Integrative Metabolomics, Enzymatic Activity, and Gene Expression Analysis Provide Insights into the Metabolic Profile Differences between the Slow-Twitch Muscle and Fast-Twitch Muscle of Pseudocaranx dentex
Abstract
1. Introduction
2. Results
2.1. The Differences in Metabolite Profile between the SM and FM of P. dentex
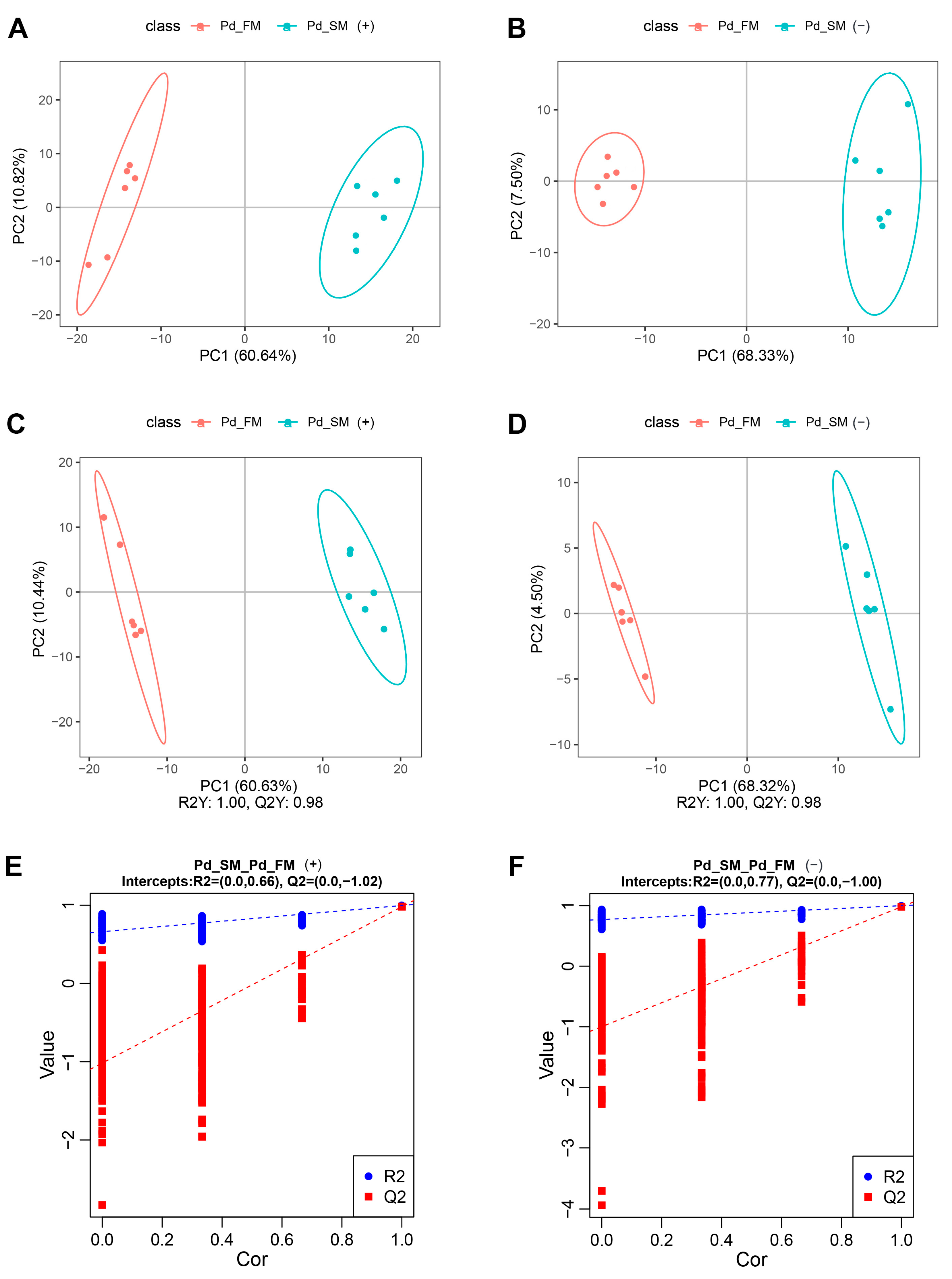
2.1.1. Overall Metabolic Profile of Muscles
2.1.2. Differences in Metabolite Profiles between SM and FM
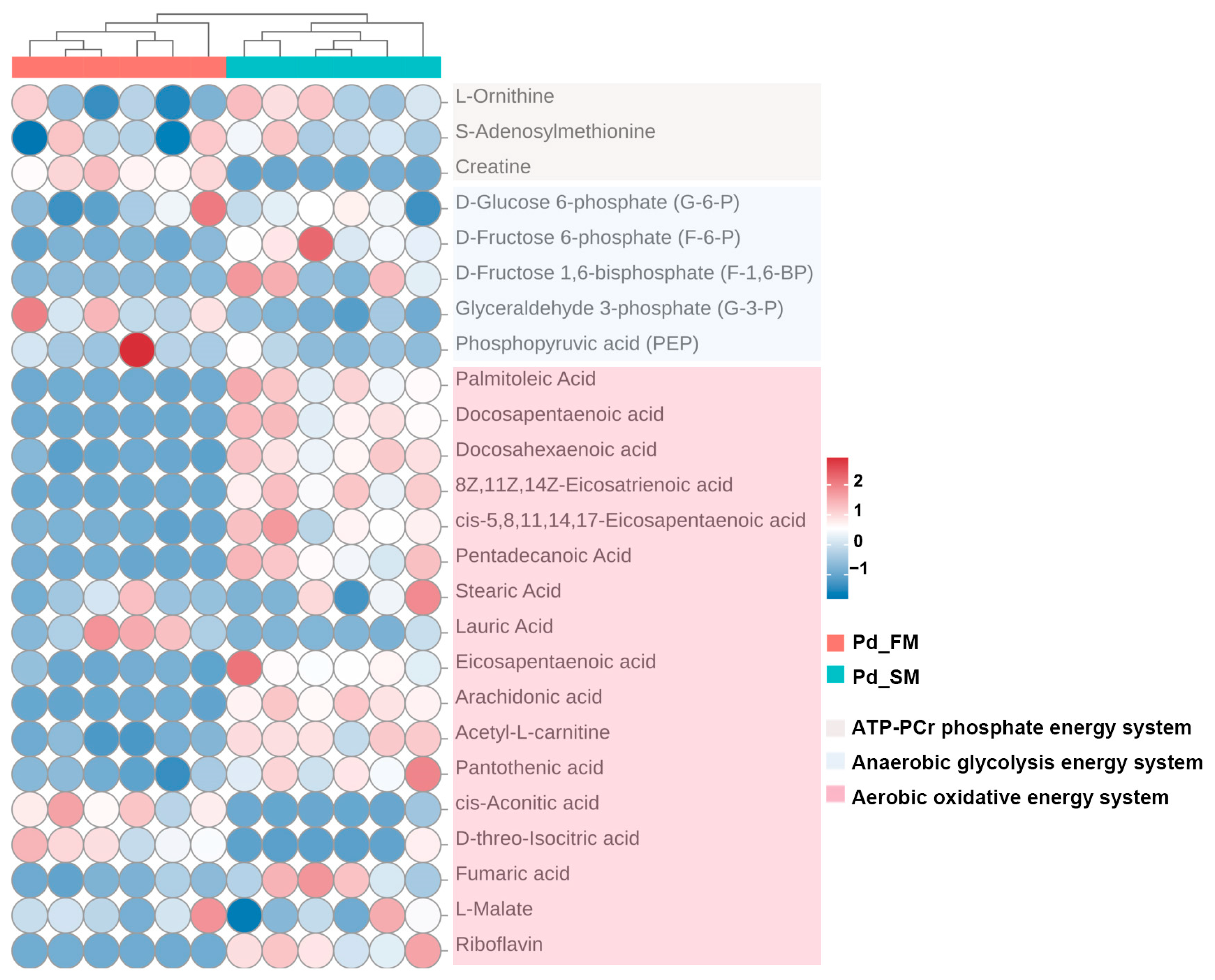
2.2. The Differential Contents of Metabolites Related to Energy Metabolism between SM and FM of P. dentex
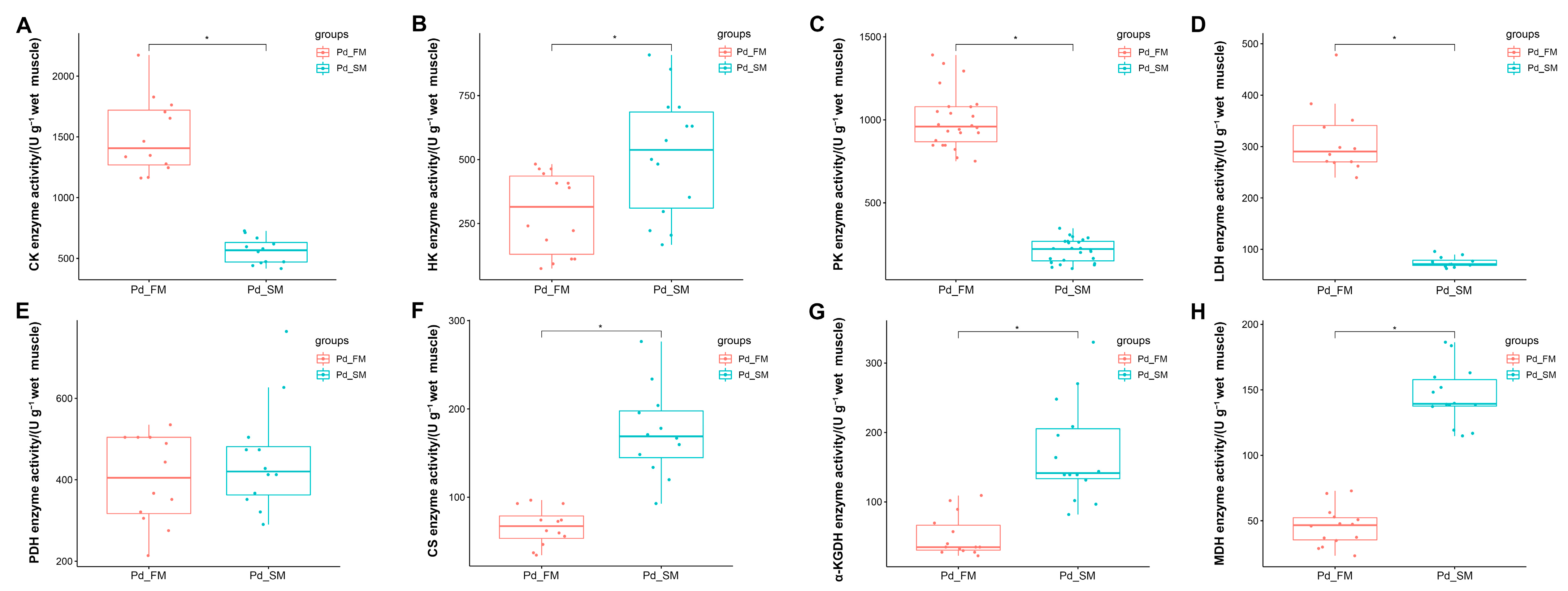
2.3. The Differential Activities of Enzymes Related to Energy Metabolism between the SM and FM of P. dentex
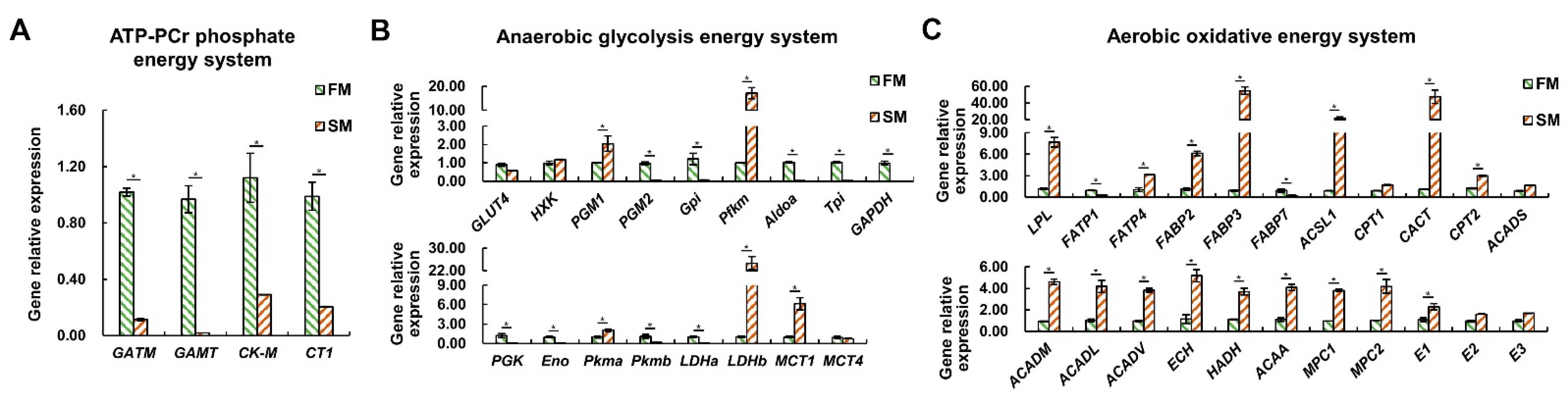
2.4. The Differential Expression Profiles of Genes Related to Energy Metabolism between the SM and FM of P. dentex
3. Discussion
4. Materials and Methods
4.1. Sample Collection
4.2. Untargeted-Metabolomics Metabolite Profiling Analysis
4.2.1. Metabolites Extraction
4.2.2. UHPLC-MS/MS Analysis
4.2.3. Data Preprocessing and Metabolite Identification
4.2.4. Multivariate Data Analysis and Univariate Statistical Analysis
4.3. Energy Metabolite Enzymatic Activity Assays
4.4. Gene Expression Analysis
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Listrat, A.; Lebret, B.; Louveau, I.; Astruc, T.; Muriel, B.; Lefaucheur, L.; Picard, B.; Bugeon, J. How Muscle Structure and Composition Influence Meat and Flesh Quality. Sci. World J. 2016, 2016, 3182746. [Google Scholar] [CrossRef]
- Jackson, H.E.; Ingham, P.W. Control of muscle fibre-type diversity during embryonic development: The zebrafish paradigm. Mech. Dev. 2013, 130, 447–457. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, H.; Shimizu, N.; Yoshikawa, N. Role of skeletal muscle glucocorticoid receptor in systemic energy homeostasis. Exp. Cell Res. 2017, 360, 24–26. [Google Scholar] [CrossRef]
- Berchtold, M.W.; Brinkmeier, H.; Müntener, M. Calcium ion in skeletal muscle: Its crucial role for muscle function, plasticity, and disease. Physiol. Rev. 2000, 80, 1215–1265. [Google Scholar] [CrossRef]
- Syme, D.A. Functional properties of skeletal muscle. Fish Biomech. 2005, 23, 179–240. [Google Scholar]
- Venugopal, V.; Shahidi, F. Structure and composition of fish muscle. Food Rev. Int. 1996, 12, 175–197. [Google Scholar] [CrossRef]
- Wu, M.P.; Chang, N.C.; Chung, C.L.; Chiu, W.C.; Hsu, C.C.; Chen, H.M.; Sheu, J.R.; Jayakumar, T.; Chou, D.S.; Fong, T.H. Analysis of Titin in Red and White Muscles: Crucial Role on Muscle Contractions Using a Fish Model. Biomed Res. Int. 2018, 2018, 5816875. [Google Scholar] [CrossRef] [PubMed]
- Dunn, J.F.; Davison, W.; Maloiy, G.M.O.; Hochachka, P.W.; Guppy, M. An ultrastructural and histochemical study of the axial musculature in the african lungfish. Cell Tissue Res. 1981, 220, 599–609. [Google Scholar] [CrossRef]
- Nag, A.C. Ultrastructure and adenosine triphosphatase activity of red and white muscle fibers of the caudal region of a fish, Salmo gairdneri. J. Cell Biol. 1972, 55, 42–57. [Google Scholar] [CrossRef]
- Leary, S.C.; Lyons, C.N.; Rosenberger, A.G.; Ballantyne, J.S.; Stillman, J.; Moyes, C.D. Fiber-type differences in muscle mitochondrial profiles. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2003, 285, R817. [Google Scholar] [CrossRef]
- Tsuyoshi, O.; Masaki, T.; Kiyo, H.; Miyuki, Y.; Sugie, H.F.; Atsushi, O. Biochemical properties of ordinary and dark muscle myosin from carp skeletal muscle. J. Biochem. 2005, 3, 255–262. [Google Scholar]
- Kiessling, A.; Ruohonen, K.; Bjørnevik, M. Muscle fibre growth and quality in fish. Archiv Tierz. /Arch. Anim. Breed. 2006, 49, 137–146. [Google Scholar]
- Teulier, L.; Thoral, E.; Queiros, Q.; McKenzie, D.J.; Roussel, D.; Dutto, G.; Gasset, E.; Bourjea, J.; Saraux, C. Muscle bioenergetics of two emblematic Mediterranean fish species: Sardina pilchardus and Sparus aurata. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2019, 235, 174–179. [Google Scholar] [CrossRef] [PubMed]
- Chaijan, M.; Klomklao, S.; Benjakul, S. Characterisation of muscles from Frigate mackerel (Auxis thazard) and catfish (Clarias macrocephalus). Food Chem. 2013, 139, 414–419. [Google Scholar] [CrossRef] [PubMed]
- Ciezarek, A.; Gardner, L.; Savolainen, V.; Block, B. Skeletal muscle and cardiac transcriptomics of a regionally endothermic fish, the Pacific bluefin tuna, Thunnus orientalis. BMC Genom. 2020, 21, 642. [Google Scholar] [CrossRef] [PubMed]
- Gao, K.; Wang, Z.; Zhou, X.; Wang, H.; Qiu, X. Comparative transcriptome analysis of fast twitch muscle and slow twitch muscle in Takifugu rubripes. Comp. Biochem. Physiol. Part D Genom. Proteom. 2017, 24, 79. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Li, B.; Yang, L.; Jiang, C.; Zhang, T.; Liu, S.; Zhuang, Z. Expression profiles and transcript properties of fast-twitch and slow-twitch muscles in a deep-sea highly migratory fish, Pseudocaranx dentex. PeerJ 2022, 10, e12720. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Li, B.; Li, J.; Jiang, C.; Liu, S.; Zhuang, Z. Label-Free Quantitative Proteomic Analysis Provides Insight Into the Differences Between Slow-Twitch Muscle and Fast-Twitch Muscle of Pseudocaranx dentex. Front. Mar. Sci. 2022, 9, 842172. [Google Scholar] [CrossRef]
- Putri, S.P.; Nakayama, Y.; Matsuda, F.; Uchikata, T.; Kobayashi, S.; Matsubara, A.; Fukusaki, E. Current metabolomics: Practical applications. J. Biosci. Bioeng. 2013, 115, 579–589. [Google Scholar] [CrossRef]
- Lulijwa, R.; Alfaro, A.C.; Young, T. Metabolomics in salmonid aquaculture research: Applications and future perspectives. Rev. Aquac. 2022, 14, 547–577. [Google Scholar] [CrossRef]
- Gastin, P.B. Energy system interaction and relative contribution during maximal exercise. Sport. Med. 2001, 31, 725–741. [Google Scholar] [CrossRef] [PubMed]
- Videler, J.J. Fish Swimming; Springer Science & Business Media: London, UK, 1993. [Google Scholar]
- Johnston, I.A.; Davison, W.; Goldspink, G. Energy metabolism of carp swimming muscles. J. Comp. Physiol. B Biochem. Syst. Environ. Physiol. 1977, 114, 203–216. [Google Scholar] [CrossRef]
- Nogueira, N.; Ferreira, M.; Cordeiro, N.; Canada, P. Quality parameters of wild white trevally (Pseudocaranx dentex) natural spawn kept in captivity. Aquaculture 2018, 495, 68–77. [Google Scholar] [CrossRef]
- Watanabe, T.; Vassallo-Agius, R. Broodstock nutrition research on marine finfish in Japan. Aquaculture 2003, 227, 35–61. [Google Scholar] [CrossRef]
- Aji, L.P. Aerobic poise of marine fish in relation to habitat and lifestyle. J. Penelit. Sains 2011, 14, 48–51. [Google Scholar]
- Muroya, S.; Ueda, S.; Komatsu, T.; Miyakawa, T.; Ertbjerg, P. MEATabolomics: Muscle and Meat Metabolomics in Domestic Animals. Metabolites 2020, 10, 188. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; You, J.; Wang, L.; Shi, L.; Liao, T.; Huang, Q.; Xiong, S.; Yin, T. Insight into the mechanism on texture change of Wuchang bream muscle during live transportation using a UPLC-QTOF-MS based metabolomics method. Food Chem. 2023, 398, 133796. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Lv, H.; Xu, Z.; Zhao, S.; Huang, T.; Manyande, A.; Xiong, S. The mechanism for improving the flesh quality of grass carp (Ctenopharyngodon idella) following the micro-flowing water treatment using a UPLC-QTOF/MS based metabolomics method. Food Chem. 2020, 327, 126777. [Google Scholar] [CrossRef] [PubMed]
- Weber, J.-M.; Choi, K.; Gonzalez, A.; Omlin, T. Metabolic fuel kinetics in fish: Swimming, hypoxia and muscle membranes. J. Exp. Biol. 2016, 219, 250–258. [Google Scholar] [CrossRef]
- Gasco, L.; Gai, F.; Maricchiolo, G.; Genovese, L.; Ragonese, S.; Bottari, T.; Caruso, G. Supplementation of vitamins, minerals, enzymes and antioxidants in fish feeds. In Feeds for the Aquaculture Sector: Current Situation and Alternative Sources; Springer: Berlin/Heidelberg, Germany, 2018; pp. 63–103. [Google Scholar]
- Hochachka, P.W.; Somero, G.N. Biochemical Adaptation; Princeton University Press: Princeton, NJ, USA, 1984. [Google Scholar]
- Abe, H. Role of histidine-related compounds as intracellular proton buffering constituents in vertebrate muscle. Biochemistry 2000, 65, 757–765. [Google Scholar]
- Schaffer, S.W.; Shimada-Takaura, K.; Jong, C.J.; Ito, T.; Takahashi, K. Impaired energy metabolism of the taurine deficient heart. Amino Acids 2016, 48, 549–558. [Google Scholar] [CrossRef] [PubMed]
- Dean, M.; Annilo, T. Evolution of the ATP-Binding Cassette (ABC) Transporter Superfamily in Vertebrates. Annu. Rev. Genom. Hum. Genet. 2005, 6, 123–142. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Ye, W.; Chen, H.; Kuang, X.; Guo, J.; Xiang, M.; Peng, C.; Chen, X.; Liu, H. Role of purines in regulation of metabolic reprogramming. Purinergic Signal. 2019, 15, 423–438. [Google Scholar] [CrossRef] [PubMed]
- Borchel, A.; Verleih, M.; Rebl, A.; Kühn, C.; Goldammer, T. Creatine metabolism differs between mammals and rainbow trout (Oncorhynchus mykiss). SpringerPlus 2014, 3, 510. [Google Scholar] [CrossRef] [PubMed]
- Snow, R.J.; Murphy, R.M. Creatine and the creatine transporter: A review. Mol. Cell. Biochem. 2001, 224, 169–181. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, K.; Yoshioka, T. Profiles of creatine kinase isoenzyme compositions in single muscle fibres of different types. J. Muscle Res. Cell Motil. 1991, 12, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Wallimann, T.; Wyss, M.; Brdiczka, D.; Nicolay, K. Intracellular compartmentation, structure and function of creatine kinase isoenzymes in tissues with high and fluctuating energy demands: The ‘phosphocreatine circuit’ for cellular energy homeostasis. Biochem. J. 1992, 281, 21–40. [Google Scholar] [CrossRef] [PubMed]
- Johnston, I.A. A comparative study of glycolysis in red and white muscles of the trout (Salmo gairdneri) and mirror carp (Cyprinus carpio). J. Fish Bio. 1977, 11, 575–588. [Google Scholar] [CrossRef]
- Washington, T.A.; Reecy, J.M.; Thompson, R.W.; Lowe, L.L.; McClung, J.M.; Carson, J.A. Lactate dehydrogenase expression at the onset of altered loading in rat soleus muscle. J. Appl. Physiol. 2004, 97, 1424–1430. [Google Scholar] [CrossRef]
- Rodnick, K.J.; Planas, J.V. The Stress and Stress Mitigation Effects of Exercise: Cardiovascular, Metabolic, and Skeletal Muscle Adjustments. Biol. Stress Fish-Fish Physiol. 2016, 35, 251–294. [Google Scholar]
- Magnoni, L.; Weber, J.M. Endurance swimming activates trout lipoprotein lipase: Plasma lipids as a fuel for muscle. J. Exp. Biol. 2007, 210, 4016–4023. [Google Scholar] [CrossRef] [PubMed]
- Magnoni, L.J.; Felip, O.; Blasco, J.; Planas, J.V. Metabolic Fuel Utilization during Swimming: Optimizing Nutritional Requirements for Enhanced Performance. In Swimming Physiology of Fish; Springer: Berlin/Heidelberg, Germany, 2013; pp. 203–235. [Google Scholar]
- Longo, N.; Frigeni, M.; Pasquali, M. Carnitine transport and fatty acid oxidation. Biochim. Biophys. Acta. 2016, 1863, 2422–2435. [Google Scholar] [CrossRef] [PubMed]
- Zangari, J.; Petrelli, F.; Maillot, B.; Martinou, J.C. The multifaceted pyruvate metabolism: Role of the mitochondrial pyruvate carrier. Biomolecules 2020, 10, 1068. [Google Scholar] [CrossRef] [PubMed]
- Hargreaves, M.; Spriet, L.L. Skeletal muscle energy metabolism during exercise. Nat. Metab. 2020, 2, 817–828. [Google Scholar] [CrossRef] [PubMed]
- Halestrap, A.P. The SLC16 gene family-structure, role and regulation in health and disease. Mol. Asp. Med. 2013, 34, 337–349. [Google Scholar] [CrossRef]
- Wen, B.; Mei, Z.; Zeng, C.; Liu, S. metaX: A flexible and comprehensive software for processing metabolomics data. BMC Bioinform. 2017, 18, 183. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; Li, B.; Li, A.; An, C.; Liu, S.; Zhuang, Z. Integrative Metabolomics, Enzymatic Activity, and Gene Expression Analysis Provide Insights into the Metabolic Profile Differences between the Slow-Twitch Muscle and Fast-Twitch Muscle of Pseudocaranx dentex. Int. J. Mol. Sci. 2024, 25, 6131. https://doi.org/10.3390/ijms25116131
Wang H, Li B, Li A, An C, Liu S, Zhuang Z. Integrative Metabolomics, Enzymatic Activity, and Gene Expression Analysis Provide Insights into the Metabolic Profile Differences between the Slow-Twitch Muscle and Fast-Twitch Muscle of Pseudocaranx dentex. International Journal of Molecular Sciences. 2024; 25(11):6131. https://doi.org/10.3390/ijms25116131
Chicago/Turabian StyleWang, Huan, Busu Li, Ang Li, Changting An, Shufang Liu, and Zhimeng Zhuang. 2024. "Integrative Metabolomics, Enzymatic Activity, and Gene Expression Analysis Provide Insights into the Metabolic Profile Differences between the Slow-Twitch Muscle and Fast-Twitch Muscle of Pseudocaranx dentex" International Journal of Molecular Sciences 25, no. 11: 6131. https://doi.org/10.3390/ijms25116131
APA StyleWang, H., Li, B., Li, A., An, C., Liu, S., & Zhuang, Z. (2024). Integrative Metabolomics, Enzymatic Activity, and Gene Expression Analysis Provide Insights into the Metabolic Profile Differences between the Slow-Twitch Muscle and Fast-Twitch Muscle of Pseudocaranx dentex. International Journal of Molecular Sciences, 25(11), 6131. https://doi.org/10.3390/ijms25116131




