipping of Murine Postn Exon 17 Partially Addresses Fibrosis in D2.mdx Mice" International Journal of Molecular Sciences 25, no. 11: 6113. https://doi.org/10.3390/ijms25116113




