Enhancing Erucic Acid and Wax Ester Production in Brassica carinata through Metabolic Engineering for Industrial Applications
Abstract
1. Introduction
2. Results
2.1. Kanamycin Resistance
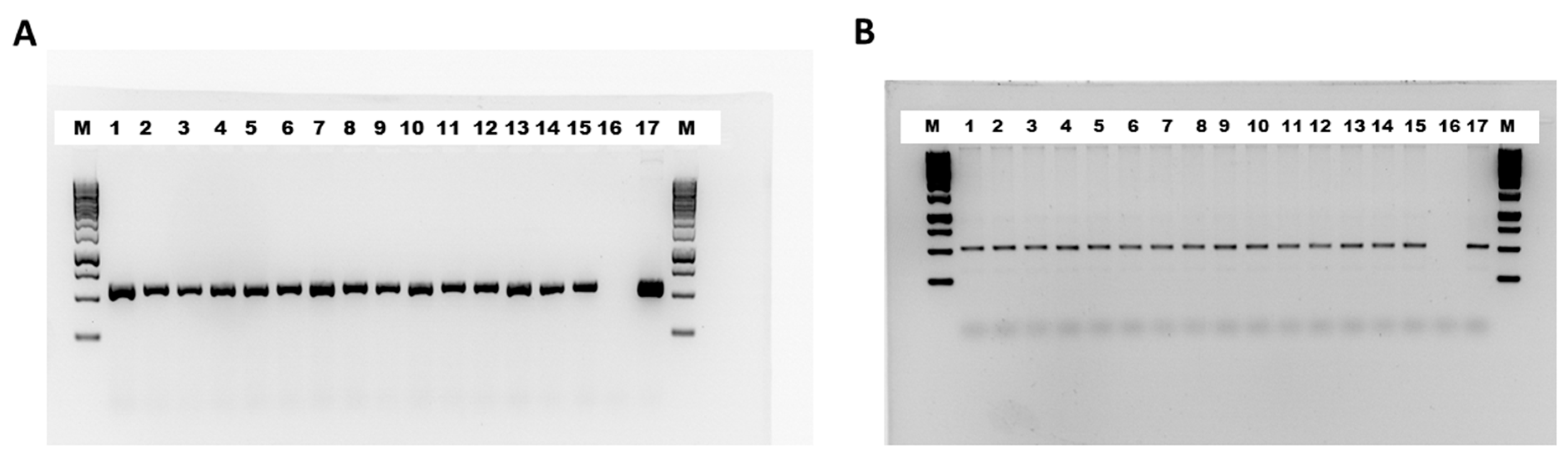
2.2. Confirmation of Transgenic Lines
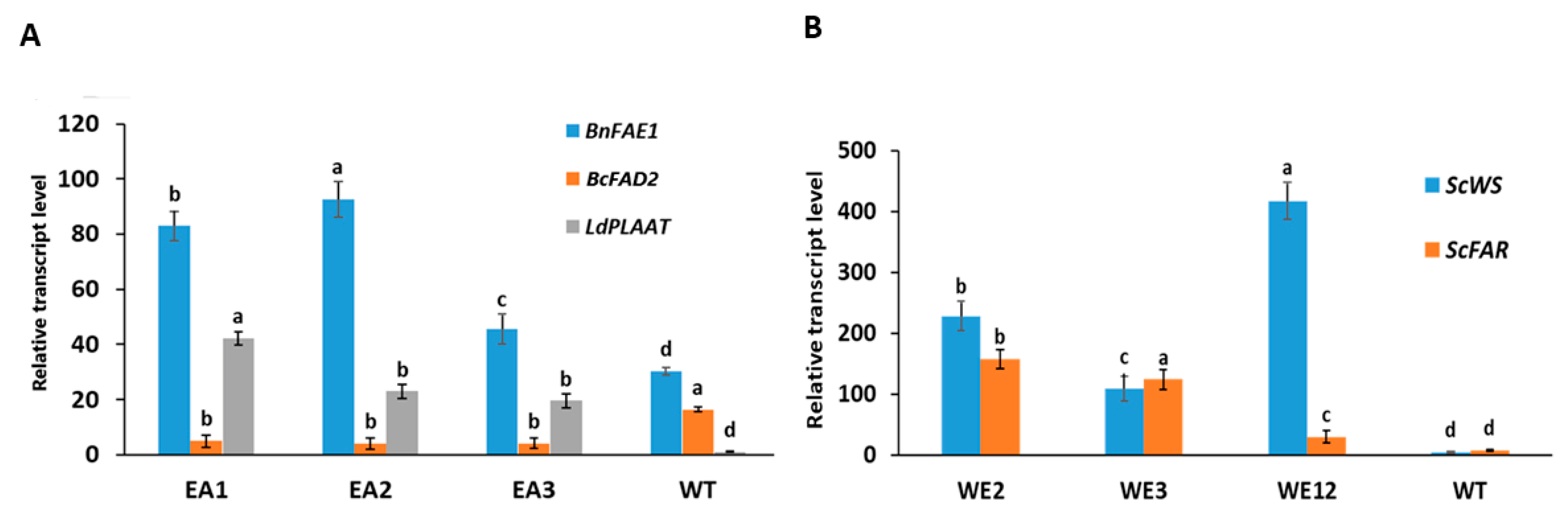
2.3. Expression Level of the Target Genes
2.4. Fatty Acid Profile
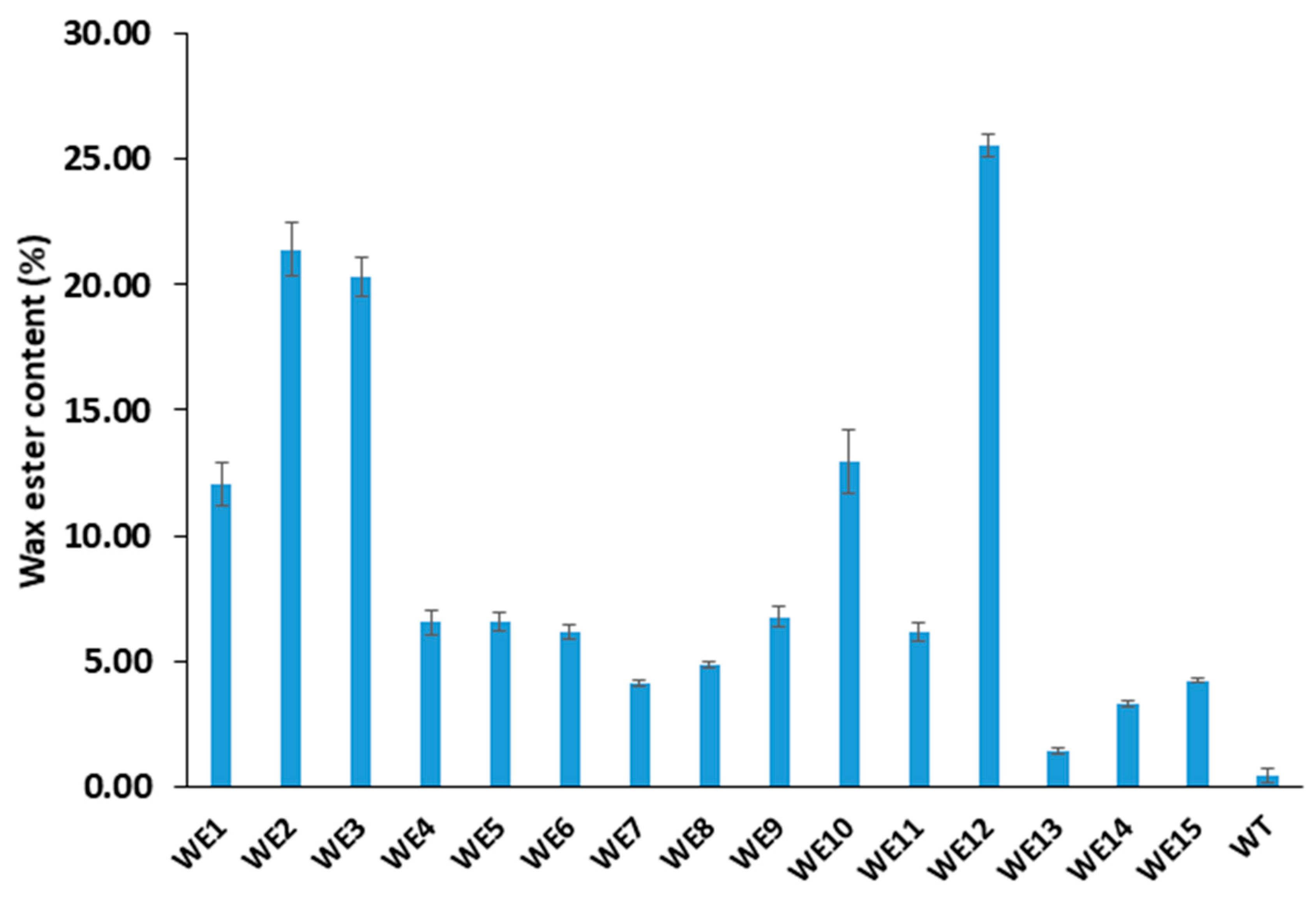
2.5. WE Content
2.6. WE Content and Species Determined by nanoESI-MS/MS
2.7. Transgenic Lines Grown in Biotron
3. Discussion
4. Materials and Methods
4.1. Plant Material and In Vitro Growth Conditions
4.2. Transformation Vectors
4.3. Transformation
4.3.1. Seed Surface Sterilization
4.3.2. Kanamycin Resistance Test
4.3.3. Agrobacterium-Mediated Transformation
4.4. Confirmation of Transgenic Lines by PCR Analysis
4.5. Quantitative Real-Time PCR (qRT-PCR) Analysis
4.6. Growth of Transgenic Lines in Biotron
4.7. Fatty Acid Profiling by Gas Chromatography (GC)
4.8. Analysis of WE Content and Profile
4.8.1. WE Analysis with GC
4.8.2. WE Profiling Analyzed by Nanoelectrospray Coupled with Tandem Mass Spectrometry (nanoESI-MS/MS)
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Durrett, T.P.; Benning, C.; Ohlrogge, J. Plant triacylglycerols as feedstocks for the production of biofuels. Plant J. 2008, 54, 593–607. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Napier, J.; Clemente, T.; Cahoon, E. New frontiers in oilseed biotechnology: Meeting the growing global demand for vegetable oils for food, feed, biofuel, and industrial uses. Curr. Opin. Biotechnol. 2011, 22, 252–259. [Google Scholar] [CrossRef] [PubMed]
- Mnzava, N.; Schippers, R. Brassica carinata A. Braun. [Internet] Record from PROTA4U; van der Vossen, H.A.M., Mkamilo, G.S., Eds.; PROTA (Plant Resources of Tropical Africa/Ressources Végétales de l’Afrique Tropicale): Wageningen, The Netherlands, 2007. [Google Scholar]
- Taylor, D.C.; Falk, K.C.; Palmer, C.D.; Hammerlindl, J.; Babic, V.; Mietkiewska, E.; Jadhav, A.; Marillia, E.-F.; Francis, T.; Hoffman, T.; et al. Brassica carinata—A new molecular farming platform for delivering bio-industrial oil feedstocks: Case studies of genetic modifications to improve very long-chain fatty acid and oil content in seeds. Biofuels Bioprod. Biorefin. 2010, 4, 538–561. [Google Scholar] [CrossRef]
- Roslinsky, V.; Falk, K.C.; Gaebelein, R.; Mason, A.S.; Eynck, C. Development of B. carinata with super-high erucic acid content through interspecific hybridization. Theor. Appl. Genet. 2021, 134, 3167–3181. [Google Scholar] [CrossRef] [PubMed]
- Derksen, J.T.; Cuperus, F.P.; Kolster, P. Paints and coatings from renewable resources. Ind. Crops Prod. 1995, 3, 225–236. [Google Scholar] [CrossRef]
- Leonard, E.C. High-erucic vegetable oils. Ind. Crops Prod. 1992, 1, 119–123. [Google Scholar] [CrossRef]
- McVetty, P.B.; Scarth, R. Breeding for improved oil quality in Brassica oilseed species. J. Crops Prod. 2002, 5, 345–369. [Google Scholar] [CrossRef]
- Semenov, V.; Semenova, D.; Slipushenko, V. Calculation of the high heat value of biofuels. Chem. Technol. Fuels Oils 2006, 42, 144–149. [Google Scholar] [CrossRef]
- Ghanevati, M.; Jaworski, J.G. Engineering and mechanistic studies of the Arabidopsis FAE1 β-ketoacyl-CoA synthase, FAE1 KCS. Eur. J. Biochem. 2002, 269, 3531–3539. [Google Scholar] [CrossRef]
- Sanyal, A.; Pinochet, X.; Merrien, A.; Laustriat, M.; Decocq, G.; Fine, F. Erucic acid rapeseed: 1. Prospects of improvements. OCL 2015, 22, D303. [Google Scholar] [CrossRef][Green Version]
- Becker, H.; Löptien, H.; Röbbelen, G. Breeding: An overview. In Biology of Brassica Coenospecies; Gómez-Campo, C., Ed.; Elsevier: Amsterdam, The Netherlands, 1999; pp. 421–425. [Google Scholar]
- Röbbelen, G. Biosynthesis of seed oil and breeding for improved oil quality of rapeseed. In Brassica Crops and Wild Allies; Japan Scientific Societies Press: Tokyo, Japan, 1980; pp. 253–283. [Google Scholar]
- Warwick, S.; Gugel, R.; McDonald, T.; Falk, K. Genetic variation and agronomic potential of Ethiopian mustard (Brassica carinata) in western Canada. Genet. Resour. Crop Evol. 2006, 53, 297–312. [Google Scholar] [CrossRef]
- Sasongko, N.D.; Möllers, C. Toward increasing erucic acid content in oilseed rape (Brassica napus L.) through the combination with genes for high oleic acid. J. Am. Oil Chem. Soc. 2005, 82, 445–449. [Google Scholar] [CrossRef]
- Zanetti, F.; Mosca, G.; Rampin, E.; Vamerali, T. Adaptability and sustainable management of high-erucic Brassicaceae in Mediterranean environment. In Oilseeds; IntechOpen Limited: London, UK, 2012; pp. 99–116. [Google Scholar]
- George, S.; Seepaul, R.; Geller, D.; Dwivedi, P.; DiLorenzo, N.; Altman, R.; Coppola, E.; Miller, S.A.; Bennett, R.; Johnston, G. A regional inter-disciplinary partnership focusing on the development of a carinata-centered bioeconomy. GCB Bioenergy 2021, 13, 1018–1029. [Google Scholar] [CrossRef]
- Domergue, F.; Miklaszewska, M. The production of wax esters in transgenic plants: Towards a sustainable source of bio-lubricants. J. Exp. Bot. 2022, 73, 2817–2834. [Google Scholar] [CrossRef] [PubMed]
- Miwa, T.K. Jojoba oil wax esters and derived fatty acids and alcohols: Gas chromatographic analyses. J. Am. Oil Chem. Soc. 1971, 48, 259–264. [Google Scholar] [CrossRef]
- Zhu, L.-H.; Krens, F.; Smith, M.A.; Li, X.; Qi, W.; Van Loo, E.N.; Iven, T.; Feussner, I.; Nazarenus, T.J.; Huai, D.; et al. Dedicated industrial oilseed crops as metabolic engineering platforms for sustainable industrial feedstock production. Sci. Rep. 2016, 6, 22181. [Google Scholar] [CrossRef]
- Wu, X.-Y.; Moreau, R.A.; Stumpf, P.K. Studies of biosynthesis of waxes by developing jojoba seed: III. Biosynthesis of wax esters from Acyl-CoA and long chain alcohols. Lipids 1981, 16, 897–902. [Google Scholar] [CrossRef]
- Vanhercke, T.; Wood, C.C.; Stymne, S.; Singh, S.P.; Green, A.G. Metabolic engineering of plant oils and waxes for use as industrial feedstocks. Plant Biotechnol. J. 2013, 11, 197–210. [Google Scholar] [CrossRef]
- Babic, V.; Datla, R.; Scoles, G.; Keller, W. Development of an efficient Agrobacterium-mediated transformation system for Brassica carinata. Plant Cell Rep. 1998, 17, 183–188. [Google Scholar] [CrossRef]
- Narasimhulu, S.B.; Kirti, P.B.; Mohapatra, T.; Prakash, S.; Chopra, V.L. Shoot regeneration in stem explants and its amenability to Agrobacterium tumefaciens mediated gene transfer in Brassica carinata. Plant Cell Rep. 1992, 11, 359–362. [Google Scholar] [CrossRef]
- Palmer, C.E.; Keller, W.A. Transgenic oilseed Brassicas. In Transgenic Plants and Crops; Khachatourians, G.C., Ed.; CRC Press: Boca Raton, FL, USA, 2002. [Google Scholar]
- Poulsen, G. Genetic transformation of Brassica. Plant Breed. 1996, 115, 209–225. [Google Scholar] [CrossRef]
- Jadhav, A.; Katavic, V.; Marillia, E.-F.; Giblin, E.M.; Barton, D.L.; Kumar, A.; Sonntag, C.; Babic, V.; Keller, W.A.; Taylor, D.C.; et al. Increased levels of erucic acid in Brassica carinata by co-suppression and antisense repression of the endogenous FAD2 gene. Metab. Eng. 2005, 7, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Mietkiewska, E.; Hoffman, T.L.; Brost, J.M.; Giblin, E.M.; Barton, D.L.; Francis, T.; Zhang, Y.; Taylor, D.C. Hairpin-RNA mediated silencing of endogenous FAD2 gene combined with heterologous expression of Crambe abyssinica FAE gene causes a dramatic increase in the level of erucic acid in transgenic Brassica carinata seeds. Mol. Breed. 2008, 22, 619–627. [Google Scholar] [CrossRef]
- Kuo, T.M.; Gardner, H.W. Lipases: Structure, function, and properties. In Lipid Biotechnology; CRC Press: Boca Raton, FL, USA, 2002; pp. 416–448. [Google Scholar]
- Nath, N.K.; Wilmer, J.A.; Wallington, E.J.; Becker, H.C.; Möllers, C. Increasing erucic acid content through combination of endogenouslow polyunsaturated fatty acids alleles withLd-LPAAT + Bn-fae1 transgenesin rapeseed (Brassica napus L.). Theor. Appl. Genet. 2009, 118, 765–773. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; van Loo, E.N.; Gruber, J.; Fan, J.; Guan, R.; Frentzen, M.; Stymne, S.; Zhu, L.H. Development of ultra-high erucic acid oil in the industrial oil crop Crambe abyssinica. Plant Biotechnol. J. 2012, 10, 862–870. [Google Scholar] [CrossRef]
- Li, X.; Guan, R.; Fan, J.; Zhu, L.-H. Development of Industrial Oil Crop Crambe abyssinica for Wax Ester Production through Metabolic Engineering and Cross Breeding. Plant Cell Physiol. 2019, 60, 1274–1283. [Google Scholar] [CrossRef] [PubMed]
- Lardizabal, K.D.; Metz, J.G.; Sakamoto, T.; Hutton, W.C.; Pollard, M.R.; Lassner, M.W. Purification of a jojoba embryo wax synthase, cloning of its cDNA, and production of high levels of wax in seeds of transgenic Arabidopsis. Plant Physiol. 2000, 122, 645–656. [Google Scholar] [CrossRef] [PubMed]
- Ivarson, E.; Iven, T.; Sturtevant, D.; Ahlman, A.; Cai, Y.; Chapman, K.; Feussner, I.; Zhu, L.-H. Production of wax esters in the wild oil species Lepidium campestre. Ind. Crops Prod. 2017, 108, 535–542. [Google Scholar] [CrossRef]
- Taylor, D.C.; Smith, M.A.; Fobert, P.; Mietkiewaska, E.; Weselake, R.J. Metabolic engineering of higher plants to produce bio-industrial oils. Compr. Biotechnol. 2011, 4, 67–85. [Google Scholar]
- Marillia, E.-F.; Francis, T.; Falk, K.C.; Smith, M.; Taylor, D.C. Palliser’s promise: Brassica carinata, an emerging western Canadian crop for delivery of new bio-industrial oil feedstocks. Biocatal. Agric. Biotechnol. 2014, 3, 65–74. [Google Scholar] [CrossRef]
- Ivarson, E.; Ahlman, A.; Li, X.; Zhu, L.-H. Development of an efficient regeneration and transformation method for the new potential oilseed crop Lepidium campestre. BMC Plant Biol. 2013, 13, 115. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ahlman, A.; Yan, X.; Lindgren, H.; Zhu, L.-H. Genetic transformation of the oilseed crop Crambe abyssinica. Plant Cell Tissue Organ Cult. 2010, 100, 149–156. [Google Scholar] [CrossRef]
- Yadav, R.C.; Singh, D. Genetic transformation in oilseed brassicas-A. Indian J. Agric. Sci. 2013, 83, 367–373. [Google Scholar]
- Wang, P.; Xiong, X.; Zhang, X.; Wu, G.; Liu, F. A review of erucic acid production in Brassicaceae oilseeds: Progress and prospects for the genetic engineering of high and low-erucic acid rapeseeds (Brassica napus). Front. Plant Sci. 2022, 13, 899076. [Google Scholar] [CrossRef] [PubMed]
- Lassner, M.W.; Levering, C.K.; Davies, H.M.; Knutzon, D.S. Lysophosphatidic acid acyltransferase from meadowfoam mediates insertion of erucic acid at the sn-2 position of triacylglycerol in transgenic rapeseed oil. Plant Physiol. 1995, 109, 1389–1394. [Google Scholar] [CrossRef]
- Brough, C.L.; Coventry, J.M.; Christie, W.W.; Kroon, J.T.M.; Brown, A.R.; Barsby, T.L.; Slabas, A.R. Towards the genetic engineering of triacylglycerols of defined fatty acid composition: Major changes in erucic acid content at the sn-2 position affected by the introduction of a 1-acyl-sn-glycerol-3-phosphate acyltransferase from Limnanthes douglasii into oil seed rape. Mol. Breed. 1996, 2, 133–142. [Google Scholar]
- Lokotsch, W.; Lang, S.; Möbius, D.; Wagner, F. Biocatalytical synthesis and monolayer studies of multiple hydroxylated wax esters. J. Am. Oil Chem. Soc. 1996, 73, 1459–1464. [Google Scholar] [CrossRef]
- Nguyen, D.H.; Raffa, G.; Morin, Y.; Desset, S.; Capet, F.; Nardello-Rataj, V.; Dumeignil, F.; Gauvin, R.M. Solvent-and base-free synthesis of wax esters from fatty acid methyl esters by consecutive one-pot, two-step catalysis. Green Chem. 2017, 19, 5665–5673. [Google Scholar] [CrossRef]
- Hagström, Å.K.; Wang, H.-L.; Liénard, M.A.; Lassance, J.-M.; Johansson, T.; Löfstedt, C. A moth pheromone brewery: Production of (Z)-11-hexadecenol by heterologous co-expression of two biosynthetic genes from a noctuid moth in a yeast cell factory. Microb. Cell Factories 2013, 12, 125. [Google Scholar] [CrossRef]
- Munkajohnpong, P.; Kesornpun, C.; Buttranon, S.; Jaroensuk, J.; Weeranoppanant, N.; Chaiyen, P. Fatty alcohol production: An opportunity of bioprocess. Biofuels Bioprod. Biorefin. 2020, 14, 986–1009. [Google Scholar] [CrossRef]
- Bansal, S.; Durrett, T.P. Camelina sativa: An ideal platform for the metabolic engineering and field production of industrial lipids. Biochimie 2016, 120, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Lazo, G.R.; Stein, P.A.; Ludwig, R.A. A DNA Transformation-Competent Arabidopsis Genomic Library in Agrobacterium. Bio-Technology 1991, 9, 963–967. [Google Scholar] [CrossRef] [PubMed]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Aldrich, J.; Cullis, C. RAPD analysis in flax: Optimization of yield and reproducibility sing Klen Taq1 DNA polymerase, Chelex 100, and gel purification of genomic DNA. Plant Mol. Biol. Rep. 1993, 11, 128–141. [Google Scholar] [CrossRef]
- Han, J.; Lühs, W.; Sonntag, K.; Zähringer, U.; Borchardt, D.S.; Wolter, F.P.; Heinz, E.; Frentzen, M. Functional characterization of â-ketoacyl-CoA synthase genes from Brassica napus L. Plant Mol. Biol. 2001, 46, 229–239. [Google Scholar] [CrossRef]
- Iven, T.; Herrfurth, C.; Hornung, E.; Heilmann, M.; Hofvander, P.; Stymne, S.; Zhu, L.H.; Feussner, I. Wax ester profiling of seed oil by nano-electrospray ionization tandem mass spectrometry. Plant Methods 2013, 9, 24. [Google Scholar] [CrossRef]


| Kanamycin Concentration (mg/L) | Shoot Regeneration % (Mean ± SD) |
|---|---|
| 0 | 20.0 ± 0.0 a |
| 15 | 18.7 ± 0.6 a |
| 25 | 14.0 ± 2.0 b |
| 35 | 11.3 ± 1.2 b |
| 50 | 4.7 ± 1.5 c |
| 75 | 0.7 ± 0.6 d |
| 100 | 0.0 ± 0.0 d |
| Lines | C18:1 | C18:2 | C18:3 | C22:1 |
|---|---|---|---|---|
| EA1 | 15.9 ± 2.2 ab | 5.7 ± 0.7 b | 8.2 ± 1.1 b | 52.7 ± 2.1 a |
| EA2 | 17.6 ± 0.8 ab | 5.9 ± 1.3 b | 8.4 ± 0.8 b | 50.8 ± 2.5 a |
| EA3 | 18.5 ± 3.2 ab | 6.9 ± 2.1 ab | 8.1 ± 1.6 b | 48.5 ± 3.3 ab |
| EA4 | 21.8 ± 1.8 a | 6.6 ± 1.6 ab | 9.4 ± 0.7 b | 44.5 ± 0.6 bc |
| EA5 | 19.5 ± 6.2 ab | 7.9 ± 3.3 ab | 10.1 ± 3.0 b | 43.4 ±2.0 bc |
| EA6 | 22.8 ± 4.0 a | 5.59 ± 1.4 b | 9.8 ± 1.6 b | 43.1 ± 1.0 bc |
| WT | 10.7 ± 0.5 b | 11.3 ± 0.2 a | 15.1 ± 1.0 a | 40.9 ± 0.9 c |
| Transgenic Line | Single Seed WE (%) | Pooled Seed WE (%) |
|---|---|---|
| WE1 | 8.1 ± 2.3 b | 12.08 ± 0.85 c |
| WE2 | 14.8 ± 2.5 b | 21.40 ± 1.1 b |
| WE3 | 13.8 ± 5.3 b | 20.34 ± 0.8 b |
| WE10 | 10.9 ± 4.0 b | 12.94 ± 1.2 c |
| WE12 | 24.3 ± 4.7 a | 25.56 ± 0.5 a |
| Line | TSW (g) | No. of Pods per Plant | No. of Seeds per Pod |
|---|---|---|---|
| EA-1 | 4.52 | 272 | 7.1 ± 1.1 d |
| EA-2 | 4.32 | 274 | 6.1 ± 1.0 e |
| EA-3 | 4.42 | 276 | 5.2 ± 1.1 f |
| EA-4 | 4.12 | 238 | 9.0 ± 0.6 b |
| EA-5 | 4.24 | 230 | 8.9 ± 1.1 b |
| EA-6 | 4.38 | 236 | 9.9 ± 1.3 a |
| WT | 5.01 | 259 | 8.0 ± 0.9 c |
| WE-1 | 5.6 | 245 | 7.1 ± 1.1 ef |
| WE-2 | 5.4 | 238 | 8.1 ± 0.9 bcd |
| WE-3 | 5.6 | 240 | 9.0 ±1.3 ab |
| WE-4 | 5.4 | 210 | 8.6 ± 1.0 bc |
| WE-5 | 5.4 | 185 | 6.0 ± 1.0 gh |
| WE-6 | 5.7 | 180 | 8.0 ± 1.0 cde |
| WE-7 | 5.4 | 265 | 5.9 ± 0.9 gh |
| WE-8 | 5.4 | 270 | 6.1 ± 1.0 fg |
| WE-9 | 5.6 | 261 | 5.17 ± 1.09 h |
| WE-10 | 5.3 | 280 | 5.9 ± 1.0 gh |
| WE-11 | 5.3 | 295 | 8.9 ± 1.1 bc |
| WE-12 | 5.4 | 310 | 9.9 ± 1.30 a |
| WE-13 | 5.6 | 258 | 8.1 ± 1.4 bcd |
| WE-14 | 5.4 | 268 | 8.1 ± 0.9 bcd |
| WE-15 | 5.5 | 260 | 9.0 ± 1.2 b |
| WT | 5.1 | 256 | 7.6 ± 1.0 de |
| Gene | Forward Primers (5′-3′) | Reverse Primers (5′-3′) |
|---|---|---|
| BnFAE1 | AATGCGTTGGTGGAAGGTAG | TGTGTAGCTCATGTCCTGGC |
| LdPLAAT | GTGGTTTTTGAGACGCAGGT | TAGACTGAATAGCCGCAGCC |
| ScFAR | AGGCATTAGGGGAGATGCTT | CCTTGAACCATTGGCAGAAT |
| ScWS | CTCTTCGCCTTTCATCTTGG | AACACAAAGAACCCCGTCAC |
| Gene | Forward Primers (5′-3′) | Reverse Primers (5′-3′) |
|---|---|---|
| BnFAE1 | CCTCCCCGGAAGACTTTTG | CATGCTTGAGTTCACCACAAG |
| BcFAD2 | CCGTGAACGTCTCCAGATAT | CGTTGACTATCAGAAGCGGA |
| LdPLAAT | AAGTAAACGCCCATCTCTCG | GGCTGCGGCTATTCAGTCTA |
| ScFAR | CTCCTCCTTCTCCACCTTCC | CCTTGAACCATTGGCAGAAT |
| ScWS | CTCTTCGCCTTTCATCTTGG | CTCGATGTGTTCCCAAACCT |
| AtUBC21 | TGCGACTCAGGGAATCTTCT | TCATCCTTTCTTAGGCATAGCG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tesfaye, M.; Wang, E.S.; Feyissa, T.; Herrfurth, C.; Haileselassie, T.; Kanagarajan, S.; Feussner, I.; Zhu, L.-H. Enhancing Erucic Acid and Wax Ester Production in Brassica carinata through Metabolic Engineering for Industrial Applications. Int. J. Mol. Sci. 2024, 25, 6322. https://doi.org/10.3390/ijms25126322
Tesfaye M, Wang ES, Feyissa T, Herrfurth C, Haileselassie T, Kanagarajan S, Feussner I, Zhu L-H. Enhancing Erucic Acid and Wax Ester Production in Brassica carinata through Metabolic Engineering for Industrial Applications. International Journal of Molecular Sciences. 2024; 25(12):6322. https://doi.org/10.3390/ijms25126322
Chicago/Turabian StyleTesfaye, Misteru, Eu Sheng Wang, Tileye Feyissa, Cornelia Herrfurth, Teklehaimanot Haileselassie, Selvaraju Kanagarajan, Ivo Feussner, and Li-Hua Zhu. 2024. "Enhancing Erucic Acid and Wax Ester Production in Brassica carinata through Metabolic Engineering for Industrial Applications" International Journal of Molecular Sciences 25, no. 12: 6322. https://doi.org/10.3390/ijms25126322
APA StyleTesfaye, M., Wang, E. S., Feyissa, T., Herrfurth, C., Haileselassie, T., Kanagarajan, S., Feussner, I., & Zhu, L.-H. (2024). Enhancing Erucic Acid and Wax Ester Production in Brassica carinata through Metabolic Engineering for Industrial Applications. International Journal of Molecular Sciences, 25(12), 6322. https://doi.org/10.3390/ijms25126322






