Rhodamine 19 Alkyl Esters as Effective Antibacterial Agents
Abstract
1. Introduction
2. Results and Discussion
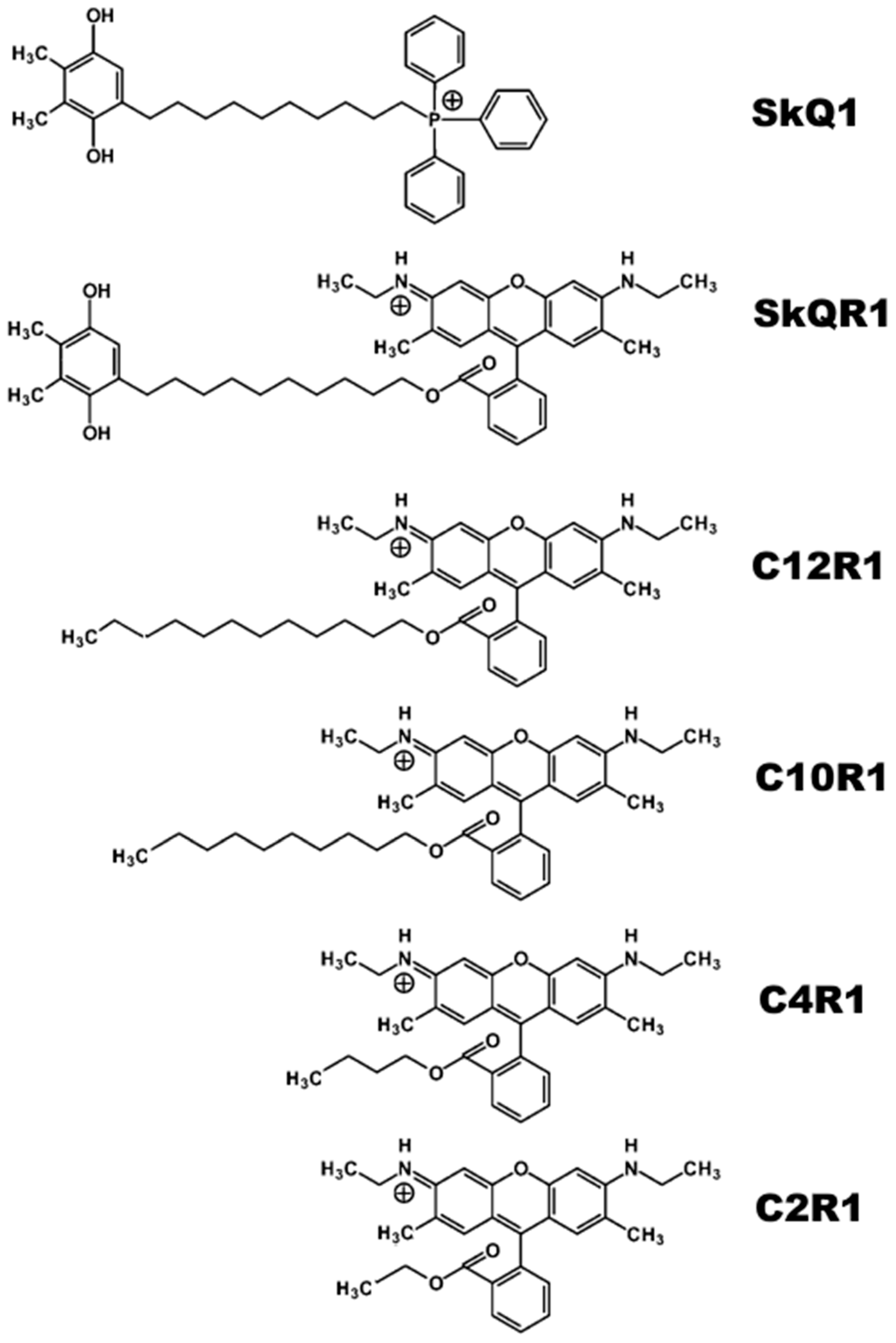
2.1. Antibacterial Activity of Rhodamine Derivatives
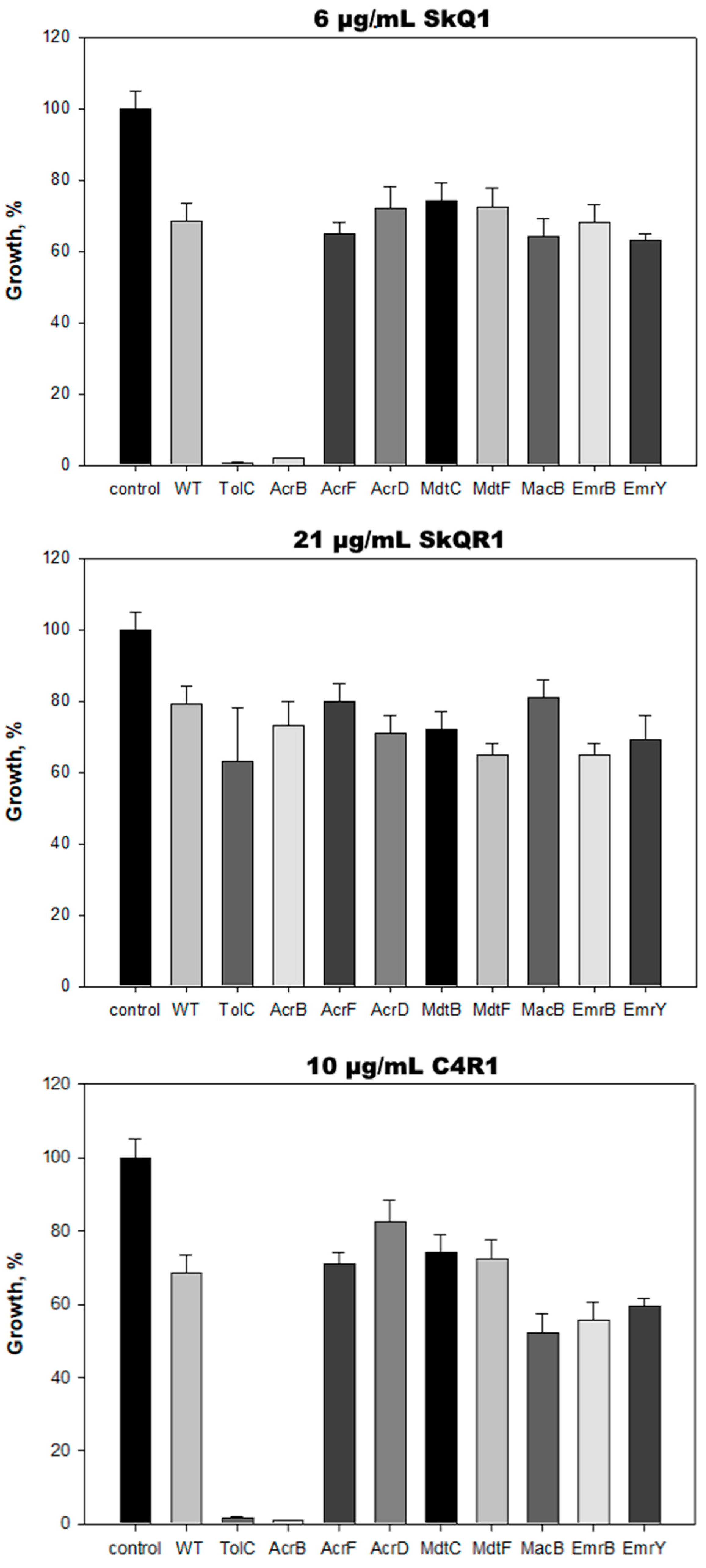
2.2. AcrAB-TolC Transporter Is Responsible for E. coli Resistance to C4R1 but Not to SkQR1
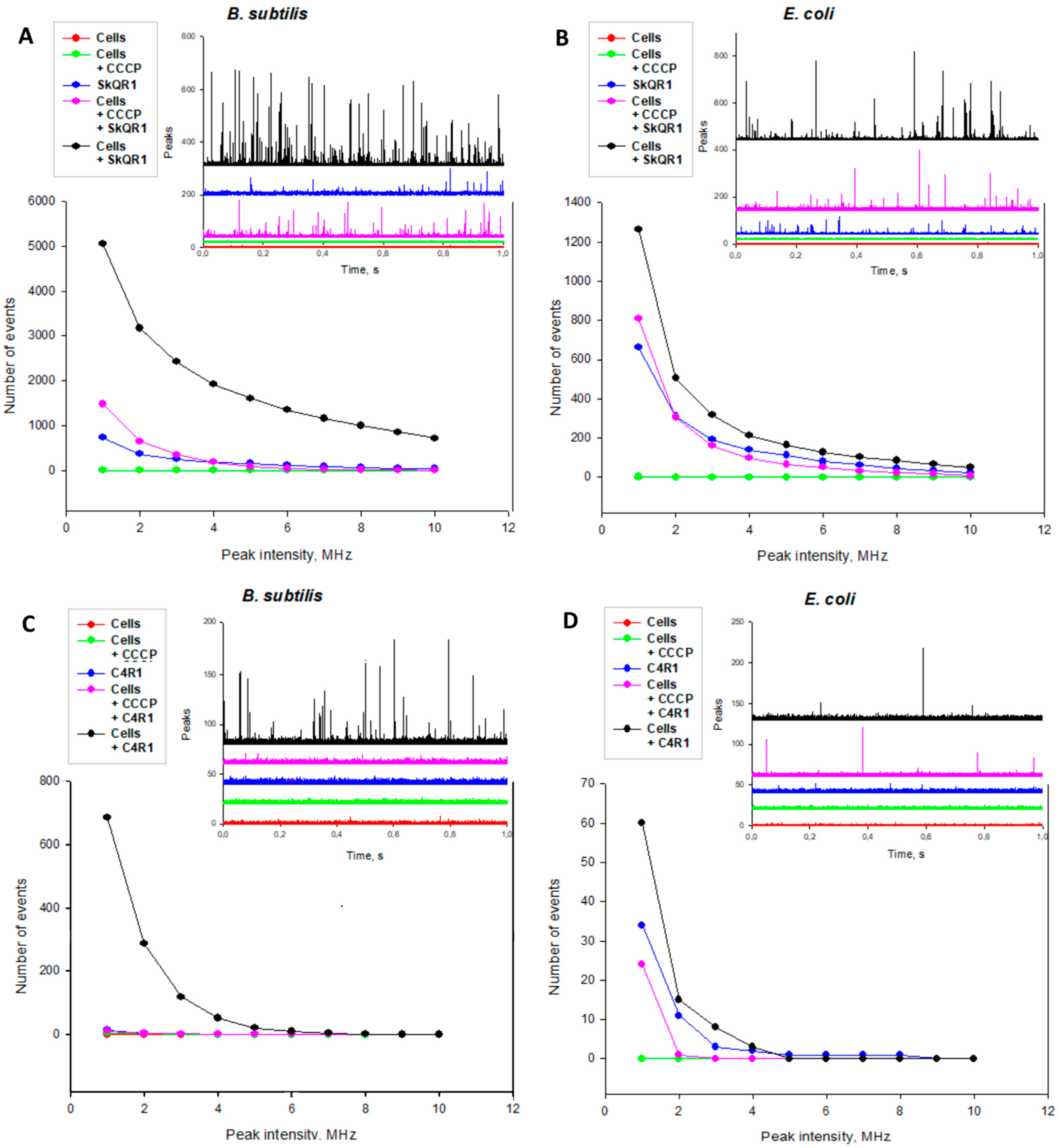
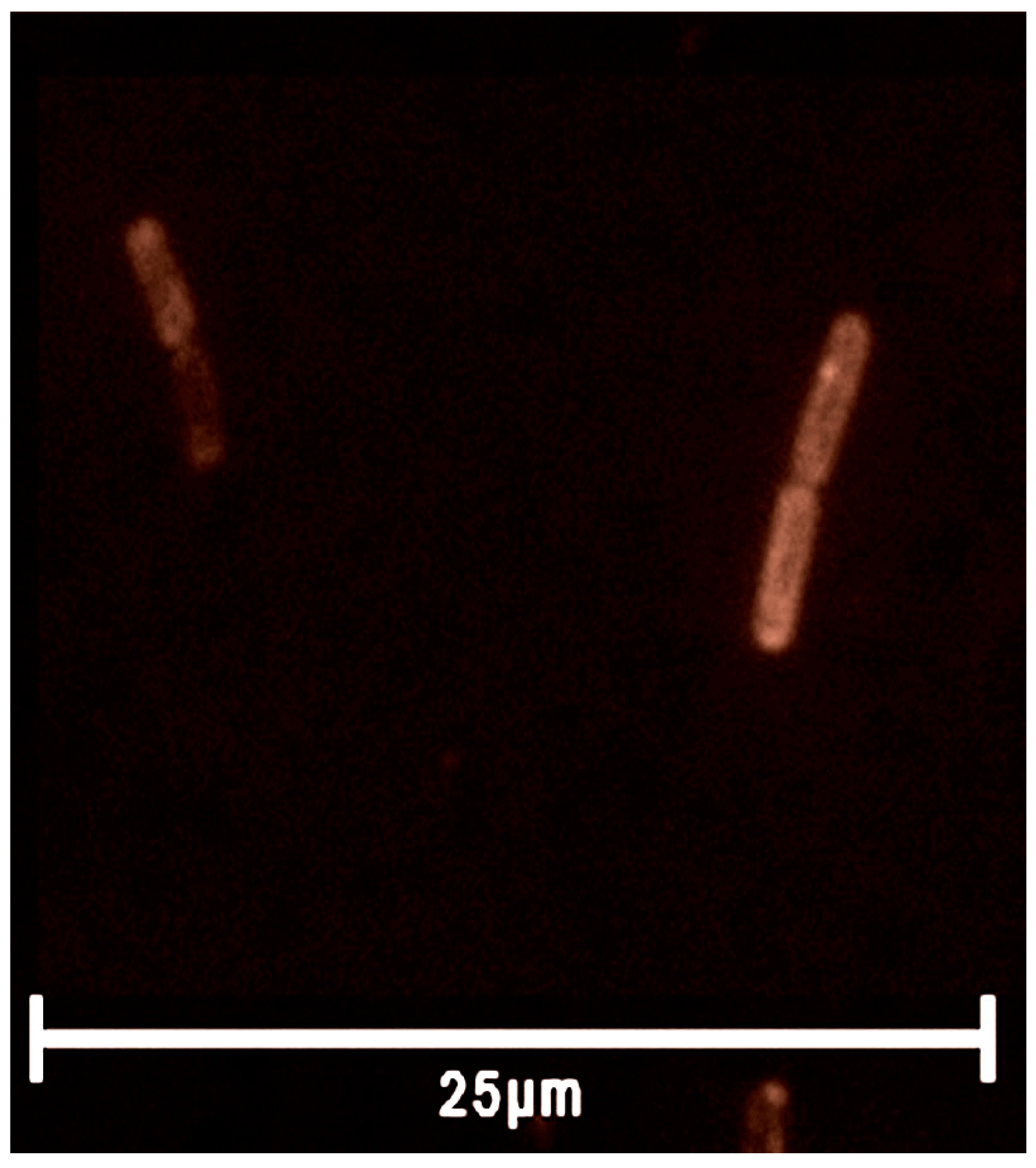
2.3. Rhodamine Derivatives Are Localized on the Bacterial Membrane
3. Materials and Methods
3.1. Materials
3.2. Bacterial Strains
3.3. Growth Suppression Assay and MIC Determination
3.4. Fluorescence Microscopy
3.4.1. Agarose Pads Preparation
3.4.2. Sample Pads Preparation
3.4.3. Equipment Setup
3.5. Fluorescence Correlation Spectroscopy
3.6. Deletion Mutants of TolC-Requiring Transporters Growth Suppression Screening
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Antonenko, Y.N.; Avetisyan, A.V.; Cherepanov, D.A.; Knorre, D.A.; Korshunova, G.A.; Markova, O.V.; Ojovan, S.M.; Perevoshchikova, I.V.; Pustovidko, A.V.; Rokitskaya, T.I.; et al. Derivatives of rhodamine 19 as mild mitochondria-targeted cationic uncouplers. J. Biol. Chem. 2011, 286, 17831–17840. [Google Scholar] [CrossRef]
- Khailova, L.S.; Silachev, D.N.; Rokitskaya, T.I.; Avetisyan, A.V.; Lyamsaev, K.G.; Severina, I.I.; Il’yasova, T.M.; Gulyaev, M.V.; Dedukhova, V.I.; Trendeleva, T.A.; et al. A short-chain alkyl derivative of Rhodamine 19 acts as a mild uncoupler of mitochondria and a neuroprotector. Biochim. Biophys. Acta 2014, 1837, 1739–1747. [Google Scholar] [CrossRef]
- Gear, A.R. Rhodamine 6G. A potent inhibitor of mitochondrial oxidative phosphorylation. J. Biol. Chem. 1974, 249, 3628–3637. [Google Scholar] [CrossRef]
- Higuti, T.; Niimi, S.; Saito, R.; Nakasima, S.; Ohe, T.; Tani, I.; Yoshimura, T. Rhodamine 6G, inhibitor of both H+-ejections from mitochondria energized with ATP and with respiratory substrates. Biochim. Biophys. Acta 1980, 593, 463–467. [Google Scholar] [CrossRef]
- Wieker, H.J.; Kuschmitz, D.; Hess, B. Inhibition of yeast mitochondrial F1-ATPase, F0F1-ATPase and submitochondrial particles by rhodamines and ethidium bromide. Biochim. Biophys. Acta 1987, 892, 108–117. [Google Scholar] [CrossRef]
- Gledhill, J.R.; Walker, J.E. Inhibition sites in F1-ATPase from bovine heart mitochondria. Biochem. J. 2005, 386 Pt 3, 591–598. [Google Scholar] [CrossRef]
- Luo, X.; Qian, L.; Xiao, Y.; Tang, Y.; Zhao, Y.; Wang, X.; Gu, L.; Lei, Z.; Bao, J.; Wu, J.; et al. A diversity-oriented rhodamine library for wide-spectrum bactericidal agents with low inducible resistance against resistant pathogens. Nat. Commun. 2019, 10, 258. [Google Scholar] [CrossRef]
- Gannon, M.K., 2nd; Holt, J.J.; Bennett, S.M.; Wetzel, B.R.; Loo, T.W.; Bartlett, M.C.; Clarke, D.M.; Sawada, G.A.; Higgins, J.W.; Tombline, G.; et al. Rhodamine inhibitors of P-glycoprotein: An amide/thioamide “switch” for ATPase activity. J. Med. Chem. 2009, 52, 3328–3341. [Google Scholar] [CrossRef]
- Miwa, S.; Takikawa, H.; Takeuchi, R.; Mizunuma, R.; Matsuoka, K.; Ogawa, H.; Kato, H.; Takasu, K. Structure-ATPase Activity Relationship of Rhodamine Derivatives as Potent Inhibitors of P-Glycoprotein CmABCB1. ACS Med. Chem. Lett. 2024, 15, 287–293. [Google Scholar] [CrossRef]
- Liberman, E.A.; Topaly, V.P.; Tsofina, L.M.; Jasaitis, A.A.; Skulachev, V.P. Mechanism of coupling of oxidative phosphorylation and the membrane potential of mitochondria. Nature 1969, 222, 1076–1078. [Google Scholar] [CrossRef]
- Liberman, E.A.; Topaly, V.P. Pronitsaemost’ bimolekuliarnykh fosfolipidnykh membran dlia zhirorastvorimykh ionov [Permeability of bimolecular phospholipid membranes for fat-soluble ions]. Biofizika 1969, 14, 452–461. (In Russian) [Google Scholar] [PubMed]
- Grinius, L.L.; Jasaitis, A.A.; Kadziauskas, Y.P.; Liberman, E.A.; Skulachev, V.P.; Topali, V.P.; Tsofina, L.M.; Vladimirova, M.A. Conversion of biomembrane-produced energy into electric form. I. Submitochondrial particles. Biochim. Biophys. Acta 1970, 216, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Liberman, E.A.; Skulachev, V.P. Conversion of biomembrane-produced energy into electric form. IV. General discussion. Biochim. Biophys. Acta 1970, 216, 30–42. [Google Scholar] [CrossRef] [PubMed]
- Burns, R.J.; Smith, R.A.; Murphy, M.P. Synthesis and characterization of thiobutyltriphenylphosphonium bromide, a novel thiol reagent targeted to the mitochondrial matrix. Arch. Biochem. Biophys. 1995, 322, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Murphy, M.P. Selective targeting of bioactive compounds to mitochondria. Trends Biotechnol. 1997, 15, 326–330. [Google Scholar] [CrossRef] [PubMed]
- Smith, R.A.; Porteous, C.M.; Coulter, C.V.; Murphy, M.P. Selective targeting of an antioxidant to mitochondria. Eur. J. Biochem. 1999, 263, 709–716. [Google Scholar] [CrossRef] [PubMed]
- Zielonka, J.; Joseph, J.; Sikora, A.; Hardy, M.; Ouari, O.; Vasquez-Vivar, J.; Cheng, G.; Lopez, M.; Kalyanaraman, B. Mitochondria-Targeted Triphenylphosphonium-Based Compounds: Syntheses, Mechanisms of Action, and Therapeutic and Diagnostic Applications. Chem. Rev. 2017, 117, 10043–10120. [Google Scholar] [CrossRef] [PubMed]
- Antonenko, Y.N.; Avetisyan, A.V.; Bakeeva, L.E.; Chernyak, B.V.; Chertkov, V.A.; Domnina, L.V.; Ivanova, O.Y.; Izyumov, D.S.; Khailova, L.S.; Klishin, S.S.; et al. Mitochondria-targeted plastoquinone derivatives as tools to interrupt execution of the aging program. 1. Cationic plastoquinone derivatives: Synthesis and in vitro studies. Biochemistry 2008, 73, 1273–1287. [Google Scholar] [CrossRef] [PubMed]
- Fetisova, E.K.; Avetisyan, A.V.; Izyumov, D.S.; Korotetskaya, M.V.; Chernyak, B.V.; Skulachev, V.P. Mitochondria-targeted antioxidant SkQR1 selectively protects MDR (Pgp 170)-negative cells against oxidative stress. FEBS Lett. 2010, 584, 562–566. [Google Scholar] [CrossRef]
- Izyumov, D.S.; Domnina, L.V.; Nepryakhina, O.K.; Avetisyan, A.V.; Golyshev, S.A.; Ivanova, O.Y.; Korotetskaya, M.V.; Lyamzaev, K.G.; Pletjushkina, O.Y.; Popova, E.N.; et al. Mitochondria as source of reactive oxygen species under oxidative stress. Study with novel mitochondria-targeted antioxidants—The “Skulachev-ion” derivatives. Biochemistry 2010, 75, 123–129. [Google Scholar] [CrossRef]
- Plotnikov, E.Y.; Morosanova, M.A.; Pevzner, I.B.; Zorova, L.D.; Manskikh, V.N.; Pulkova, N.V.; Galkina, S.I.; Skulachev, V.P.; Zorov, D.B. Protective effect of mitochondria-targeted antioxidants in an acute bacterial infection. Proc. Natl. Acad. Sci. USA 2013, 110, E3100–E3108. [Google Scholar] [CrossRef] [PubMed]
- Nazarov, P.A.; Osterman, I.A.; Tokarchuk, A.V.; Karakozova, M.V.; Korshunova, G.A.; Lyamzaev, K.G.; Skulachev, M.V.; Kotova, E.A.; Skulachev, V.P.; Antonenko, Y.N. Mitochondria-targeted antioxidants as highly effective antibiotics. Sci. Rep. 2017, 7, 1394. [Google Scholar] [CrossRef] [PubMed]
- Nazarov, P.A.; Kotova, E.A.; Skulachev, V.P.; Antonenko, Y.N. Genetic Variability of the AcrAB-TolC Multidrug Efflux Pump Underlies SkQ1 Resistance in Gram-Negative Bacteria. Acta Nat. 2019, 11, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Nazarov, P.A.; Sorochkina, A.I.; Karakozova, M.V. New Functional Criterion for Evaluation of Homologous MDR Pumps. Front. Microbiol. 2020, 11, 592283. [Google Scholar] [CrossRef] [PubMed]
- Nishino, K.; Yamaguchi, A. EvgA of the two-component signal transduction system modulates production of the yhiUV multidrug transporter in Escherichia coli. J. Bacteriol. 2002, 184, 2319–2323. [Google Scholar] [CrossRef] [PubMed]
- Yu, E.W.; McDermott, G.; Zgurskaya, H.I.; Nikaido, H.; Koshland, D.E., Jr. Structural basis of multiple drug-binding capacity of the AcrB multidrug efflux pump. Science 2003, 300, 976–980. [Google Scholar] [CrossRef] [PubMed]
- Sjuts, H.; Vargiu, A.V.; Kwasny, S.M.; Nguyen, S.T.; Kim, H.S.; Ding, X.; Ornik, A.R.; Ruggerone, P.; Bowlin, T.L.; Nikaido, H.; et al. Molecular basis for inhibition of AcrB multidrug efflux pump by novel and powerful pyranopyridine derivatives. Proc. Natl. Acad. Sci. USA 2016, 113, 3509–3514. [Google Scholar] [CrossRef] [PubMed]
- Zwama, M.; Yamasaki, S.; Nakashima, R.; Sakurai, K.; Nishino, K.; Yamaguchi, A. Multiple entry pathways within the efflux transporter AcrB contribute to multidrug recognition. Nat. Commun. 2018, 9, 124. [Google Scholar] [CrossRef] [PubMed]
- Rajapaksha, P.; Ojo, I.; Yang, L.; Pandeya, A.; Abeywansha, T.; Wei, Y. Insight into the AcrAB-TolC Complex Assembly Process Learned from Competition Studies. Antibiotics 2021, 10, 830. [Google Scholar] [CrossRef]
- Sulavik, M.C.; Houseweart, C.; Cramer, C.; Jiwani, N.; Murgolo, N.; Greene, J.; DiDomenico, B.; Shaw, K.J.; Miller, G.H.; Hare, R.; et al. Antibiotic susceptibility profiles of Escherichia coli strains lacking multidrug efflux pump genes. Antimicrob. Agents Chemother. 2001, 45, 1126–1136. [Google Scholar] [CrossRef]
- Lu, M.; Symersky, J.; Radchenko, M.; Koide, A.; Guo, Y.; Nie, R.; Koide, S. Structures of a Na+-coupled, substrate-bound MATE multidrug transporter. Proc. Natl. Acad. Sci. USA 2013, 110, 2099–2104. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, W.; Onishi, M.; Ni, R.; Tsuchiya, T.; Kuroda, T. Functional study of the novel multidrug efflux pump KexD from Klebsiella pneumoniae. Gene 2012, 498, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Sekiya, H.; Mima, T.; Morita, Y.; Kuroda, T.; Mizushima, T.; Tsuchiya, T. Functional cloning and characterization of a multidrug efflux pump, mexHI-opmD, from a Pseudomonas aeruginosa mutant. Antimicrob. Agents Chemother. 2003, 47, 2990–2992. [Google Scholar] [CrossRef] [PubMed]
- Morita, Y.; Kodama, K.; Shiota, S.; Mine, T.; Kataoka, A.; Mizushima, T.; Tsuchiya, T. NorM, a putative multidrug efflux protein, of Vibrio parahaemolyticus and its homolog in Escherichia coli. Antimicrob. Agents Chemother. 1998, 42, 1778–1782. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.; Borsch, C.M.; Taylor, S.S.; Vázquez-Laslop, N.; Neyfakh, A.A. A protein that activates expression of a multidrug efflux transporter upon binding the transporter substrates. J. Biol. Chem. 1994, 269, 28506–28513. [Google Scholar] [CrossRef] [PubMed]
- Zheleznova, E.E.; Markham, P.N.; Neyfakh, A.A.; Brennan, R.G. Structural basis of multidrug recognition by BmrR, a transcription activator of a multidrug transporter. Cell 1999, 96, 353–362. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, M.A.; Brennan, R.G. Deciphering the molecular basis of multidrug recognition: Crystal structures of the Staphylococcus aureus multidrug binding transcription regulator QacR. Res. Microbiol. 2003, 154, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Neyfakh, A.A.; Bidnenko, V.E.; Chen, L.B. Efflux-mediated multidrug resistance in Bacillus subtilis: Similarities and dissimilarities with the mammalian system. Proc. Natl. Acad. Sci. USA 1991, 88, 4781–4785. [Google Scholar] [CrossRef] [PubMed]
- Plé, C.; Tam, H.K.; Vieira Da Cruz, A.; Compagne, N.; Jiménez-Castellanos, J.C.; Müller, R.T.; Pradel, E.; Foong, W.E.; Malloci, G.; Ballée, A.; et al. Pyridylpiperazine-based allosteric inhibitors of RND-type multidrug efflux pumps. Nat. Commun. 2022, 13, 115. [Google Scholar] [CrossRef]
- Zorova, L.D.; Pevzner, I.B.; Khailova, L.S.; Korshunova, G.A.; Kovaleva, M.A.; Kovalev, L.I.; Serebryakova, M.V.; Silachev, D.N.; Sudakov, R.V.; Zorov, S.D.; et al. Mitochondrial ATP Synthase and Mild Uncoupling by Butyl Ester of Rhodamine 19, C4R1. Antioxidants 2023, 12, 646. [Google Scholar] [CrossRef]
- Nazarov, P.A.; Majorov, K.B.; Apt, A.S.; Skulachev, M.V. Penetration of Triphenylphosphonium Derivatives through the Cell Envelope of Bacteria of Mycobacteriales Order. Pharmaceuticals 2023, 16, 688. [Google Scholar] [CrossRef] [PubMed]
- Sander, P.; De Rossi, E.; Böddinghaus, B.; Cantoni, R.; Branzoni, M.; Böttger, E.C.; Takiff, H.; Rodriquez, R.; Lopez, G.; Riccardi, G. Con-tribution of the multidrug efflux pump LfrA to innate mycobacterial drug resistance. FEMS Microbiol. Lett. 2000, 193, 19–23. [Google Scholar] [CrossRef] [PubMed]
- Niederweis, M. Mycobacterial porins–new channel proteins in unique outer membranes. Mol. Microbiol. 2003, 49, 1167–1177. [Google Scholar] [CrossRef] [PubMed]
- Brennan, P.J.; Nikaido, H. The envelope of mycobacteria. Annu. Rev. Biochem. 1995, 64, 29–63. [Google Scholar] [CrossRef] [PubMed]
- Remm, S.; Earp, J.C.; Dick, T.; Dartois, V.; Seeger, M.A. Critical discussion on drug efflux in Mycobacterium tuberculosis. FEMS Microbiol. Rev. 2022, 46, fuab050. [Google Scholar] [CrossRef] [PubMed]
- Fishbein, S.R.S.; Tomasi, F.G.; Wolf, I.D.; Dulberger, C.L.; Wang, A.; Keshishian, H.; Wallace, L.; Carr, S.A.; Ioerger, T.R.; Rego, E.H.; et al. The conserved translation factor LepA is required for optimal synthesis of a porin family in Mycobacterium smegmatis. J. Bacteriol. 2020, 203, e00604-20. [Google Scholar] [CrossRef] [PubMed]
- Nazarov, P.A. MDR Pumps as Crossroads of Resistance: Antibiotics and Bacteriophages. Antibiotics 2022, 11, 734. [Google Scholar] [CrossRef] [PubMed]
- Rokitskaya, T.I.; Nazarov, P.A.; Golovin, A.V.; Antonenko, Y.N. Blocking of Single α-Hemolysin Pore by Rhodamine Derivatives. Biophys. J. 2017, 112, 2327–2335. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute (CLSI). Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically, 9th ed.; CLSI Document M07-A9, Approved Standard; CLSI: Wayne, PA, USA, 2012; Volume 32, ISBN 1562387839. [Google Scholar]
- Young, J.W.; Locke, J.C.; Altinok, A.; Rosenfeld, N.; Bacarian, T.; Swain, P.S.; Mjolsness, E.; Elowitz, M.B. Measuring single-cell gene expression dynamics in bacteria using fluorescence time-lapse microscopy. Nat. Protoc. 2011, 7, 80–88. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Schindelin, J.; Rueden, C.T.; Hiner, M.C.; Eliceiri, K.W. The ImageJ ecosystem: An open platform for biomedical image analysis. Mol. Reprod. Dev. 2015, 82, 518–529. [Google Scholar] [CrossRef] [PubMed]
- Perevoshchikova, I.V.; Zorov, D.B.; Antonenko, Y.N. Peak intensity analysis as a method for estimation of fluorescent probe binding to artificial and natural nanoparticles: Tetramethylrhodamine uptake by isolated mitochondria. Biochim. Biophys. Acta 2008, 1778, 2182–2190. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K. At the Crossroads of Bioenergetics and Antibiotic Discovery. Biochemistry 2020, 85, 1469–1483. [Google Scholar] [CrossRef] [PubMed]
- Hards, K.; Cook, G.M. Targeting bacterial energetics to produce new antimicrobials. Drug Resist. Updat. 2018, 36, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Keller, M.R.; Dörr, T. Bacterial metabolism and susceptibility to cell wall-active antibiotics. Adv. Microb. Physiol. 2023, 83, 181–219. [Google Scholar] [CrossRef] [PubMed]
- Geng, Z.; Cao, Z.; Liu, J. Recent advances in targeted antibacterial therapy basing on nanomaterials. Exploration 2023, 3, 20210117. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K. Platforms for antibiotic discovery. Nat. Rev. Drug Discov. 2013, 12, 371–387. [Google Scholar] [CrossRef] [PubMed]
- Halawa, E.M.; Fadel, M.; Al-Rabia, M.W.; Behairy, A.; Nouh, N.A.; Abdo, M.; Olga, R.; Fericean, L.; Atwa, A.M.; El-Nablaway, M.; et al. Antibiotic action and resistance: Updated review of mechanisms, spread, influencing factors, and alternative approaches for combating resistance. Front. Pharmacol. 2024, 14, 1305294. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Yao, F.; Zhao, J.; Zhang, W.; Chen, L.; Wang, X.; Yang, P.; Tang, J.; Chi, Y. Unraveling mitochondria-targeting reactive oxygen species modulation and their implementations in cancer therapy by nanomaterials. Exploration 2023, 3, 20220115. [Google Scholar] [CrossRef]
- Ibrahim, M.K.; Haria, A.; Mehta, N.V.; Degani, M.S. Antimicrobial potential of quaternary phosphonium salt compounds: A review. Future Med. Chem. 2023, 15, 2113–2141. [Google Scholar] [CrossRef]
- Skulachev, V.P. Cationic antioxidants as a powerful tool against mitochondrial oxidative stress. Biochem. Biophys. Res. Commun. 2013, 441, 275–279. [Google Scholar] [CrossRef]
- Kumawat, M.; Nabi, B.; Daswani, M.; Viquar, I.; Pal, N.; Sharma, P.; Tiwari, S.; Sarma, D.K.; Shubham, S.; Kumar, M. Role of bacterial efflux pump proteins in antibiotic resistance across microbial species. Microb. Pathog. 2023, 181, 106182. [Google Scholar] [CrossRef]
- Nazarov, P.A.; Zinovkina, L.A.; Brezgunova, A.A.; Lyamzaev, K.G.; Golovin, A.V.; Karakozova, M.V.; Kotova, E.A.; Plotnikov, E.Y.; Zinovkin, R.A.; Skulachev, M.V.; et al. Relationship of Cytotoxic and Antimicrobial Effects of Triphenylphosphonium Conjugates with Various Quinone Derivatives. Biochemistry 2024, 89, 212–222. [Google Scholar] [CrossRef]
- Pavlova, J.A.; Khairullina, Z.Z.; Tereshchenkov, A.G.; Nazarov, P.A.; Lukianov, D.A.; Volynkina, I.A.; Skvortsov, D.A.; Makarov, G.I.; Abad, E.; Murayama, S.Y.; et al. Triphenilphosphonium Analogs of Chloramphenicol as Dual-Acting Antimicrobial and Antiproliferating Agents. Antibiotics 2021, 10, 489. [Google Scholar] [CrossRef] [PubMed]
- Morris, M.A.; Malek, M.; Hashemian, M.H.; Nguyen, B.T.; Manuse, S.; Lewis, K.; Nowick, J.S. A Fluorescent Teixobactin Analogue. ACS Chem. Biol. 2020, 15, 1222–1231. [Google Scholar] [CrossRef]
- Nazarov, P.A.; Kirsanov, R.S.; Denisov, S.S.; Khailova, L.S.; Karakozova, M.V.; Lyamzaev, K.G.; Korshunova, G.A.; Lukyanov, K.A.; Kotova, E.A.; Antonenko, Y.N. Fluorescein Derivatives as Antibacterial Agents Acting via Membrane Depolarization. Biomolecules 2020, 10, 309. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.; Sunwoo, K.; Jung, Y.; Hur, J.K.; Park, K.H.; Kim, J.S.; Kim, D. Membrane-Targeting Triphenylphosphonium Functionalized Ciprofloxacin for Methicillin-Resistant Staphylococcus aureus (MRSA). Antibiotics 2020, 9, 758. [Google Scholar] [CrossRef] [PubMed]
- Goleva, T.N.; Lyamzaev, K.G.; Rogov, A.G.; Khailova, L.S.; Epremyan, K.K.; Shumakovich, G.P.; Domnina, L.V.; Ivanova, O.Y.; Marmiy, N.V.; Zinevich, T.V.; et al. Mitochondria-targeted 1,4-naphthoquinone (SkQN) is a powerful prooxidant and cytotoxic agent. Biochim. Biophys. Acta Bioenerg. 2020, 1861, 148210. [Google Scholar] [CrossRef]
- Nunes, B.; Cagide, F.; Fernandes, C.; Borges, A.; Borges, F.; Simões, M. Efficacy of Novel Quaternary Ammonium and Phosphonium Salts Differing in Cation Type and Alkyl Chain Length against Antibiotic-Resistant Staphylococcus aureus. Int. J. Mol. Sci. 2023, 25, 504. [Google Scholar] [CrossRef]
- Meier, G.; Thavarasah, S.; Ehrenbolger, K.; Hutter, C.A.J.; Hürlimann, L.M.; Barandun, J.; Seeger, M.A. Deep mutational scan of a drug efflux pump reveals its structure-function landscape. Nat. Chem. Biol. 2023, 19, 440–450. [Google Scholar] [CrossRef]
- Seeger, M.A.; Diederichs, K.; Eicher, T.; Brandstätter, L.; Schiefner, A.; Verrey, F.; Pos, K.M. The AcrB efflux pump: Conformational cycling and peristalsis lead to multidrug resistance. Curr. Drug Targets 2008, 9, 729–749. [Google Scholar] [CrossRef] [PubMed]
- Pos, K.M. Drug transport mechanism of the AcrB efflux pump. Biochim. Biophys. Acta 2009, 1794, 782–793. [Google Scholar] [CrossRef] [PubMed]
- Rybenkov, V.V.; Zgurskaya, H.I.; Ganguly, C.; Leus, I.V.; Zhang, Z.; Moniruzzaman, M. The Whole Is Bigger than the Sum of Its Parts: Drug Transport in the Context of Two Membranes with Active Efflux. Chem. Rev. 2021, 121, 5597–5631. [Google Scholar] [CrossRef] [PubMed]
- Manrique, P.D.; López, C.A.; Gnanakaran, S.; Rybenkov, V.V.; Zgurskaya, H.I. New understanding of multidrug efflux and permea-tion in antibiotic resistance, persistence, and heteroresistance. Ann. N. Y. Acad. Sci. 2023, 1519, 46–62. [Google Scholar] [CrossRef]
- Pos, K.M. RND multidrug efflux transporters: Similar appearances, diverse actions. J. Bacteriol. 2024, 206, e0040323. [Google Scholar] [CrossRef]

| Bacillus subtilis | Escherichia coli | Escherichia coli ΔtolC | Staphylococcus aureus | Mycobacterium smegmatis | |
|---|---|---|---|---|---|
| SkQ1 | 0.6–1.2 | 21 | 1.2 | 0.6–1.2 | 0.6 |
| SkQR1 | 2.8 | 89.6< | 89.6 | 2.8 | 1.4 |
| C12R1 | 2.4 | 76< | 76 | 2.4 | 1.2 |
| C10R1 | 1.2 | 75< | 38 | 1.2 | 1.2 |
| C4R1 | 0.5–1 | 64< | 4 | 0.5 | 0.125 |
| C2R1 | 0.48–0.96 | 61< | 3.8 | 0.96 | 0.48 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nazarov, P.A.; Maximov, V.S.; Firsov, A.M.; Karakozova, M.V.; Panfilova, V.; Kotova, E.A.; Skulachev, M.V.; Antonenko, Y.N. Rhodamine 19 Alkyl Esters as Effective Antibacterial Agents. Int. J. Mol. Sci. 2024, 25, 6137. https://doi.org/10.3390/ijms25116137
Nazarov PA, Maximov VS, Firsov AM, Karakozova MV, Panfilova V, Kotova EA, Skulachev MV, Antonenko YN. Rhodamine 19 Alkyl Esters as Effective Antibacterial Agents. International Journal of Molecular Sciences. 2024; 25(11):6137. https://doi.org/10.3390/ijms25116137
Chicago/Turabian StyleNazarov, Pavel A., Vladislav S. Maximov, Alexander M. Firsov, Marina V. Karakozova, Veronika Panfilova, Elena A. Kotova, Maxim V. Skulachev, and Yuri N. Antonenko. 2024. "Rhodamine 19 Alkyl Esters as Effective Antibacterial Agents" International Journal of Molecular Sciences 25, no. 11: 6137. https://doi.org/10.3390/ijms25116137
APA StyleNazarov, P. A., Maximov, V. S., Firsov, A. M., Karakozova, M. V., Panfilova, V., Kotova, E. A., Skulachev, M. V., & Antonenko, Y. N. (2024). Rhodamine 19 Alkyl Esters as Effective Antibacterial Agents. International Journal of Molecular Sciences, 25(11), 6137. https://doi.org/10.3390/ijms25116137







