Methionine Sulfoxide Speciation in Mouse Hippocampus Revealed by Global Proteomics Exhibits Age- and Alzheimer’s Disease-Dependent Changes Targeted to Mitochondrial and Glycolytic Pathways
Abstract
1. Introduction
- Examine the early aging-, sex-, and disease-dependent changes in MSox levels;
- Identify peptide and protein targets specifically sensitive and resistant to the oxidation of methionine;
- Validate key changes with peptide-based absolute quantification; and
- Explore the stability of key MSox sites to handling and storage.
2. Results
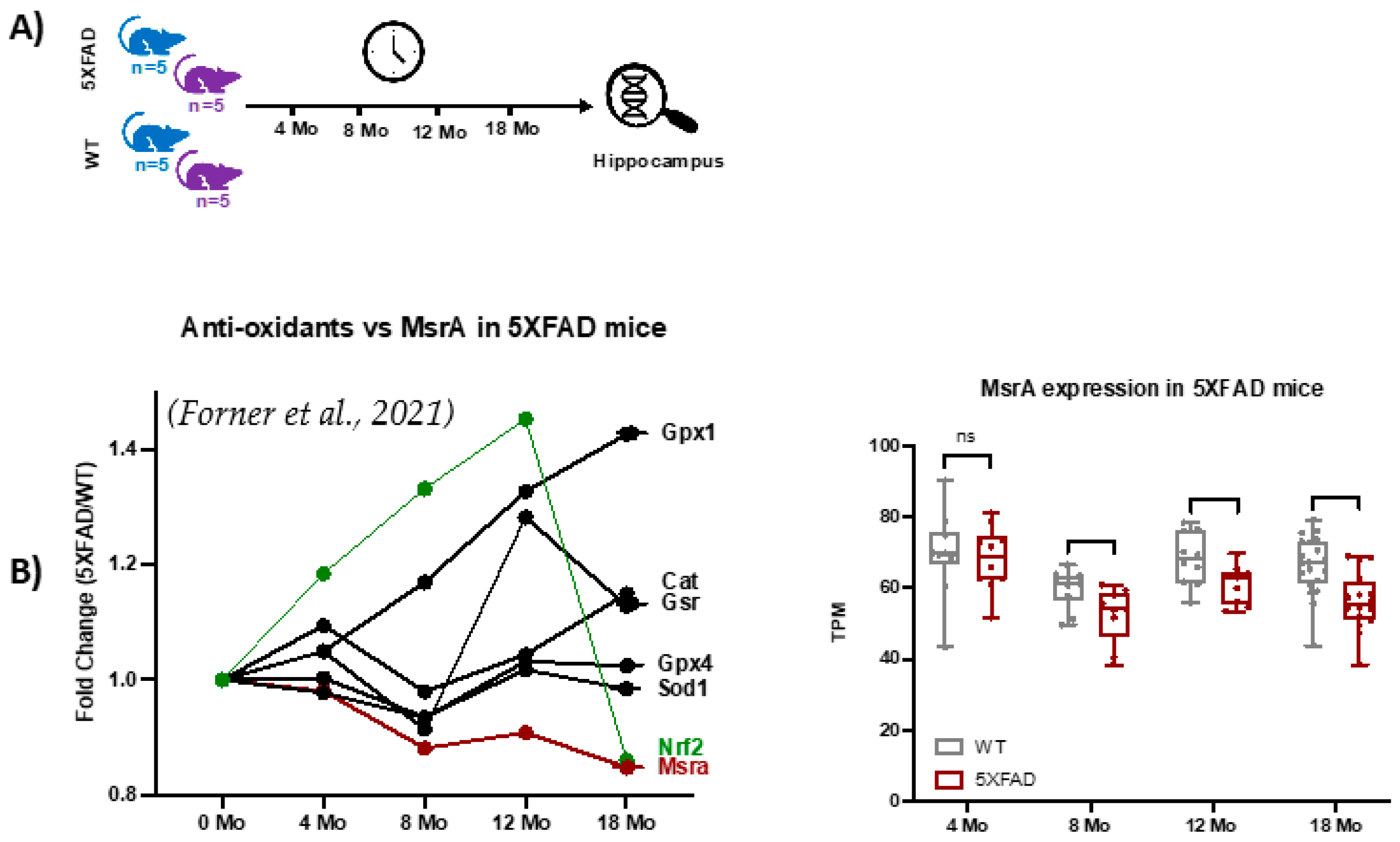
2.1. Experimental Design
2.2. Age-Dependent Increase in Global MSox
2.3. Pathway Analysis Reveals Distinct Age-Dependent Signatures
2.4. High-Abundance Proteins Are Susceptible to MSox Accumulation
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Label-Free Quantitative Proteomic Analysis
5.2. PRM
5.2.1. Sample Preparation for PRM Assay
5.2.2. Development and Analytical Validation of Targeted MS Assays/Measurements
5.3. Enrichment Pathway Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Levine, R.L.; Moskovitz, J.; Stadtman, E.R. Oxidation of methionine in proteins: Roles in antioxidant defense and cellular regulation. IUBMB Life 2000, 50, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Stadtman, E.R.; Van Remmen, H.; Richardson, A.; Wehr, N.B.; Levine, R.L. Methionine oxidation and aging. Biochim. Biophys. Acta 2005, 1703, 135–140. [Google Scholar] [CrossRef] [PubMed]
- Schoneich, C. Redox processes of methionine relevant to beta-amyloid oxidation and Alzheimer’s disease. Arch. Biochem. Biophys. 2002, 397, 370–376. [Google Scholar] [CrossRef] [PubMed]
- Shringarpure, R.; Davies, K.J. Protein turnover by the proteasome in aging and disease. Free Radic. Biol. Med. 2002, 32, 1084–1089. [Google Scholar] [CrossRef] [PubMed]
- Auten, R.L.; Davis, J.M. Oxygen toxicity and reactive oxygen species: The devil is in the details. Pediatr. Res. 2009, 66, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Chance, M.R. Hydroxyl radical-mediated modification of proteins as probes for structural proteomics. Chem. Rev. 2007, 107, 3514–3543. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Guo, C.; Kong, J. Oxidative stress in neurodegenerative diseases. Neural Regen. Res. 2012, 7, 376–385. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Barnham, K.J.; Masters, C.L.; Bush, A.I. Neurodegenerative diseases and oxidative stress. Nat. Rev. Drug Discov. 2004, 3, 205–214. [Google Scholar] [CrossRef] [PubMed]
- Sies, H.; Jones, D.P. Reactive oxygen species (ROS) as pleiotropic physiological signalling agents. Nat. Rev. Mol. Cell. Biol. 2020, 21, 363–383. [Google Scholar] [CrossRef] [PubMed]
- Taungjaruwinai, W.M.; Bhawan, J.; Keady, M.; Thiele, J.J. Differential expression of the antioxidant repair enzyme methionine sulfoxide reductase (MSRA and MSRB) in human skin. Am. J. Dermatopathol. 2009, 31, 427–431. [Google Scholar] [CrossRef] [PubMed]
- Soriani, F.M.; Kress, M.R.; Fagundes de Gouvea, P.; Malavazi, I.; Savoldi, M.; Gallmetzer, A.; Strauss, J.; Goldman, M.H.; Goldman, G.H. Functional characterization of the Aspergillus nidulans methionine sulfoxide reductases (msrA and msrB). Fungal Genet. Biol. 2009, 46, 410–417. [Google Scholar] [CrossRef] [PubMed]
- Rocco-Machado, N.; Lai, L.; Kim, G.; He, Y.; Luczak, E.D.; Anderson, M.E.; Levine, R.L. Oxidative stress-induced autonomous activation of the calcium/calmodulin-dependent kinase II involves disulfide formation in the regulatory domain. J. Biol. Chem. 2022, 298, 102579. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Luczak, E.D.; Anderson, M.E. CaMKII oxidative activation and the pathogenesis of cardiac disease. J. Mol. Cell. Cardiol. 2014, 73, 112–116. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Fremont, S.; Romet-Lemonne, G.; Houdusse, A.; Echard, A. Emerging roles of MICAL family proteins—From actin oxidation to membrane trafficking during cytokinesis. J. Cell Sci. 2017, 130, 1509–1517. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.; Terman, J.R. MICAL redox enzymes and actin remodeling: New links to classical tumorigenic and cancer pathways. Mol. Cell. Oncol. 2018, 5, e1384881. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Andrieu, C.; Vergnes, A.; Loiseau, L.; Aussel, L.; Ezraty, B. Characterisation of the periplasmic methionine sulfoxide reductase (MsrP) from Salmonella Typhimurium. Free Radic. Biol. Med. 2020, 160, 506–512. [Google Scholar] [CrossRef] [PubMed]
- Arias, D.G.; Cabeza, M.S.; Echarren, M.L.; Faral-Tello, P.; Iglesias, A.A.; Robello, C.; Guerrero, S.A. On the functionality of a methionine sulfoxide reductase B from Trypanosoma cruzi. Free Radic. Biol. Med. 2020, 158, 96–114. [Google Scholar] [CrossRef] [PubMed]
- Cha, H.N.; Woo, C.H.; Kim, H.Y.; Park, S.Y. Methionine sulfoxide reductase B3 deficiency inhibits the development of diet-induced insulin resistance in mice. Redox Biol. 2021, 38, 101823. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Das, K.; Garnica, O.; Flores, J.; Dhandayuthapani, S. Methionine sulfoxide reductase A (MsrA) modulates cells and protects against Mycoplasma genitalium induced cytotoxicity. Free Radic. Biol. Med. 2020, 152, 323–335. [Google Scholar] [CrossRef] [PubMed]
- Javitt, G.; Cao, Z.; Resnick, E.; Gabizon, R.; Bulleid, N.J.; Fass, D. Structure and Electron-Transfer Pathway of the Human Methionine Sulfoxide Reductase MsrB3. Antioxid. Redox Signal. 2020, 33, 665–678. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jiang, B.; Adams, Z.; Moonah, S.; Shi, H.; Maupin-Furlow, J.; Moskovitz, J. The Antioxidant Enzyme Methionine Sulfoxide Reductase A (MsrA) Interacts with Jab1/CSN5 and Regulates Its Function. Antioxidants 2020, 9, 452. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Li, H.; Liang, M.; Wang, Z.; Zhang, Y.; Wu, Q.; Yang, L. Rice Protein Exerts Endogenous Antioxidant Capacity via Methionine Sulfoxide Reductase and the Nrf2 Antioxidant System Independent of Age. J. Med. Food 2020, 23, 565–574. [Google Scholar] [CrossRef] [PubMed]
- Nasreen, M.; Dhouib, R.; Hosmer, J.; Wijesinghe, H.G.S.; Fletcher, A.; Mahawar, M.; Essilfie, A.T.; Blackall, P.J.; McEwan, A.G.; Kappler, U. Peptide Methionine Sulfoxide Reductase from Haemophilus influenzae Is Required for Protection against HOCl and Affects the Host Response to Infection. ACS Infect Dis. 2020, 6, 1928–1939. [Google Scholar] [CrossRef] [PubMed]
- Salmon, A.B.; Kim, G.; Liu, C.; Wren, J.D.; Georgescu, C.; Richardson, A.; Levine, R.L. Effects of transgenic methionine sulfoxide reductase A (MsrA) expression on lifespan and age-dependent changes in metabolic function in mice. Redox Biol. 2016, 10, 251–256. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chevion, M.; Berenshtein, E.; Stadtman, E.R. Human studies related to protein oxidation: Protein carbonyl content as a marker of damage. Free Radic Res. 2000, 33, S99–S108. [Google Scholar] [PubMed]
- Lee, B.C.; Lee, Y.K.; Lee, H.J.; Stadtman, E.R.; Lee, K.H.; Chung, N. Cloning and characterization of antioxidant enzyme methionine sulfoxide-S-reductase from Caenorhabditis elegans. Arch. Biochem. Biophys. 2005, 434, 275–281. [Google Scholar] [CrossRef] [PubMed]
- Levine, R.L.; Wehr, N.; Williams, J.A.; Stadtman, E.R.; Shacter, E. Determination of carbonyl groups in oxidized proteins. Methods Mol Biol. 2000, 99, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Moskovitz, J.; Poston, J.M.; Berlett, B.S.; Nosworthy, N.J.; Szczepanowski, R.; Stadtman, E.R. Identification and characterization of a putative active site for peptide methionine sulfoxide reductase (MsrA) and its substrate stereospecificity. J. Biol. Chem. 2000, 275, 14167–14172. [Google Scholar] [CrossRef] [PubMed]
- Stadtman, E.R.; Levine, R.L. Protein oxidation. Ann. N. Y. Acad. Sci. 2000, 899, 191–208. [Google Scholar] [CrossRef] [PubMed]
- Gella, A.; Durany, N. Oxidative stress in Alzheimer disease. Cell. Adh. Migr. 2009, 3, 88–93. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Varadarajan, S.; Yatin, S.; Kanski, J.; Jahanshahi, F.; Butterfield, D.A. Methionine residue 35 is important in amyloid beta-peptide-associated free radical oxidative stress. Brain Res. Bull. 1999, 50, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Schoneich, C. Methionine oxidation by reactive oxygen species: Reaction mechanisms and relevance to Alzheimer’s disease. Biochim. Biophys. Acta 2005, 1703, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Gabbita, S.P.; Aksenov, M.Y.; Lovell, M.A.; Markesbery, W.R. Decrease in peptide methionine sulfoxide reductase in Alzheimer’s disease brain. J. Neurochem. 1999, 73, 1660–1666. [Google Scholar] [CrossRef] [PubMed]
- Xiang, X.J.; Song, L.; Deng, X.J.; Tang, Y.; Min, Z.; Luo, B.; Wen, Q.X.; Li, K.Y.; Chen, J.; Ma, Y.L.; et al. Mitochondrial methionine sulfoxide reductase B2 links oxidative stress to Alzheimer’s disease-like pathology. Exp. Neurol. 2019, 318, 145–156. [Google Scholar] [CrossRef] [PubMed]
- Oblak, A.L.; Lin, P.B.; Kotredes, K.P.; Pandey, R.S.; Garceau, D.; Williams, H.M.; Uyar, A.; O‘Rourke, R.; O‘Rourke, S.; Ingraham, C.; et al. Comprehensive Evaluation of the 5XFAD Mouse Model for Preclinical Testing Applications: A MODEL-AD Study. Front. Aging Neurosci. 2021, 13, 713726. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Gurel, B.; Cansev, M.; Koc, C.; Ocalan, B.; Cakir, A.; Aydin, S.; Kahveci, N.; Ulus, I.H.; Sahin, B.; Basar, M.K.; et al. Proteomics Analysis of CA1 Region of the Hippocampus in Pre-, Progression and Pathological Stages in a Mouse Model of the Alzheimer’s Disease. Curr. Alzheimer Res. 2019, 16, 613–621. [Google Scholar] [CrossRef] [PubMed]
- Lopes, F.B.T.P.; Schlatzer, D.; Wang, R.; Li, X.; Feng, E.; Koyuturk, M.; Qi, X.; Chance, M.R. Temporal and sex-linked protein expression dynamics in a familial model of Alzheimer’s Disease. Mol. Cell. Proteom. 2022, 21, 100280. [Google Scholar] [CrossRef] [PubMed]
- Jackson, S.J.; Andrews, N.; Ball, D.; Bellantuono, I.; Gray, J.; Hachoumi, L.; Holmes, A.; Latcham, J.; Petrie, A.; Potter, P.; et al. Does age matter? The impact of rodent age on study outcomes. Lab. Anim. 2017, 51, 160–169. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Forner, S.; Kawauchi, S.; Balderrama-Gutierrez, G.; Kramár, E.A.; Matheos, D.P.; Phan, J.; Javonillo, D.I.; Tran, K.M.; Hingco, E.; Da Cunha, C.; et al. Systematic phenotyping and characterization of the 5xFAD mouse model of Alzheimer’s disease. Sci. Data 2021, 8, 270. [Google Scholar] [CrossRef] [PubMed]
- Serrano-Pozo, A.; Das, S.; Hyman, B.T. APOE and Alzheimer’s disease: Advances in genetics, pathophysiology, and therapeutic approaches. Lancet Neurol. 2021, 20, 68–80, Erratum in Lancet Neurol. 2021, 20, e2. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tran, N.H.; Qiao, R.; Xin, L.; Chen, X.; Liu, C.; Zhang, X.; Shan, B.; Ghodsi, A.; Li, M. Deep learning enables de novo peptide sequencing from data-independent-acquisition mass spectrometry. Nat. Methods 2019, 16, 63–66. [Google Scholar] [CrossRef] [PubMed]
- Tran, N.H.; Zhang, X.; Xin, L.; Shan, B.; Li, M. De novo peptide sequencing by deep learning. Proc. Natl. Acad. Sci. USA 2017, 114, 8247–8252. [Google Scholar] [CrossRef] [PubMed]
- Deutsch, E.W.; Bandeira, N.; Sharma, V.; Perez-Riverol, Y.; Carver, J.J.; Kundu, D.J.; García-Seisdedos, D.; Jarnuczak, A.F.; Hewapathirana, S.; Pullman, B.S.; et al. The ProteomeXchange consortium in 2020: Enabling ‘big data’ approaches in proteomics. Nucleic Acids Res. 2020, 48, D1145–D1152. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Thomas, P.D.; Campbell, M.J.; Kejariwal, A.; Mi, H.; Karlak, B.; Daverman, R.; Diemer, K.; Muruganujan, A.; Narechania, A. PANTHER: A library of protein families and subfamilies indexed by function. Genome Res. 2003, 13, 2129–2141. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
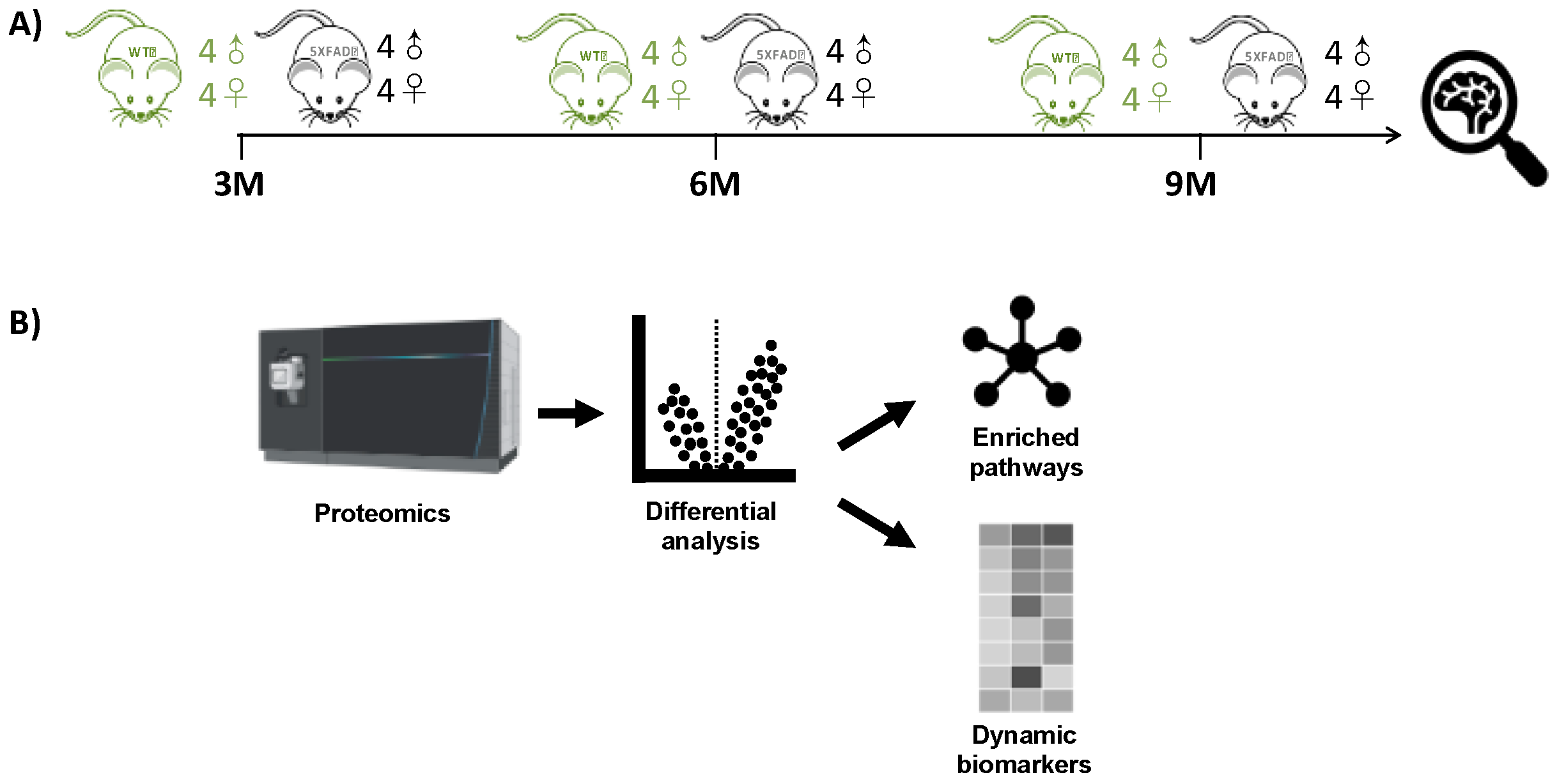
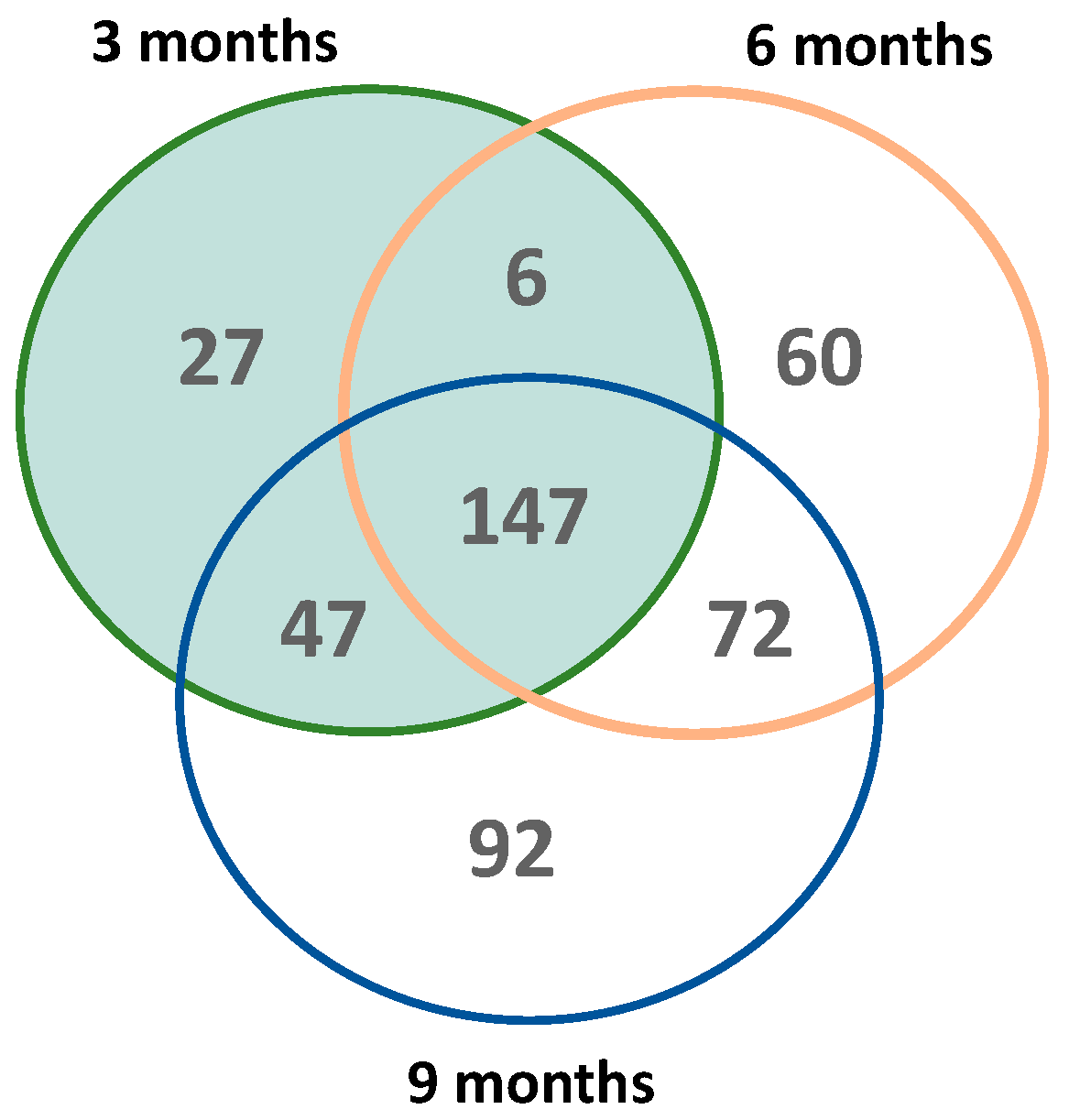
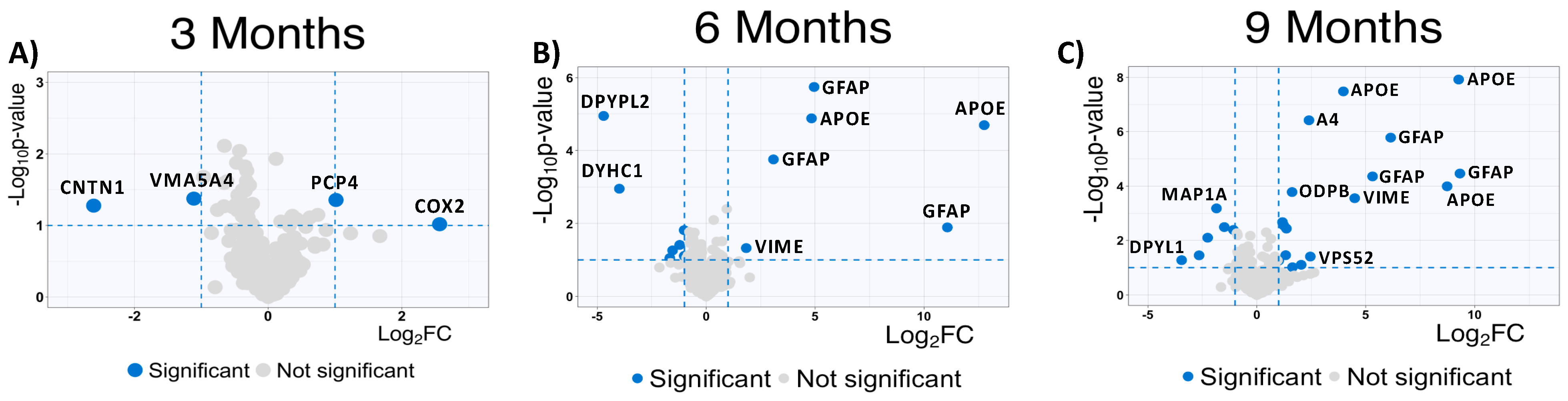
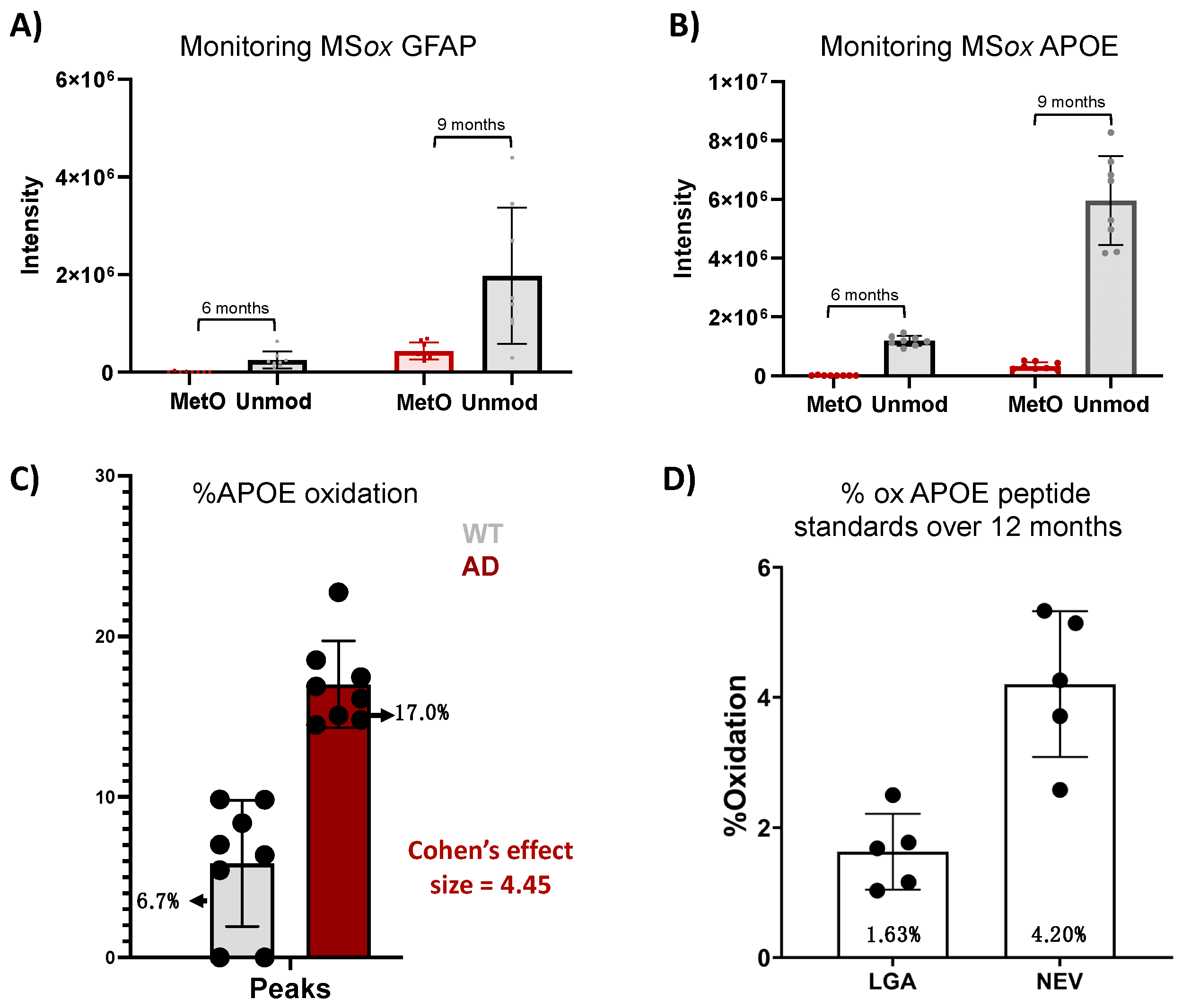
| (A) | 3 Months | 6 Months | 9 Months | |||
| WT | 5XFAD | WT | 5XFAD | WT | 5XFAD | |
| MSox peptides | 396 | 396 | 603 | 606 | 819 | 822 |
| MSox proteins | 228 | 228 | 282 | 285 | 358 | 358 |
| MSox % | 3.59 | 3.59 | 4.20 | 4.22 | 4.48 | 4.48 |
| (B) | 3 Months | 5XFAD Mice 6 Months | 9 Months | |||
| Female | Male | Female | Male | Female | Male | |
| MSox peptides | 396 | 395 | 597 | 602 | 821 | 820 |
| MSox proteins | 228 | 228 | 283 | 284 | 358 | 357 |
| MSox % | 3.59 | 3.58 | 4.16 | 4.19 | 4.48 | 4.47 |
| MSox | Proteome | ||||||
|---|---|---|---|---|---|---|---|
| Enriched Terms | 3 M | 6 M | 9 M | 3 M | 6 M | 9 M | |
| Reactome Pathways | Glucose metabolism | 1.76 × 10−2 | 7.05 × 10−5 | 5.92 × 10−5 | 1.00 × 100 | 1.00 × 100 | 5.56 × 10−1 |
| Immune system | 1.05 × 10−2 | 3.90 × 10−3 | 9.34 × 10−5 | 4.28 × 10−1 | 1.76 × 10−5 | 1.31 × 10−11 | |
| Gluconeogenesis | 1.31 × 10−2 | 3.32 × 10−4 | 1.22 × 10−4 | 6.58 × 10−1 | 1.00 × 100 | 1.00 × 100 | |
| Organelle biogen and maintenance | 1.17 × 10−2 | 4.46 × 10−3 | 4.07 × 10−4 | 1.00 × 100 | 2.99 × 10−1 | 7.69 × 10−2 | |
| Transport of small molecules | ns | 6.46 × 10−4 | 4.29 × 10−4 | 4.53 × 10−1 | 7.23 × 10−1 | 1.33 × 10−1 | |
| Transmission chemical synapses | 9.80 × 10−3 | 5.43 × 10−4 | 6.07 × 10−4 | 1.00 × 100 | 7.11 × 10−1 | 2.47 × 10−2 | |
| Innate immune system | ns | 1.49 × 10−2 | 8.32 × 10−4 | 4.47 × 10−1 | 9.58 × 10−6 | 1.25 × 10−11 | |
| Signal transduction | 3.65 × 10−2 | 2.43 × 10−2 | 8.37 × 10−4 | 3.64 × 10−1 | 4.04 × 10−1 | 2.89 × 10−4 | |
| L1CAM interactions | 6.47 × 10−3 | 6.29 × 10−4 | 5.59 × 10−3 | 4.64 × 10−2 | 1.70 × 10−1 | 1.14 × 10−4 | |
| Recycling pathway of L1 | 1.05 × 10−2 | 6.32 × 10−4 | 1.27 × 10−3 | 2.44 × 10−1 | 4.15 × 10−1 | 1.15 × 10−3 | |
| Glycolysis | 1.18 × 10−2 | 7.92 × 10−4 | 2.44 × 10−3 | 1.00 × 100 | 1.00 × 100 | 8.12 × 10−1 | |
| Neurotransmitter release cycle | 1.03 × 10−2 | 8.38 × 10−4 | 2.66 × 10−3 | 1.00 × 100 | ns | 9.68 × 10−2 | |
| Cellular Component | Cytoplasm | ns | ns | ns | 1.41 × 10−215 | 1.49 × 10−243 | 9.63 × 10−263 |
| Intracellular anatomical structure | ns | ns | ns | 3.51 × 10−152 | 1.19 × 10−166 | 6.39 × 10−186 | |
| Cytosol | ns | ns | ns | 8.87 × 10−98 | 4.40 × 10−103 | 4.91 × 10−107 | |
| Organelle | 7.42 × 10−3 | ns | 5.02 × 10−3 | 3.12 × 10−113 | 3.30 × 10−124 | 9.44 × 10−128 | |
| Postsynapse | 3.53 × 10−2 | 5.53 × 10−3 | 6.98 × 10−3 | 2.48 × 10−112 | 5.86 × 10−117 | 4.81 × 10−112 | |
| Axon | 3.05 × 10−9 | 2.80 × 10−7 | 4.35 × 10−6 | 5.43 × 10−58 | 2.93 × 10−64 | 2.52 × 10−62 | |
| Myelin sheath | 2.94 × 10−29 | 1.76 × 10−30 | 5.49 × 10−30 | 7.26 × 10−79 | 2.27 × 10−72 | 1.72 × 10−68 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lopes, F.B.T.P.; Schlatzer, D.; Li, M.; Yilmaz, S.; Wang, R.; Qi, X.; Ayati, M.; Koyutürk, M.; Chance, M.R. Methionine Sulfoxide Speciation in Mouse Hippocampus Revealed by Global Proteomics Exhibits Age- and Alzheimer’s Disease-Dependent Changes Targeted to Mitochondrial and Glycolytic Pathways. Int. J. Mol. Sci. 2024, 25, 6516. https://doi.org/10.3390/ijms25126516
Lopes FBTP, Schlatzer D, Li M, Yilmaz S, Wang R, Qi X, Ayati M, Koyutürk M, Chance MR. Methionine Sulfoxide Speciation in Mouse Hippocampus Revealed by Global Proteomics Exhibits Age- and Alzheimer’s Disease-Dependent Changes Targeted to Mitochondrial and Glycolytic Pathways. International Journal of Molecular Sciences. 2024; 25(12):6516. https://doi.org/10.3390/ijms25126516
Chicago/Turabian StyleLopes, Filipa Blasco Tavares Pereira, Daniela Schlatzer, Mengzhen Li, Serhan Yilmaz, Rihua Wang, Xin Qi, Marzieh Ayati, Mehmet Koyutürk, and Mark R. Chance. 2024. "Methionine Sulfoxide Speciation in Mouse Hippocampus Revealed by Global Proteomics Exhibits Age- and Alzheimer’s Disease-Dependent Changes Targeted to Mitochondrial and Glycolytic Pathways" International Journal of Molecular Sciences 25, no. 12: 6516. https://doi.org/10.3390/ijms25126516
APA StyleLopes, F. B. T. P., Schlatzer, D., Li, M., Yilmaz, S., Wang, R., Qi, X., Ayati, M., Koyutürk, M., & Chance, M. R. (2024). Methionine Sulfoxide Speciation in Mouse Hippocampus Revealed by Global Proteomics Exhibits Age- and Alzheimer’s Disease-Dependent Changes Targeted to Mitochondrial and Glycolytic Pathways. International Journal of Molecular Sciences, 25(12), 6516. https://doi.org/10.3390/ijms25126516







