Exploring the Ocular Surface Microbiome and Tear Proteome in Glaucoma
Abstract
1. Introduction
2. Results
2.1. Demographic Data
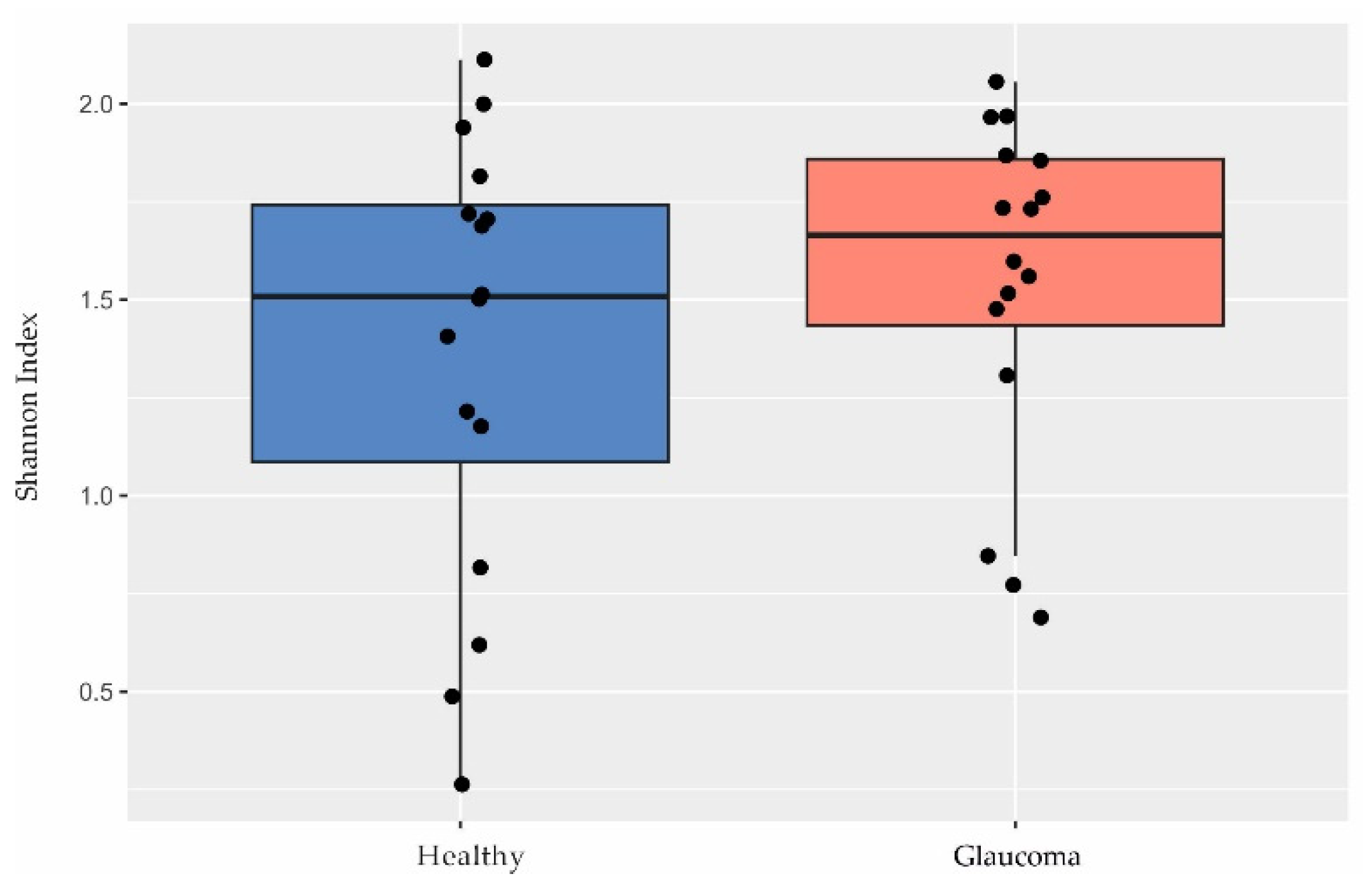
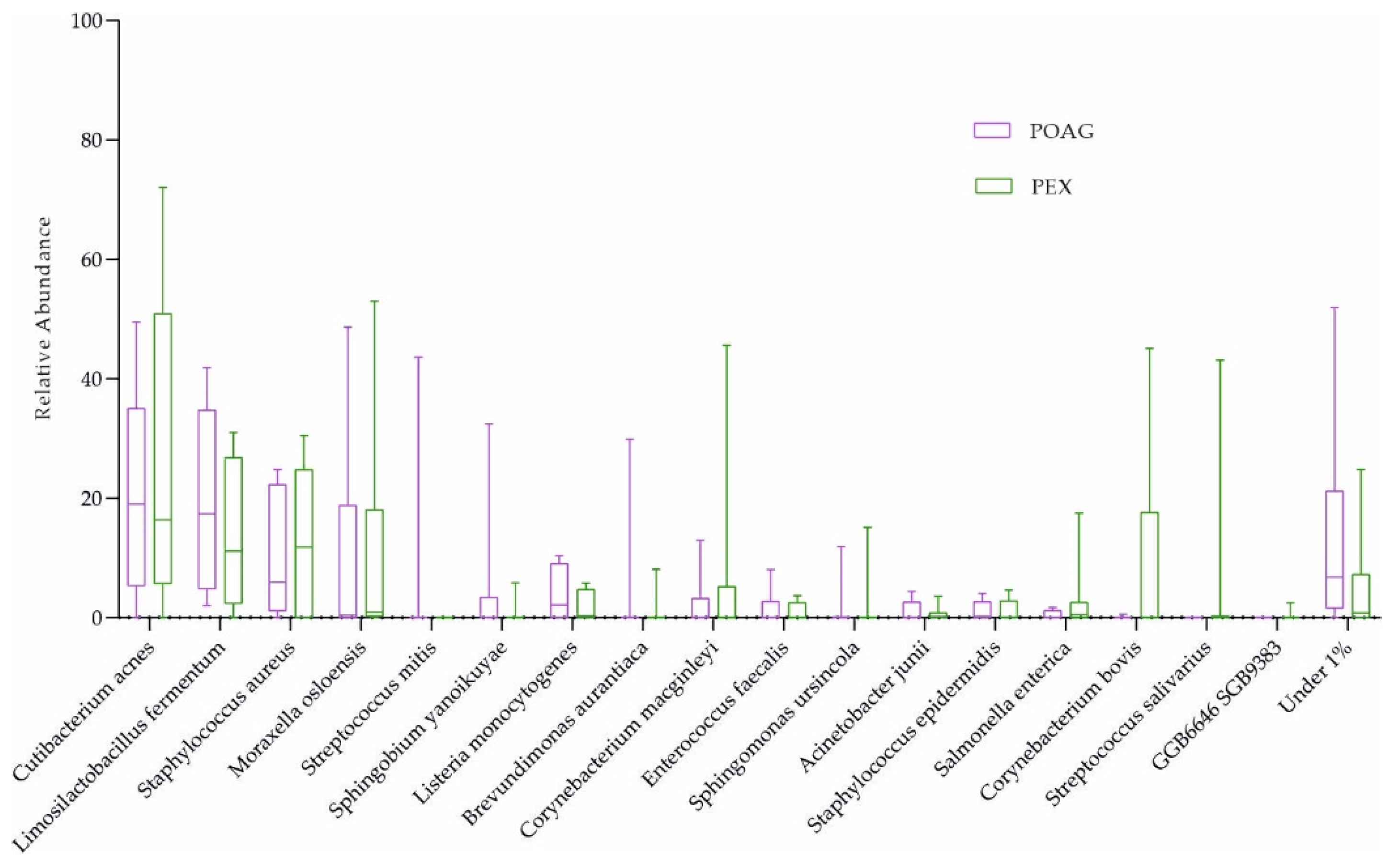
2.2. Characterization of the Ocular Surface Microbiome in Glaucoma Patients
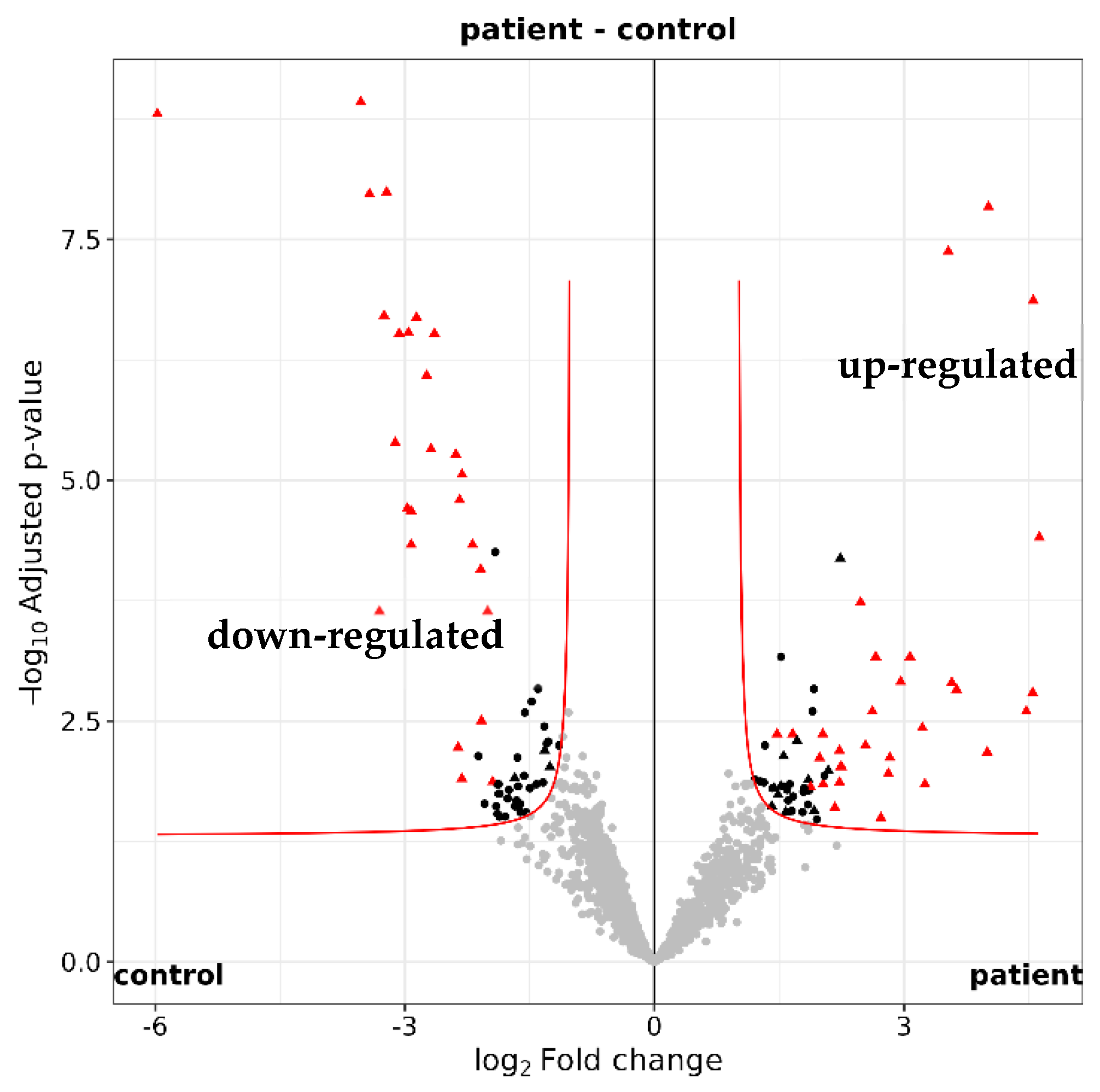
2.3. Functional Annotation of the Tear Proteome in Glaucoma
3. Discussion
3.1. Implications of the Ocular Surface Microbiome in Glaucoma
3.2. The Influence of Intraocular Pressur-Lowering Eye Drops on the Ocular Microbiome
3.3. Implications of the Tear Proteome and the Immune System in Glaucoma
3.4. Conclusions
4. Materials and Methods
4.1. Study Design and Recruitment
4.2. Sample Collection
4.3. Metagenomic DNA Sequencing and Analysis
4.4. Tear Fluid Processing and Analysis [102,103,104,105,106,107,108,109,110,111]
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Weinreb, R.N.; Aung, T.; Medeiros, F.A. The pathophysiology and treatment of glaucoma: A review. JAMA 2014, 311, 1901–1911. [Google Scholar] [CrossRef]
- Agarwal, R.; Gupta, S.K.; Agarwal, P.; Saxena, R.; Agrawal, S.S. Current concepts in the pathophysiology of glaucoma. Indian J. Ophthalmol. 2009, 57, 257–266. [Google Scholar]
- Shon, K.; Wollstein, G.; Schuman, J.S.; Sung, K.R. Prediction of glaucomatous visual field progression: Pointwise analysis. Curr. Eye Res. 2014, 39, 705–710. [Google Scholar] [CrossRef] [PubMed]
- Tham, Y.C.; Li, X.; Wong, T.Y.; Quigley, H.A.; Aung, T.; Cheng, C.Y. Global prevalence of glaucoma and projections of glaucoma burden through 2040: A systematic review and meta-analysis. Ophthalmology 2014, 121, 2081–2090. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Hong, Y.; Fu, X.; Tan, H.; Chen, Y.; Wang, Y.; Chen, D. The role of the microbiota in glaucoma. Mol. Asp. Med. 2023, 94, 101221. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Wang, N.; Xiong, S.; Xia, X. The correlation between primary open-angle glaucoma (POAG) and gut microbiota: A pilot study towards predictive, preventive, and personalized medicine. EPMA J 2023, 14, 539–552. [Google Scholar] [CrossRef] [PubMed]
- Padhy, B.; Alone, D. Is pseudoexfoliation glaucoma a neurodegenerative disorder? J. Biosci. 2021, 46. [Google Scholar] [CrossRef]
- Sihota, R.; Angmo, D.; Ramaswamy, D.; Dada, T. Simplifying “target” intraocular pressure for different stages of primary open-angle glaucoma and primary angle-closure glaucoma. Indian J. Ophthalmol. 2018, 66, 495–505. [Google Scholar] [CrossRef] [PubMed]
- Sambhara, D.; Aref, A.A. Glaucoma management: Relative value and place in therapy of available drug treatments. Ther. Adv. Chronic Dis. 2014, 5, 30–43. [Google Scholar] [CrossRef]
- Peirce, J.M.; Alviña, K.A.-O. The role of inflammation and the gut microbiome in depression and anxiety. J. Neurosci. Res. 2019, 97, 1223–1241. [Google Scholar] [CrossRef]
- Cresci, G.A.; Bawden, E. Gut Microbiome: What We Do and Don’t Know. Nutr. Clin. Pract. 2015, 30, 734–746. [Google Scholar] [CrossRef]
- Gancz, N.N.; Levinson, J.A.; Callaghan, B.L. Sex and gender as critical and distinct contributors to the human brain-gut-microbiome axis. Brain Res. Bull. 2023, 199, 110665. [Google Scholar] [CrossRef] [PubMed]
- Asadi, A.; Shadab Mehr, N.; Mohamadi, M.H.; Shokri, F.; Heidary, M.A.-O.; Sadeghifard, N.; Khoshnood, S. Obesity and gut-microbiota-brain axis: A narrative review. J. Clin. Lab. Anal. 2022, 36, e24420. [Google Scholar] [CrossRef] [PubMed]
- Trakman, G.A.-O.; Fehily, S.A.-O.; Basnayake, C.A.-O.X.; Hamilton, A.A.-O.; Russell, E.; Wilson-O’Brien, A.; Kamm, M.A.-O. Diet and gut microbiome in gastrointestinal disease. J. Gastroenterol. Hepatol. 2022, 37, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Simpson, C.A.; Diaz-Arteche, C.; Eliby, D.; Schwartz, O.S.; Simmons, J.G.; Cowan, C.S.M. The gut microbiota in anxiety and depression—A systematic review. Clin. Psychol. Rev. 2021, 83, 101943. [Google Scholar] [CrossRef] [PubMed]
- Krilis, M.; Fry, L.; Ngo, P.; Goldberg, I. The gut microbiome and primary open angle glaucoma: Evidence for a ‘gut-glaucoma’ axis? Eur. J. Ophthalmol. 2023, 11206721231219147. [Google Scholar] [CrossRef] [PubMed]
- Abraham, P.; Pratap, N. Dysbiosis in Irritable Bowel Syndrome. J. Assoc. Physicians India 2023, 79, 75–81. [Google Scholar]
- Li, J.; Wei, J.; Wang, J.; Xu, T.; Wu, B.; Yang, S.; Jing, S.; Wu, H.; Hao, H. Association between gut microbiota and spinal stenosis: A two-sample mendelian randomization study. Front. Immunol. 2024, 15, 1360132. [Google Scholar] [CrossRef] [PubMed]
- Pheiffer, C.A.-O.; Riedel, S.A.-O.; Dias, S.A.-O.; Adam, S.A.-O. Gestational Diabetes and the Gut Microbiota: Fibre and Polyphenol Supplementation as a Therapeutic Strategy. Microorganisms 2024, 12, 633. [Google Scholar] [CrossRef]
- Omar, W.E.W.; Singh, G.; McBain, A.J.; Cruickshank, F.; Radhakrishnan, H. Gut Microbiota Profiles in Myopes and Nonmyopes. Investig. Ophthalmol. Vis. Sci. 2024, 65, 2. [Google Scholar] [CrossRef]
- Yao, S.Q.; Yang, X.; Cen, L.P.; Tan, S. The Role of Gut Microbiota in Neuromyelitis Optica Spectrum Disorder. Int. J. Mol. Sci. 2024, 25, 3179. [Google Scholar] [CrossRef] [PubMed]
- Bron, A.J.; de Paiva, C.S.; Chauhan, S.K.; Bonini, S.; Gabison, E.E.; Jain, S.; Knop, E.; Markoulli, M.; Ogawa, Y.; Perez, V.; et al. TFOS DEWS II pathophysiology report. Ocul. Surf. 2017, 15, 438–510. [Google Scholar] [PubMed]
- Kalyana Chakravarthy, S.; Jayasudha, R.; Ranjith, K.; Dutta, A.; Pinna, N.K.; Mande, S.S.; Sharma, S.; Garg, P.; Murthy, S.I.; Shivaji, S. Alterations in the gut bacterial microbiome in fungal Keratitis patients. PLoS ONE 2018, 13, e0199640. [Google Scholar] [CrossRef] [PubMed]
- Petrillo, F.; Pignataro, D.; Lavano, M.A.; Santella, B.; Folliero, V.; Zannella, C.; Astarita, C.; Gagliano, C.; Franci, G.; Avitabile, T.; et al. Current Evidence on the Ocular Surface Microbiota and Related Diseases. Microorganisms 2020, 8, 1033. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Yang, B.; Li, W. Defining the normal core microbiome of conjunctival microbial communities. Clin. Microbiol. Infect. 2016, 22, 643.e7–643.e12. [Google Scholar] [CrossRef] [PubMed]
- Peter, V.G.; Morandi, S.C.; Herzog, E.L.; Zinkernagel, M.S.; Zysset-Burri, D.C. Investigating the Ocular Surface Microbiome: What Can It Tell Us? Clin. Ophthalmol. 2023, 17, 259–271. [Google Scholar] [CrossRef] [PubMed]
- Zysset-Burri, D.C.; Schlegel, I.; Lincke, J.B.; Jaggi, D.; Keller, I.; Heller, M.; Lagache, S.B.; Wolf, S.; Zinkernagel, M.S. Understanding the Interactions Between the Ocular Surface Microbiome and the Tear Proteome. Investig. Ophthalmol. Vis. Sci. 2021, 62, 8. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Ley, R.E.; Hamady, M.; Fraser-Liggett, C.M.; Knight, R.; Gordon, J.I. The human microbiome project. Hum. Microbiome Project. Nat. 2007, 449, 804–810. [Google Scholar] [CrossRef] [PubMed]
- Schlegel, I.; De Gouyon Matignon de Pontourade, C.M.F.; Lincke, J.B.; Keller, I.; Zinkernagel, M.S.; Zysset-Burri, D.C. The Human Ocular Surface Microbiome and Its Associations with the Tear Proteome in Dry Eye Disease. Int. J. Mol. Sci. 2023, 24, 14091. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Fang, X.; Wang, X.; Lin, Y.A.; Wu, H.; Li, C. Effects of Sodium Hyaluronate Eye Drops with or without Preservatives on Ocular Surface Bacterial Microbiota. Front. Med. 2022, 9, 793565. [Google Scholar] [CrossRef]
- Hotta, F.; Eguchi, H.; Kuwahara, T.; Nakayama-Imaohji, H.; Shimomura, Y.; Kusaka, S. Disturbances in the ocular surface microbiome by perioperative antimicrobial eye drops. Front. Cell. Infect. Microbiol. 2023, 13, 1172345. [Google Scholar] [CrossRef] [PubMed]
- Labetoulle, M.; Baudouin, C.; Benitez Del Castillo, J.M.; Rolando, M.; Rescigno, M.; Messmer, E.M.; Aragona, P. How gut microbiota may impact ocular surface homeostasis and related disorders. Prog. Retin. Eye Res. 2024, 100, 101250. [Google Scholar] [CrossRef] [PubMed]
- Tezel, G. A proteomics view of the molecular mechanisms and biomarkers of glaucomatous neurodegeneration. Prog. Retin. Eye Res. 2013, 35, 18–43. [Google Scholar] [CrossRef] [PubMed]
- Wen, X.; Miao, L.; Deng, Y.; Bible, P.W.; Hu, X.; Zou, Y.; Liu, Y.; Guo, S.; Liang, J.; Chen, T.; et al. The Influence of Age and Sex on Ocular Surface Microbiota in Healthy Adults. Investig. Ophthalmol. Vis. Sci. 2017, 58, 6030–6037. [Google Scholar] [CrossRef] [PubMed]
- Katzka, W.; Dong, T.S.; Luu, K.; Lagishetty, V.; Sedighian, F.; Arias-Jayo, N.; Jacobs, J.P.; Hsu, H.Y. The Ocular Microbiome Is Altered by Sampling Modality and Age. Transl. Vis. Sci. Technol. 2021, 10, 24. [Google Scholar] [CrossRef] [PubMed]
- Nattinen, J.; Jylha, A.; Aapola, U.; Makinen, P.; Beuerman, R.; Pietila, J.; Vaajanen, A.; Uusitalo, H. Age-associated changes in human tear proteome. Clin. Proteom. 2019, 16, 11. [Google Scholar] [CrossRef] [PubMed]
- Mallick, H.; Rahnavard, A.; McIver, L.J.; Ma, S.; Zhang, Y.; Nguyen, L.H.; Tickle, T.L.; Weingart, G.; Ren, B.; Schwager, E.H.; et al. Multivariable association discovery in population-scale meta-omics studies. PLoS Comput. Biol. 2021, 17, e1009442. [Google Scholar] [CrossRef] [PubMed]
- Herzog, E.L.; Kreuzer, M.; Zinkernagel, M.S.; Zysset-Burri, D.C. Challenges and insights in the exploration of the low abundance human ocular surface microbiome. Front. Cell. Infect. Microbiol. 2023, 13, 1232147. [Google Scholar] [CrossRef] [PubMed]
- Gomes, J.A.P.; Frizon, L.; Demeda, V.F. Ocular Surface Microbiome in Health and Disease. Asia-Pac. J. Ophthalmol. 2020, 9, 505–511. [Google Scholar] [CrossRef] [PubMed]
- Grzybowski, A.; Brona, P.; Kim, S.J. Microbial flora and resistance in ophthalmology: A review. Graefes Arch. Clin. Exp. Ophthalmol. 2017, 255, 851–862. [Google Scholar] [CrossRef]
- Honda, R.; Toshida, H.; Suto, C.; Fujimaki, T.; Kimura, T.; Ohta, T.; Murakami, A. Effect of long-term treatment with eyedrops for glaucoma on conjunctival bacterial flora. Infect. Drug Resist. 2011, 4, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Xue, W.; Li, J.J.; Zou, Y.; Zou, B.; Wei, L. Microbiota and Ocular Diseases. Front. Cell. Infect. Microbiol. 2021, 11, 759333. [Google Scholar] [CrossRef] [PubMed]
- Nayyar, A.; Gindina, S.; Barron, A.; Hu, Y.; Danias, J. Do epigenetic changes caused by commensal microbiota contribute to development of ocular disease? A review of evidence. Hum. Genom. 2020, 14, 11. [Google Scholar] [CrossRef] [PubMed]
- Guedes, G.; CTsai, J.; ALoewen, N. Glaucoma and aging. Glaucoma and aging. Curr. Aging Sci. 2011, 4, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Pezzino, S.A.-O.; Sofia, M.; Greco, L.P.; Litrico, G.; Filippello, G.; Sarvà, I.; La Greca, G.; Latteri, S. Microbiome Dysbiosis: A Pathological Mechanism at the Intersection of Obesity and Glaucoma. Int. J. Mol. Sci. 2023, 24, 1166. [Google Scholar] [CrossRef] [PubMed]
- Shin, D.Y.; Jung, K.I.; Park, H.Y.L.; Park, C.K. The effect of anxiety and depression on progression of glaucoma. Sci. Rep. 2021, 11, 1769. [Google Scholar] [CrossRef] [PubMed]
- Goyal, D.; Ali, S.A.; Singh, R.K. Emerging role of gut microbiota in modulation of neuroinflammation and neurodegeneration with emphasis on Alzheimer’s disease. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2021, 106, 110112. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.J.; Gerasimidis, K.; Edwards, C.A.; Shaikh, M.G. Role of Gut Microbiota in the Aetiology of Obesity: Proposed Mechanisms and Review of the Literature. J. Obes. 2016, 2016, 7353642. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Wu, J.; Wang, D.; Li, T.; Shi, X.; Li, L.; Zhu, M.; Zhang, Z.; Yu, X.; Dai, Q. Metagenomic profiling of ocular surface microbiome changes in Demodex blepharitis patients. Front. Cell. Infect. Microbiol. 2022, 12, 922753. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zou, X.; Xue, W.; Zhang, P.; Wang, S.; Zou, H. Ocular Surface Microbiota in Diabetic Patients with Dry Eye Disease. Investig. Ophthalmol. Vis. Sci. 2021, 62, 13. [Google Scholar] [CrossRef]
- Liang, X.; Li, Y.; Xiong, K.; Chen, S.; Li, Z.; Zhang, Z.; Xia, Z.; Yi, G.; Fu, M. Demodex Infection Changes Ocular Surface Microbial Communities, in Which Meibomian Gland Dysfunction May Play a Role. Ophthalmol. Ther. 2021, 10, 601–617. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Ge, X.; Li, Y.; Zou, B.; Wen, X.; Chen, W.; Lu, L.; Zhang, M.; Zhang, X.; Li, C.; et al. Identification of an intraocular microbiota. Cell Discov. 2021, 7, 13. [Google Scholar] [CrossRef] [PubMed]
- Kittipibul, T.; Puangsricharern, V.; Chatsuwan, T. Comparison of the ocular microbiome between chronic Stevens-Johnson syndrome patients and healthy subjects. Sci. Rep. 2020, 10, 4353. [Google Scholar] [CrossRef] [PubMed]
- Cheleuitte-Nieves, C.; Gulvik, C.A.; Humrighouse, B.W.; Bell, M.E.; Villarma, A.; Westblade, L.F.; Lipman, N.S.; Fischetti, V.A.; McQuiston, J.R. Draft Reference Genome Sequence of Corynebacterium mastitidis 16-1433, Isolated from a Mouse. Genome Announc. 2018, 6. [Google Scholar] [CrossRef] [PubMed]
- St Leger, A.J.; Caspi, R.R. Visions of Eye Commensals: The Known and the Unknown about How the Microbiome Affects Eye Disease. BioEssays News Rev. Mol. Cell. Dev. Biol. 2018, 40, e1800046. [Google Scholar] [CrossRef] [PubMed]
- St Leger, A.J.; Desai, J.V.; Drummond, R.A.; Kugadas, A.; Almaghrabi, F.; Silver, P.; Raychaudhuri, K.; Gadjeva, M.; Iwakura, Y.; Lionakis, M.S.; et al. An Ocular Commensal Protects against Corneal Infection by Driving an Interleukin-17 Response from Mucosal gammadelta T Cells. Immunity 2017, 47, 148–158.e5. [Google Scholar] [CrossRef] [PubMed]
- Mandal, A.; Varghese, M.V.; James, J. Chapter 22—Clinical significance of phospholipase A2 in glaucoma. In Phospholipases in Physiology and Pathology; Chakraborti, S., Ed.; Academic Press: Cambridge, MA, USA, 2023; pp. 331–341. [Google Scholar] [CrossRef]
- Zhou, Y.; Sidhu, G.S.; Whitlock, J.A.; Abdelmalik, B.; Mayer, Z.; Li, Y.; Wang, G.P.; Steigleman, W.A. Effects of Carboxymethylcellulose Artificial Tears on Ocular Surface Microbiome Diversity and Composition, A Randomized Controlled Trial. Transl. Vis. Sci. Technol. 2023, 12, 5. [Google Scholar] [CrossRef] [PubMed]
- Ohtani, S.; Shimizu, K.; Nejima, R.; Kagaya, F.; Aihara, M.; Iwasaki, T.; Shoji, N.; Miyata, K. Conjunctival Bacteria Flora of Glaucoma Patients During Long-Term Administration of Prostaglandin Analog Drops. Investig. Ophthalmol. Vis. Sci. 2017, 58, 3991–3996. [Google Scholar] [CrossRef] [PubMed]
- Priluck, A.; Ramulu, P.; Dosto, N.; Quigley, H.; Abraham, A. Validation of 16S rRNA Gene Sequencing of the Periocular Microbiome and Lack of Alteration by Topical Eyedrops. Transl. Vis. Sci. Technol. 2023, 12, 32. [Google Scholar] [CrossRef]
- Ekici Gok, Z.; Gunduz, A.; Bozgül, P.G. Evaluation of the effects of mono or combined use of topical antiglaucomatous drops on conjunctival flora and antibiotic susceptibility. Eur. J. Ophthalmol. 2023, 11206721231219275. [Google Scholar] [CrossRef]
- de Kaspar, H.M.; Kreidl, K.O.; Singh, K.; Ta, C.N. Comparison of preoperative conjunctival bacterial flora in patients undergoing glaucoma or cataract surgery. J. Glaucoma 2004, 13, 507–509. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.J.; Somohano, K.; Zemsky, C.; Uhlemann, A.C.; Liebmann, J.; Cioffi, G.A.; Al-Aswad, L.A.; Lynch, S.V.; Winn, B.J. Topical Glaucoma Therapy Is Associated with Alterations of the Ocular Surface Microbiome. Investig. Ophthalmol. Vis. Sci. 2022, 63, 32. [Google Scholar] [CrossRef]
- Jiang, S.; Kametani, M.; Chen, D.F. Adaptive Immunity: New Aspects of Pathogenesis Underlying Neurodegeneration in Glaucoma and Optic Neuropathy. Front. Immunol. 2020, 11, 65. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Xiu, W.; Chen, Q.; Peng, K.; Zhu, X.; Wang, Z.; Xu, X.; Chen, Y.; Zhang, G.; Fu, J.; et al. Gut-licensed β7+ CD4+ T cells contribute to progressive retinal ganglion cell damage in glaucoma. Sci. Transl. Med. 2023, 15, eadg1656. [Google Scholar] [CrossRef] [PubMed]
- Cong, H.; Jiang, H.; Peng, J.; Cui, S.; Liu, L.; Wang, J.; Zhang, X. Change of Th17 Lymphocytes and Treg/Th17 in Typical and Atypical Optic Neuritis. PLoS ONE 2016, 11, e0146270. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Li, Z.; van Rooijen, N.; Wang, N.; Pang, C.P.; Cui, Q. Different responses of macrophages in retinal ganglion cell survival after acute ocular hypertension in rats with different autoimmune backgrounds. Exp. Eye Res. 2007, 85, 659–666. [Google Scholar] [CrossRef] [PubMed]
- Mick, A.B. A Few Steps Toward Improving Glaucoma Diagnostic Accuracy and Understanding Intraocular Pressure. Optom. Vis. Sci. 2018, 95, 86–87. [Google Scholar] [CrossRef] [PubMed]
- Zukerman, R.; Harris, A.; Oddone, F.; Siesky, B.; Verticchio Vercellin, A.; Ciulla, T.A. Glaucoma Heritability: Molecular Mechanisms of Disease. Genes 2021, 12, 1135. [Google Scholar] [CrossRef] [PubMed]
- DeMaio, A.; Mehrotra, S.; Sambamurti, K.; Husain, S. The role of the adaptive immune system and T cell dysfunction in neurodegenerative diseases. J. Neuroinflammation 2022, 19, 251. [Google Scholar] [CrossRef]
- Kermer, P.; Klocker, N.; Labes, M.; Thomsen, S.; Srinivasan, A.; Bahr, M. Activation of caspase-3 in axotomized rat retinal ganglion cells in vivo. FEBS Lett. 1999, 453, 361–364. [Google Scholar] [CrossRef]
- Husain, S.; Abdul, Y.; Crosson, C.E. Preservation of retina ganglion cell function by morphine in a chronic ocular-hypertensive rat model. Investig. Ophthalmol. Vis. Sci. 2012, 53, 4289–4298. [Google Scholar] [CrossRef] [PubMed]
- Almasieh, M.; Wilson, A.M.; Morquette, B.; Vargas, J.L.C.; Di Polo, A. The molecular basis of retinal ganglion cell death in glaucoma. Prog. Retin. Eye Res. 2012, 31, 152–181. [Google Scholar] [CrossRef] [PubMed]
- Himori, N.; Yamamoto, K.; Maruyama, K.; Ryu, M.; Taguchi, K.; Yamamoto, M.; Nakazawa, T. Critical role of Nrf2 in oxidative stress-induced retinal ganglion cell death. J. Neurochem. 2013, 127, 669–680. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.C.; Huang, B.Q.; Yang, J. Regulatory mechanisms of retinal ganglion cell death in normal tension glaucoma and potential therapies. Neural Regen. Res. 2023, 18, 87–93. [Google Scholar] [PubMed]
- Coleman-Belin, J.A.-O.; Harris, A.; Chen, B.; Zhou, J.; Ciulla, T.A.-O.; Verticchio, A.; Antman, G.; Chang, M.; Siesky, B. Aging Effects on Optic Nerve Neurodegeneration. Int. J. Mol. Sci. 2023, 24, 2573. [Google Scholar] [CrossRef] [PubMed]
- Sivakumar, V.; Foulds, W.S.; Luu, C.D.; Ling, E.A.; Kaur, C. Retinal ganglion cell death is induced by microglia derived pro-inflammatory cytokines in the hypoxic neonatal retina. J. Pathol. 2011, 224, 245–260. [Google Scholar] [CrossRef] [PubMed]
- Claes, M.; De Groef, L.A.-O.; Moons, L.A.-O. Target-Derived Neurotrophic Factor Deprivation Puts Retinal Ganglion Cells on Death Row: Cold Hard Evidence and Caveats. Int. J. Mol. Sci. 2019, 20, 4314. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Hou, X.; Ni, Y.; Du, W.; Han, H.; Yu, Y.; Shi, G. The Imbalance of FOXP3/GATA3 in Regulatory T Cells from the Peripheral Blood of Asthmatic Patients. J. Immunol. Res. 2018, 2018, 3096183. [Google Scholar] [CrossRef] [PubMed]
- Joachim, S.C.; Grus, F.H.; Pfeiffer, N. Analysis of autoantibody repertoires in sera of patients with glaucoma. Eur. J. Ophthalmol. 2003, 13, 752–758. [Google Scholar] [CrossRef]
- Dutta Majumder, P.; Marchese, A.; Pichi, F.; Garg, I.; Agarwal, A. An update on autoimmune retinopathy. Indian J. Ophthalmol. 2020, 68, 1829–1837. [Google Scholar] [CrossRef]
- Joachim, S.C.; Bruns, K.; Lackner, K.J.; Pfeiffer, N.; Grus, F.H. Antibodies to α B-Crystallin, Vimentin, and Heat Shock Protein 70 in Aqueous Humor of Patients with Normal Tension Glaucoma and IgG Antibody Patterns against Retinal Antigen in Aqueous Humor. Curr. Eye Res. 2007, 32, 501–509. [Google Scholar] [CrossRef] [PubMed]
- Tezel, G.; Seigel, G.M.; Wax, M.B. Autoantibodies to small heat shock proteins in glaucoma. Investig. Ophthalmol. Vis. Sci. 1998, 39, 2277–2287. [Google Scholar]
- Loones, M.T.; Chang, Y.; Morange, M. The distribution of heat shock proteins in the nervous system of the unstressed mouse embryo suggests a role in neuronal and non-neuronal differentiation. Cell Stress Chaperones 2000, 5, 291. [Google Scholar] [CrossRef] [PubMed]
- Grus, F.H.; Joachim, S.C.; Wuenschig, D.; Rieck, J.; Pfeiffer, N. Autoimmunity and glaucoma. J. Glaucoma 2008, 17, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Wakefield, D.; Wildner, G. Is glaucoma an autoimmune disease? Clin. Transl. Immunol. 2020, 9, e1180. [Google Scholar] [CrossRef] [PubMed]
- Laspas, P.; Gramlich, O.W.; Muller, H.D.; Cuny, C.S.; Gottschling, P.F.; Pfeiffer, N.; Dick, H.B.; Joachim, S.C.; Grus, F.H. Autoreactive antibodies and loss of retinal ganglion cells in rats induced by immunization with ocular antigens. Investig. Ophthalmol. Vis. Sci. 2011, 52, 8835–8848. [Google Scholar] [CrossRef] [PubMed]
- Qing, G.; Duan, X.; Jiang, Y. Heat shock protein 72 protects retinal ganglion cells in rat model of acute glaucoma. Yan Ke Xue Bao 2005, 21, 163–168. [Google Scholar] [PubMed]
- Ishii, Y.; Kwong, J.M.; Caprioli, J. Retinal ganglion cell protection with geranylgeranylacetone, a heat shock protein inducer, in a rat glaucoma model. Investig. Ophthalmol. Vis. Sci. 2003, 44, 1982–1992. [Google Scholar] [CrossRef]
- Park, K.H.; Cozier, F.; Ong, O.C.; Caprioli, J. Induction of heat shock protein 72 protects retinal ganglion cells in a rat glaucoma model. Investig. Ophthalmol. Vis. Sci. 2001, 42, 1522–1530. [Google Scholar]
- Harada, C.; Kimura, A.; Guo, X.; Namekata, K.; Harada, T. Recent advances in genetically modified animal models of glaucoma and their roles in drug repositioning. Br. J. Ophthalmol. 2019, 103, 161–166. [Google Scholar] [CrossRef]
- Ishikawa, M.; Yoshitomi, T.; Zorumski, C.F.; Izumi, Y. Experimentally Induced Mammalian Models of Glaucoma. Biomed. Res. Int. 2015, 2015, 281214. [Google Scholar] [CrossRef]
- Yang, Q.; Cho, K.S.; Chen, H.; Yu, D.; Wang, W.H.; Luo, G.; Pang, I.H.; Guo, W.; Chen, D.F. Microbead-induced ocular hypertensive mouse model for screening and testing of aqueous production suppressants for glaucoma. Investig. Ophthalmol. Vis. Sci. 2012, 53, 3733–3741. [Google Scholar] [CrossRef]
- Hao, H.J.; Li, Y.H.; Yu, B.; Liu, X.; Zhang, Y.; Xing, X.L. Neuroprotective effects of acteoside in a glaucoma mouse model by targeting Serta domain-containing protein 4. Int. J. Ophthalmol. 2024, 17, 625–637. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.; Heger, A.; Sudbery, I. UMI-tools: Modeling sequencing errors in Unique Molecular Identifiers to improve quantification accuracy. Genome Res. 2017, 27, 491–499. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Genome Project Data Processing, S. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Miguez, A.; Beghini, F.; Cumbo, F.; McIver, L.J.; Thompson, K.N.; Zolfo, M.; Manghi, P.; Dubois, L.; Huang, K.D.; Thomas, A.M.; et al. Extending and improving metagenomic taxonomic profiling with uncharacterized species with MetaPhlAn 4. Nat. Biotechnol. 2023, 41, 1633–1644. [Google Scholar] [CrossRef]
- Truong, D.T.; Tett, A.; Pasolli, E.; Huttenhower, C.; Segata, N. Microbial strain-level population structure and genetic diversity from metagenomes. Genome Res. 2017, 27, 626–638. [Google Scholar] [CrossRef]
- Beghini, F.; McIver, L.J.; Blanco-Míguez, A.; Dubois, L.; Asnicar, F.; Maharjan, S.; Mailyan, A.; Manghi, P.; Scholz, M.; Thomas, A.M.; et al. Integrating taxonomic, functional, and strain-level profiling of diverse microbial communities with bioBakery 3. eLife 2021, 10, e65088. [Google Scholar] [CrossRef]
- UniProt Consortium. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef]
- Yu, F.; Haynes, S.E.; Teo, G.C.; Avtonomov, D.M.; Polasky, D.A.; Nesvizhskii, A.I. Fast Quantitative Analysis of timsTOF PASEF Data with MSFragger and IonQuant. Mol. Cell. Proteom. 2020, 19, 1575–1585. [Google Scholar] [CrossRef] [PubMed]
- Kong, A.T.; Leprevost, F.A.-O.; Avtonomov, D.M.; Mellacheruvu, D.A.-O.; Nesvizhskii, A.A.-O. MSFragger: Ultrafast and comprehensive peptide identification in mass spectrometry-based proteomics. Nat. Methods 2017, 14, 513–520. [Google Scholar] [CrossRef] [PubMed]
- da Veiga Leprevost, F.; Haynes, S.E.; Avtonomov, D.M.; Chang, H.Y.; Shanmugam, A.K.; Mellacheruvu, D.; Kong, A.T.; Nesvizhskii, A.I. Philosopher: A versatile toolkit for shotgun proteomics data analysis. Nat. Methods 2020, 17, 869–870. [Google Scholar] [CrossRef]
- Yu, F.; Haynes, S.E.; Nesvizhskii, A.I. IonQuant Enables Accurate and Sensitive Label-Free Quantification with FDR-Controlled Match-Between-Runs. Mol. Cell. Proteom. 2021, 20, 100077. [Google Scholar] [CrossRef]
- Silva, J.C.; Gorenstein, M.V.; Li, G.Z.; Vissers, J.P.; Geromanos, S.J. Absolute quantification of proteins by LCMSE: A virtue of parallel MS acquisition. Mol. Cell. Proteom. 2006, 5, 144–156. [Google Scholar] [CrossRef]
- Huber, W.; Von Heydebreck, A.; Sültmann, H.; Poustka, A.; Vingron, M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 2002, 18, S96–S104. [Google Scholar] [CrossRef]
- Kammers, K.; Cole, R.N.; Tiengwe, C.; Ruczinski, I. Detecting Significant Changes in Protein Abundance. EuPA Open Proteom. 2015, 7, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Uldry, A.A.-O.; Maciel-Dominguez, A.; Jornod, M.; Buchs, N.; Braga-Lagache, S.; Brodard, J.A.-O.; Jankovic, J.; Bonadies, N.A.-O.; Heller, M.A.-O. Effect of Sample Transportation on the Proteome of Human Circulating Blood Extracellular Vesicles. Int. J. Mol. Sci. 2022, 23, 4515. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Posa, A.; Bräuer, L.; Schicht, M.; Garreis, F.; Beileke, S.; Paulsen, F. Schirmer strip vs. capillary tube method: Non-invasive methods of obtaining proteins from tear fluid. Ann. Anat.-Anat. Anz. 2013, 195, 137–142. [Google Scholar] [CrossRef] [PubMed]
- Gunasekera, K.; Wüthrich, D.; Braga-Lagache, S.; Heller, M.; Ochsenreiter, T. Proteome remodelling during development from blood to insect-form Trypanosoma brucei quantified by SILAC and mass spectrometry. BMC Genom. 2012, 13, 556. [Google Scholar] [CrossRef] [PubMed]


| Total | Female | Male | Age Mean | SD | Variance (Age) | Chi-Squared Test (Sex) | |
|---|---|---|---|---|---|---|---|
| Healthy | 16 | 10 | 6 | 68.1 | 7.3 | 56.3 | 0.54 |
| Glaucoma | 16 | 7 | 9 | 76.5 | 7.7 | 63.2 |
| Patient | Prostaglandin Analogs | Carbonic Anhydrase | Beta-Blockers | Alpha-Agonists |
|---|---|---|---|---|
| 1 | x | x | x | |
| 2 | x | x | x | x |
| 3 | x | x | ||
| 4 | x | x | x | x |
| 5 | x | x | ||
| 6 | x | x | x | |
| 7 | x | x | x | x |
| 8 | x | x | x | x |
| 9 | x | x | x | |
| 10 | x | x | x | x |
| 11 | x | x | x | x |
| 12 | x | x | x | |
| 13 | x | x | ||
| 14 | ||||
| 15 | x | x | x | |
| 16 | x | x | x |
| C. Mastitidis | Positive | Negative |
|---|---|---|
| glaucoma | 0 | 16 |
| controls | 7 | 9 |
| GO Term ID | Biological Process | Genes Mapped | Enrichment Score | False Discovery Rate |
|---|---|---|---|---|
| GO:0006952 | Defense response | 153 | 1.62573 | 1.28 × 10−12 |
| GO:0006959 | Humoral immune response | 59 | 2.35722 | 1.28 × 10−12 |
| GO:0006955 | Immune response | 141 | 1.49942 | 5.32 × 10−12 |
| GO:0098542 | Defense response to other organism | 123 | 1.61702 | 3.21 × 10−10 |
| GO:0042742 | Defense response to bacterium | 45 | 2.36236 | 3.48 × 10−8 |
| GO:0045087 | Innate immune response | 98 | 1.52697 | 2.90 × 10−17 |
| GO:0006954 | Inflammatory response | 55 | 1.65598 | 6.60 × 10−7 |
| GO:0006956 | Complement activation | 27 | 1.24414 | 1.04× 10−6 |
| GO:0002682 | Regulation of immune system process | 133 | 2.08046 | 1.27 × 10−6 |
| GO:0019730 | Antimicrobial humoral response | 30 | 2.82318 | 1.27 × 10−12 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Spörri, L.; Uldry, A.-C.; Kreuzer, M.; Herzog, E.L.; Zinkernagel, M.S.; Unterlauft, J.D.; Zysset-Burri, D.C. Exploring the Ocular Surface Microbiome and Tear Proteome in Glaucoma. Int. J. Mol. Sci. 2024, 25, 6257. https://doi.org/10.3390/ijms25116257
Spörri L, Uldry A-C, Kreuzer M, Herzog EL, Zinkernagel MS, Unterlauft JD, Zysset-Burri DC. Exploring the Ocular Surface Microbiome and Tear Proteome in Glaucoma. International Journal of Molecular Sciences. 2024; 25(11):6257. https://doi.org/10.3390/ijms25116257
Chicago/Turabian StyleSpörri, Livia, Anne-Christine Uldry, Marco Kreuzer, Elio L. Herzog, Martin S. Zinkernagel, Jan D. Unterlauft, and Denise C. Zysset-Burri. 2024. "Exploring the Ocular Surface Microbiome and Tear Proteome in Glaucoma" International Journal of Molecular Sciences 25, no. 11: 6257. https://doi.org/10.3390/ijms25116257
APA StyleSpörri, L., Uldry, A.-C., Kreuzer, M., Herzog, E. L., Zinkernagel, M. S., Unterlauft, J. D., & Zysset-Burri, D. C. (2024). Exploring the Ocular Surface Microbiome and Tear Proteome in Glaucoma. International Journal of Molecular Sciences, 25(11), 6257. https://doi.org/10.3390/ijms25116257






