Methylation-Based Characterization of a New IDH2 Mutation in Sinonasal Undifferentiated Carcinoma
Abstract
1. Introduction
2. Results and Discussion
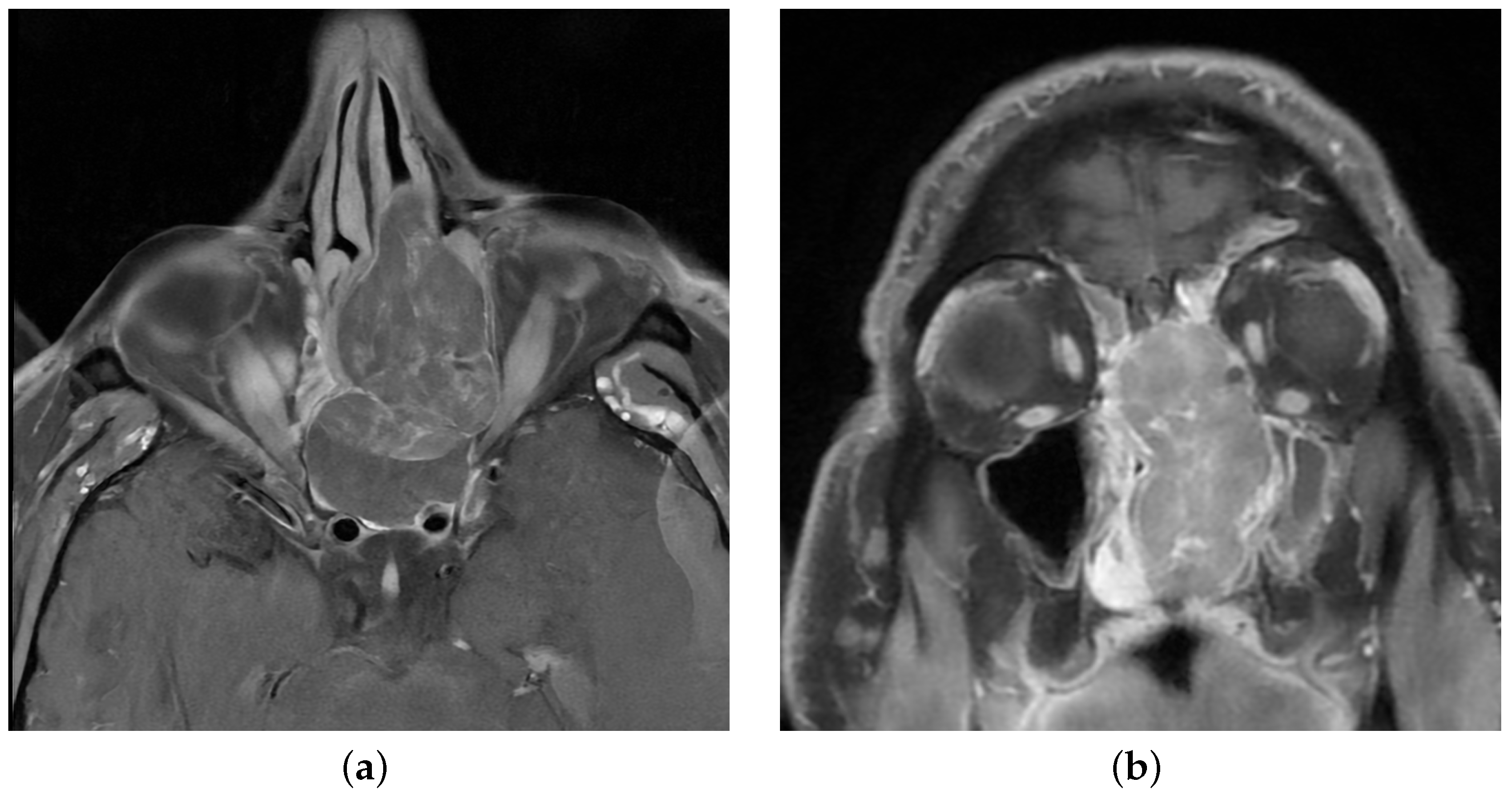
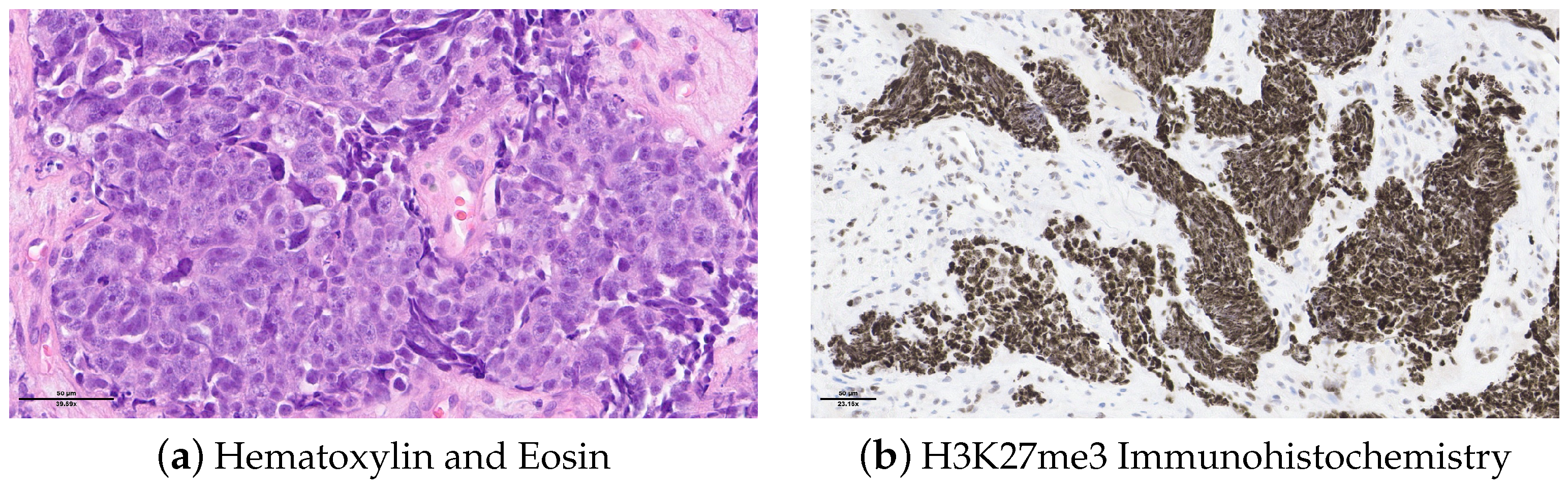
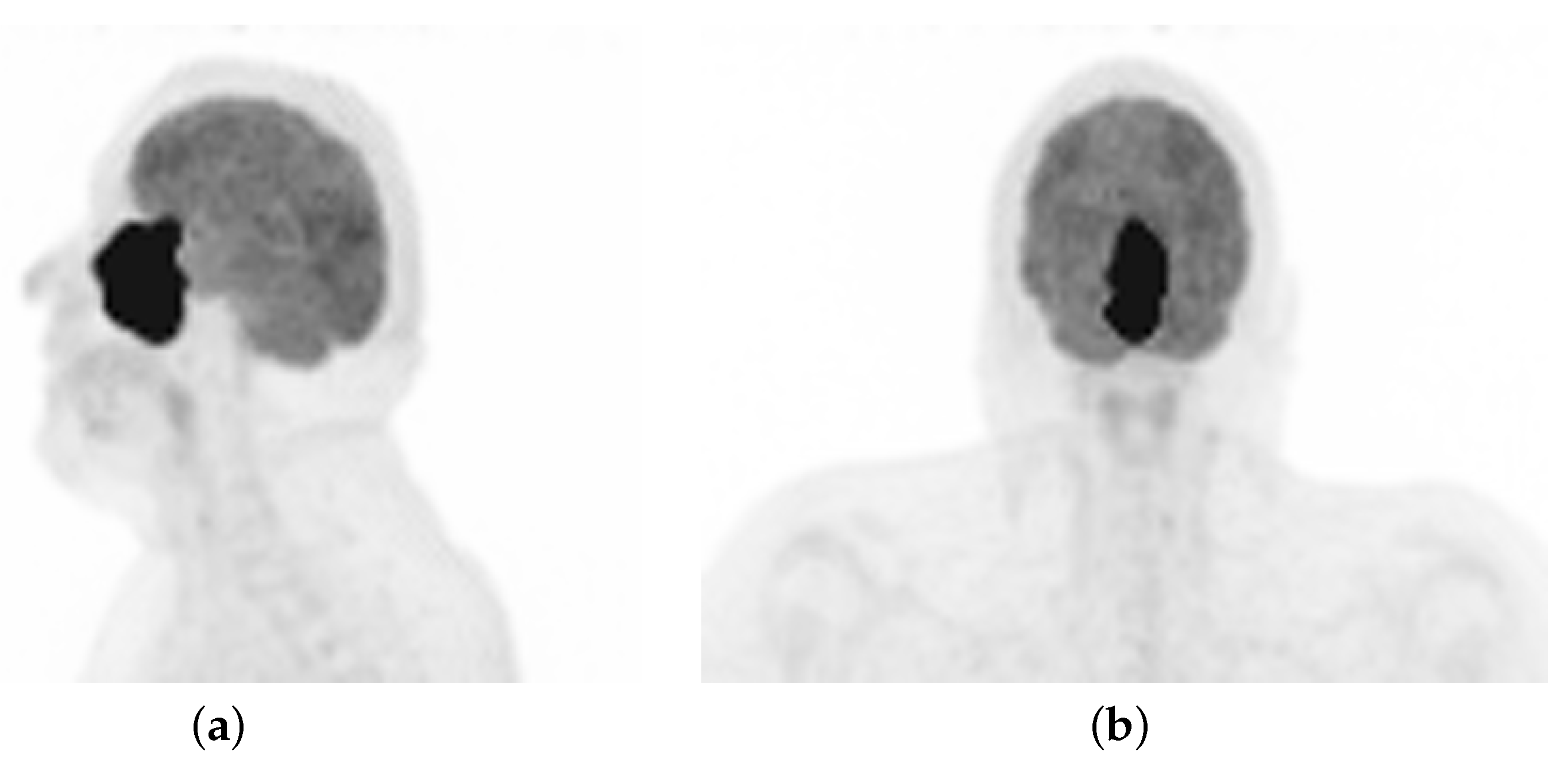
2.1. Case Description
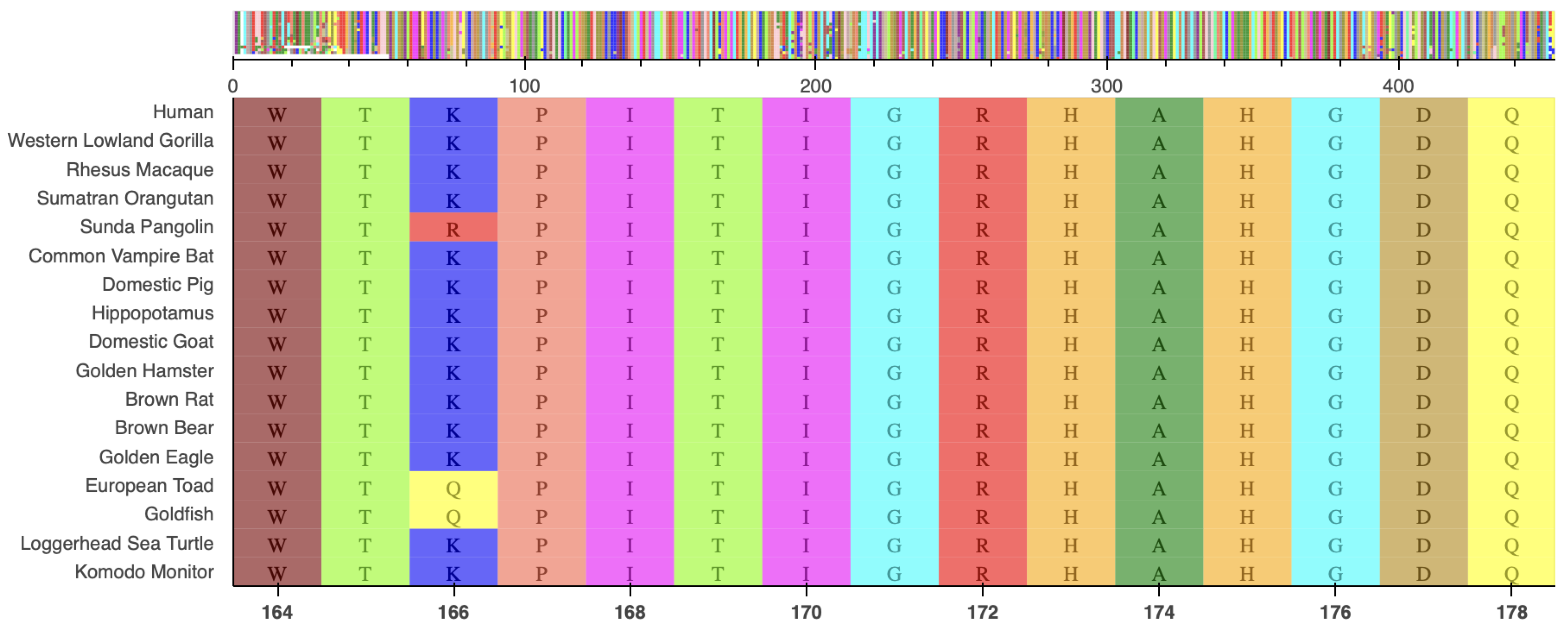
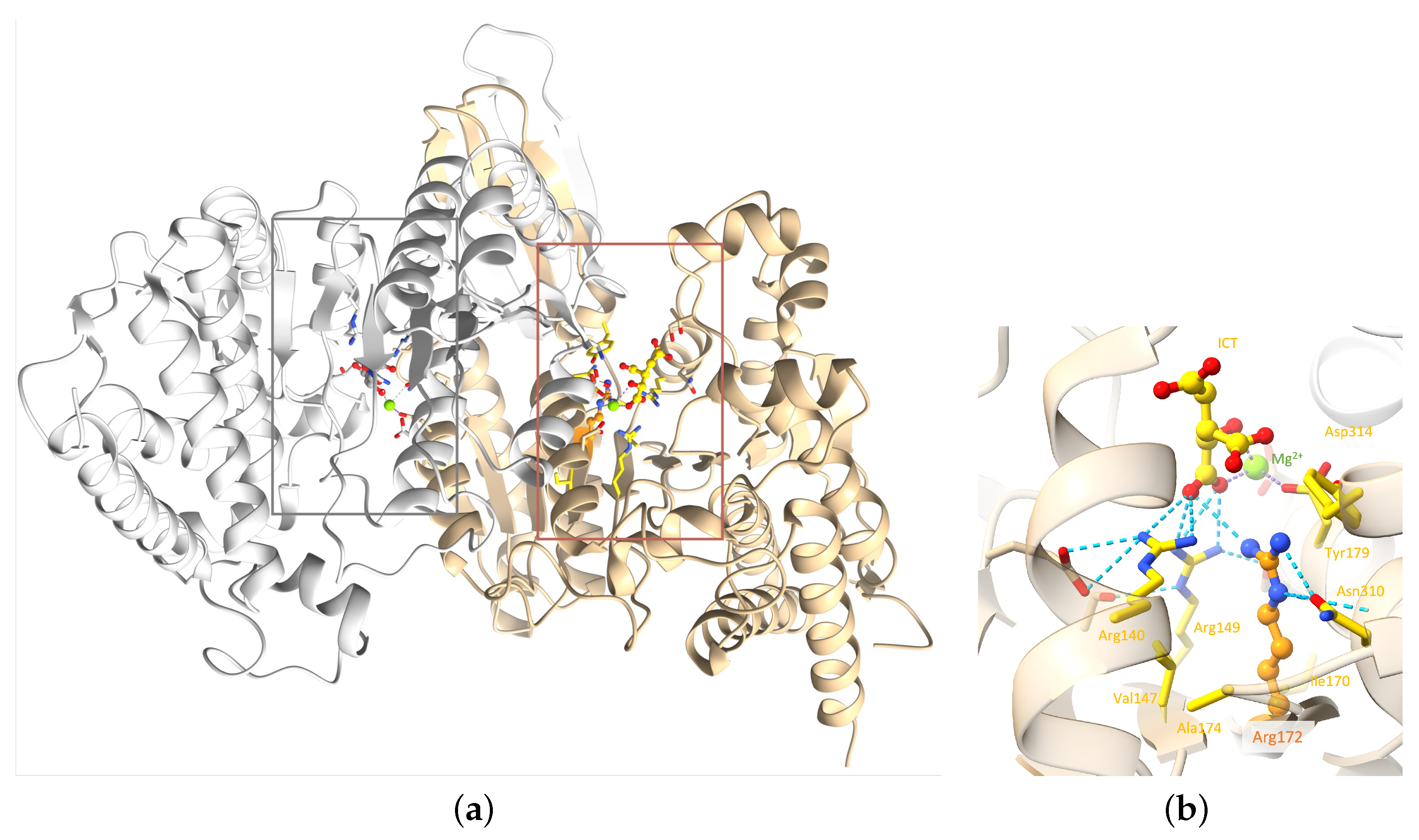
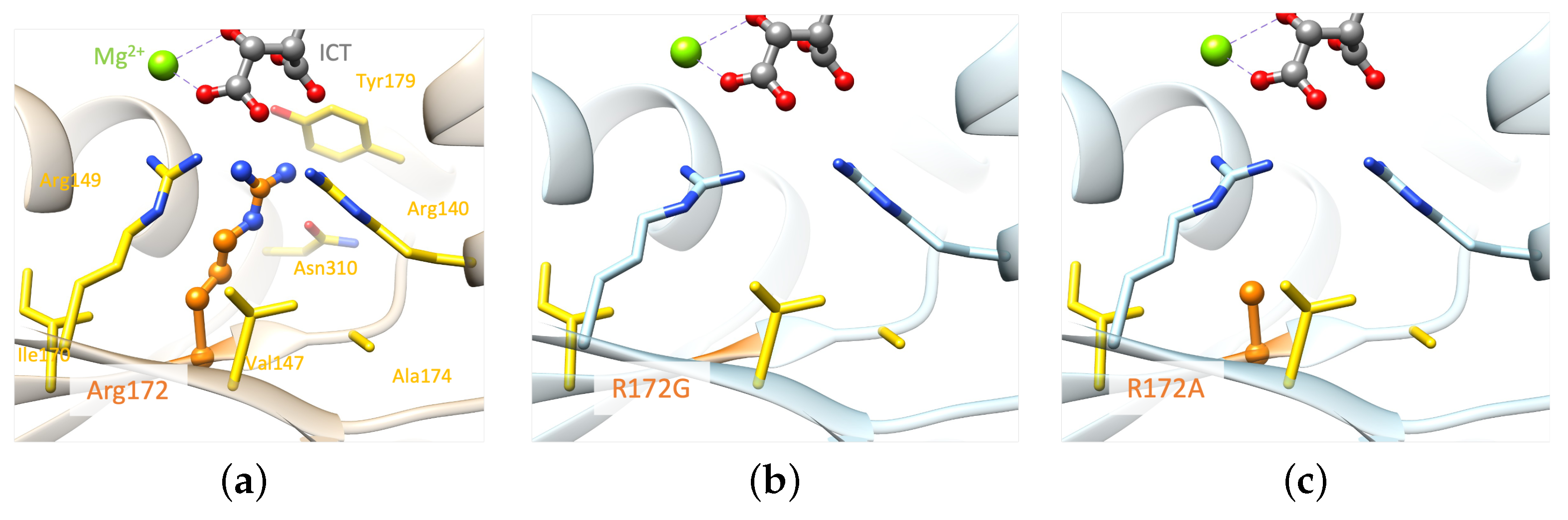
2.2. Molecular Analysis
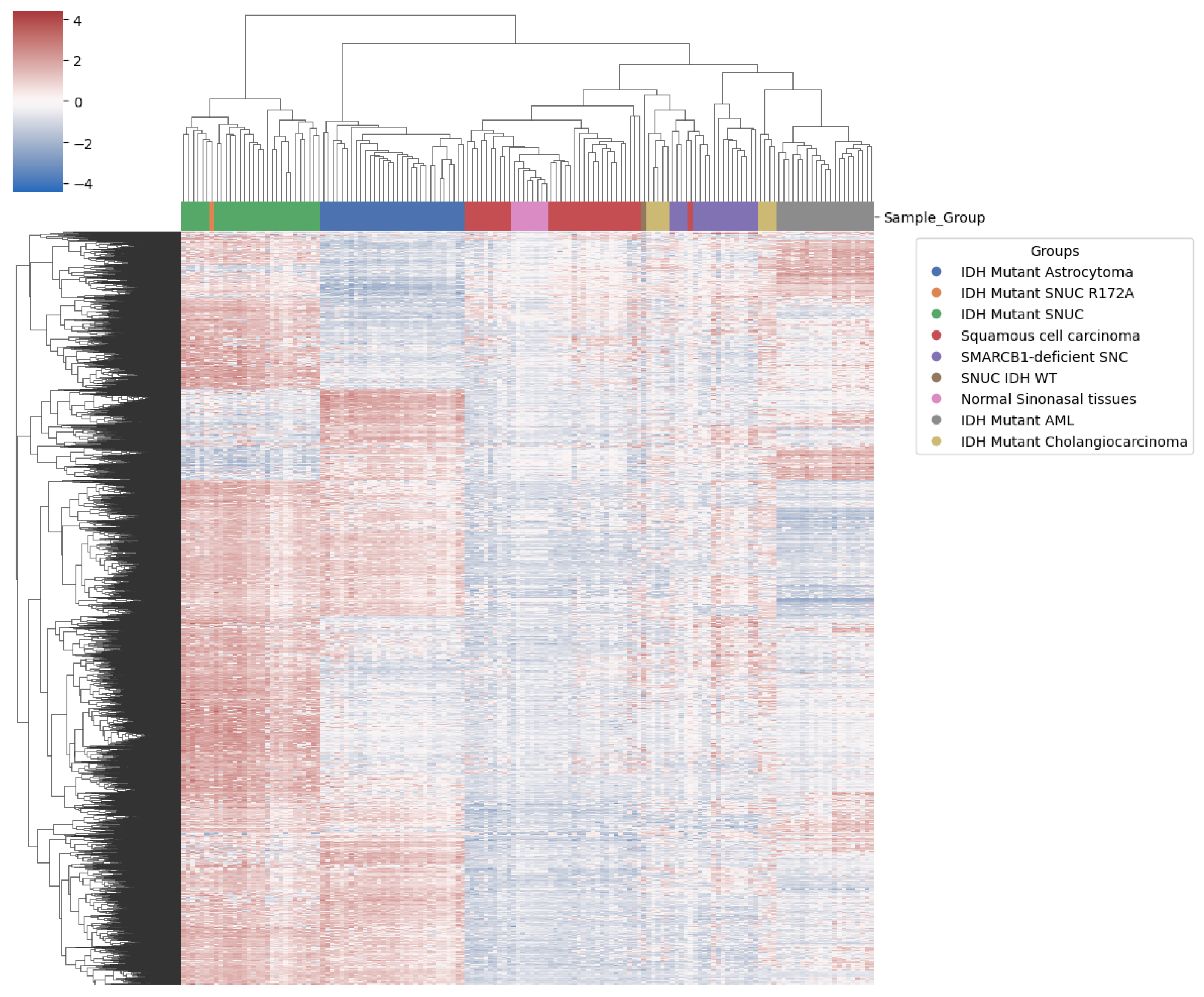
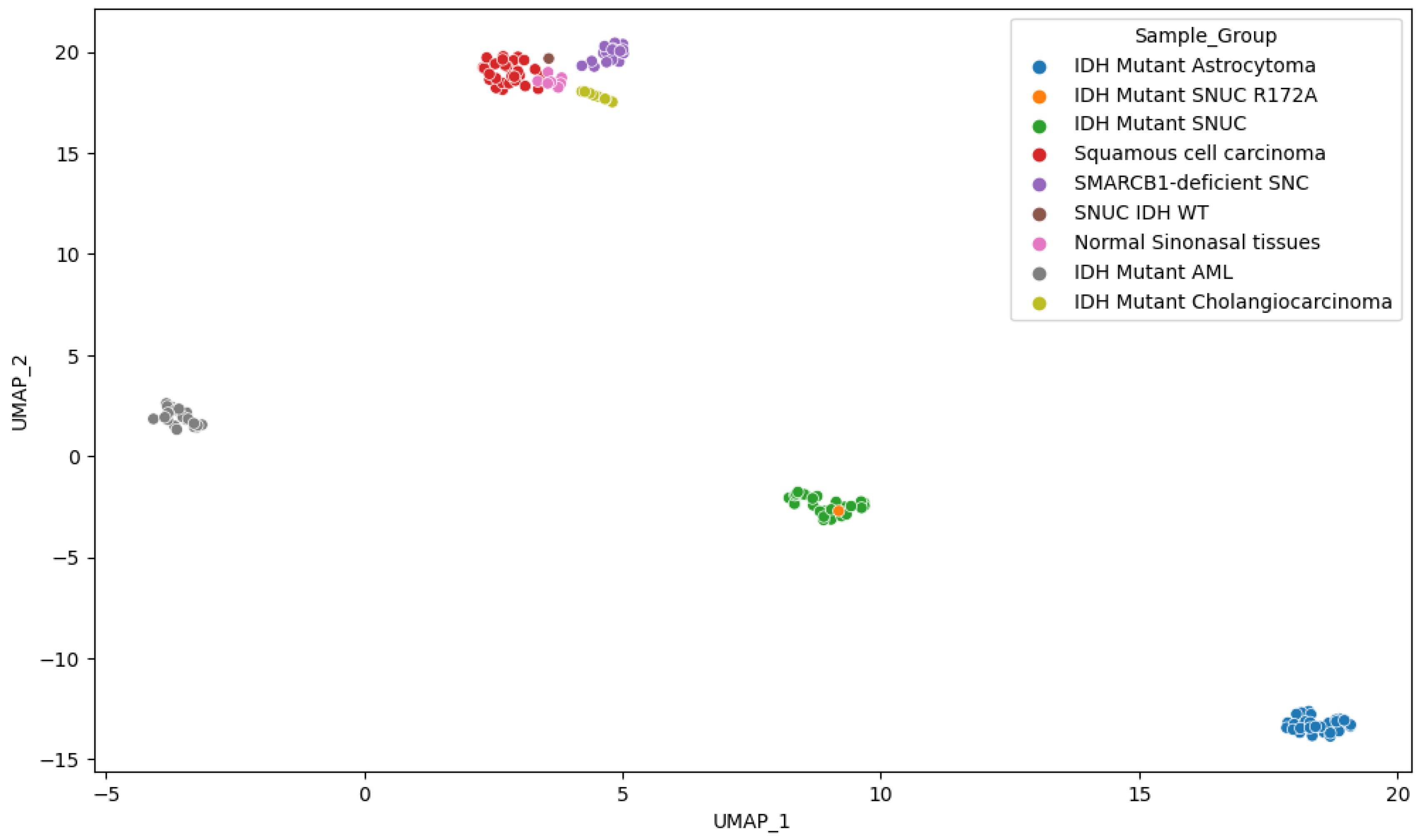
2.3. Methylation Analysis
3. Materials and Methods
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| IDH | Isocitrate dehydrogenase |
| ICT | Isocitrate |
| -KG | Alpha-Ketoglutarate |
| 2-HG | 2-Hydroxyglutarate |
| SNUC | Sinonasal undifferentiated carcinoma |
| SNC | Sinonasal carcinoma |
| GEO | Gene Expression Omnibus |
| PCA | Principal component analysis |
| UMAP | Uniform manifold approximation and projection |
| NGS | Next-generation sequencing; |
| VAF | Variant allele frequency |
| MRI | Magnetic resonance imaging |
| PET | Positron emission tomography |
Appendix A
| Sample Type | Number of Cases | GEO IDs | Sources |
|---|---|---|---|
| IDH mutant SNUC | 29 | GSE124617 & GSE196228 | Bledea et al. [29] Jurmeister et al. [30] |
| IDH wild type SNUC | 1 | GSE196228 | Jurmeister et al. [30] |
| SMARCB1-deficient SNC | 18 | GSE196228 | Jurmeister et al. [30] |
| Normal sinonasal tissues | 8 | GSE196228 | Jurmeister et al. [30] |
| Sinonasal squamous cell carcinomas | 31 | GSE196228 | Jurmeister et al. [30] |
| IDH mutant Cholangiocarcinoma | 9 | GSE124617 | Bledea et al. [29] |
| IDH mutant Acute Myeloid Leukaemia | 21 | GSE124617 | Bledea et al. [29] |
| IDH mutant Astrocytoma | 31 | GSE124617 | Bledea et al. [29] |
References
- Dogan, S.; Vasudevaraja, V.; Xu, B.; Serrano, J.; Ptashkin, R.N.; Jung, H.J.; Chiang, S.; Jungbluth, A.A.; Cohen, M.A.; Ganly, I.; et al. DNA methylation-based classification of sinonasal undifferentiated carcinoma. Mod. Pathol. 2019, 32, 1447–1459. [Google Scholar] [CrossRef] [PubMed]
- Losman, J.; Kaelin, W.J. What a difference a hydroxyl makes: Mutant IDH, (R)-2-hydroxyglutarate, and cancer. Genes Dev. 2013, 27, 836–852. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Zhang, R.; Yang, Z.; Duan, Z.; Yin, D.; Zhou, Y. Biological Roles and Therapeutic Applications of IDH2 Mutations in Human Cancer. Front. Oncol. 2021, 26, 644857. [Google Scholar] [CrossRef] [PubMed]
- Showalter, M.R.; Hatakeyama, J.; Cajka, T.; VanderVorst, K.; Carraway, K.L.; Fiehn, O.; Reproducibility Project: Cancer Biology. Replication Study: The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. eLife 2017, 6, e26030. [Google Scholar] [CrossRef] [PubMed]
- Nakazawa, S.; Sakata, K.I.; Liang, S.; Yoshikawa, K.; Iizasa, H.; Tada, M.; Hamada, J.I.; Kashiwazaki, H.; Kitagawa, Y.; Yamazaki, Y. Dominant-negative p53 mutant R248Q increases the motile and invasive activities of oral squamous cell carcinoma cells. Biomed Res. 2019, 40, 37–49. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, K.; Hamada, J.; Tada, M.; Kameyama, T.; Nakagawa, K.; Suzuki, Y.; Ikawa, M.; Hassan, N.M.; Kitagawa, Y.; Moriuchi, T. Mutant p53 R248Q but not R248W enhances in vitro invasiveness of human lung cancer NCI-H1299 cells. Biomed Res. 2010, 31, 401–411. [Google Scholar] [CrossRef] [PubMed]
- Monticone, M.; Biollo, E.; Maffei, M.; Donadini, A.; Romeo, F.; Storlazzi, C.T.; Giaretti, W.; Castagnola, P. Gene expression deregulation by KRAS G12D and G12V in a BRAF V600E context. Mol. Cancer 2008, 7, 92. [Google Scholar] [CrossRef]
- Tan, B.S.; Tiong, K.H.; Choo, H.L.; Chung, F.F.; Hii, L.W.; Tan, S.H.; Yap, I.K.; Pani, S.; Khor, N.T.; Wong, S.F.; et al. Mutant p53-R273H mediates cancer cell survival and anoikis resistance through AKT-dependent suppression of BCL2-modifying factor (BMF). Cell Death Dis. 2015, 6, e1826. [Google Scholar] [CrossRef] [PubMed]
- Dong, P.; Tada, M.; Hamada, J.; Nakamura, A.; Moriuchi, T.; Sakuragi, N. p53 dominant-negative mutant R273H promotes invasion and migration of human endometrial cancer HHUA cells. Clin. Exp. Metastasis 2007, 24, 471–483. [Google Scholar] [CrossRef]
- Tate, J.G.; Bamford, S.; Jubb, H.C.; Sondka, Z.; Beare, D.M.; Bindal, N.; Boutselakis, H.; Cole, C.G.; Creatore, C.; Dawson, E.; et al. COSMIC: The Catalogue of Somatic Mutations in Cancer. Nucl. Acids Res. 2019, 47, D941–D947. [Google Scholar] [CrossRef]
- de Bruijn, I.; Kundra, R.; Mastrogiacomo, B.; Tran, T.N.; Sikina, L.; Mazor, T.; Li, X.; Ochoa, A.; Zhao, G.; Lai, B.; et al. Analysis and Visualization of Longitudinal Genomic and Clinical Data from the AACR Project GENIE Biopharma Collaborative in cBioPortal. Cancer Res. 2023, 83, 3861–3867. [Google Scholar] [CrossRef]
- Landrum, M.J.; Lee, J.M.; Benson, M.; Brown, G.R.; Chao, C.; Chitipiralla, S.; Gu, B.; Hart, J.; Hoffman, D.; Jang, W.; et al. ClinVar: Improving access to variant interpretations and supporting evidence. Nucl Acids Res. 2018, 46, D1062–D1067. [Google Scholar] [CrossRef] [PubMed]
- Chakravarty, D.; Gao, J.; Phillips, S.M.; Kundra, R.; Zhang, H.; Wang, J.; Rudolph, J.E.; Yaeger, R.; Soumerai, T.; Nissan, M.H.; et al. OncoKB: A Precision Oncology Knowledge Base. JCO Precis Oncol. 2017, 2017, 2473–4284. [Google Scholar] [CrossRef] [PubMed]
- Patterson, S.E.; Liu, R.; Statz, C.M.; Durkin, D.; Lakshminarayana, A.; Mockus, S.M. The clinical trial landscape in oncology and connectivity of somatic mutational profiles to targeted therapies. Hum. Genom. 2016, 4, 1479–7364. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gillil, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucl. Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Liu, L.; Nakamura, A.; Someya, S.; Miyakawa, T.; Tanokura, M. Studies on the regulatory mechanism of isocitrate dehydrogenase 2 using acetylation mimics. Sci. Rep. 2017, 7, 9785. [Google Scholar] [CrossRef] [PubMed]
- Consortium, U. UniProt Consortium. UniProt: The Universal Protein Knowledgebase in 2023. Nucl. Acids Res. 2023, 6, D523–D531. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 11, 539. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Yang, H.; Ye, D.; Guan, K.L.; Xiong, Y. IDH1 and IDH2 mutations in tumorigenesis: Mechanistic insights and clinical perspectives. Clin. Cancer Res. 2012, 18, 5562–5571. [Google Scholar] [CrossRef]
- Mondesir, J.; Willekens, C.; Touat, M.; de Botton, S. IDH1 and IDH2 mutations as novel therapeutic targets: Current perspectives. J. Blood Med. 2016, 7, 171–180. [Google Scholar] [PubMed]
- Koh, J.; Cho, H.; Kim, H.; Kim, S.I.; Yun, S.; Park, C.K.; Lee, S.H.; Choi, S.H.; Park, S.H. IDH2 mutation in gliomas including novel mutation. Neuropathology 2015, 35, 236–244. [Google Scholar] [CrossRef]
- Ward, P.S.; Patel, J.; Wise, D.R.; Abdel-Wahab, O.; Bennett, B.D.; Coller, H.A.; Cross, J.R.; Fantin, V.R.; Hedvat, C.V.; Perl, A.E. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell 2010, 17, 225–234. [Google Scholar] [CrossRef]
- Jin, G.; Reitman, Z.J.; Spasojevic, I.; Batinic-Haberle, I.; Yang, J.; Schmidt-Kittler, O.; Bigner, D.D.; Yan, H. 2-hydroxyglutarate production, but not dominant negative function, is conferred by glioma-derived NADP-dependent isocitrate dehydrogenase mutations. PLoS ONE 2011, 6, e16812. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.Y.; Lai, Y.S.; Tsai, H.J.; Kuo, C.C.; Yen, B.L.; Yeh, S.P.; Sun, H.S.; Hung, W.C. The oncometabolite R-2-hydroxyglutarate activates NF-κB-dependent tumor-promoting stromal niche for acute myeloid leukemia cells. Sci. Rep. 2016, 6, 32428. [Google Scholar] [CrossRef] [PubMed]
- Schymkowitz, J.; Borg, J.; Stricher, F.; Nys, R.; Rousseau, F.; Serrano, L. The FoldX web server: An online force field. Nucl. Acids Res. 2005, 33, W382–W388. [Google Scholar] [CrossRef] [PubMed]
- Capper, D.; Jones, D.T.W.; Sill, M.; Hovestadt, V.; Schrimpf, D.; Sturm, D.; Koelsche, C.; Sahm, F.; Chavez, L.; Reuss, D.E.; et al. DNA methylation-based classification of central nervous system tumours. Nature 2018, 555, 469–474. [Google Scholar] [CrossRef] [PubMed]
- Molecular Neuropathology. Available online: https://www.molecularneuropathology.org/mnp/classifiers/11 (accessed on 2 May 2023).
- Bledea, R.; Vasudevaraja, V.; Patel, S.; Stafford, J.; Serrano, J.; Esposito, G.; Tredwin, L.M.; Goodman, N.; Kloetgen, A.; Golfinos, J.G.; et al. Functional and topographic effects on DNA methylation in IDH1/2 mutant cancers. Sci. Rep. 2019, 9, 16830. [Google Scholar] [CrossRef] [PubMed]
- Jurmeister, P.; Glöß, S.; Roller, R.; Leitheiser, M.; Schmid, S.; Mochmann, L.H.; Payá Capilla, E.; Fritz, R.; Dittmayer, C.; Friedrich, C.; et al. DNA methylation-based classification of sinonasal tumors. Nat. Commun. 2022, 13, 7148. [Google Scholar] [CrossRef] [PubMed]
- Sciarra, A.; Missiaglia, E.; Trimech, M.; Melloul, E.; Broul, J.P.; Sempoux, C.; La Rosa, S. Gallbladder Mixed Neuroendocrine-Non-neuroendocrine Neoplasm (MiNEN) Arising in Intracholecystic Papillary Neoplasm: Clinicopathologic and Molecular Analysis of a Case and Review of the Literature. Endocr. Pathol. 2020, 31, 84–93. [Google Scholar] [CrossRef]
- Aryee, M.J.; Jaffe, A.E.; Corrada-Bravo, H.; Ladd-Acosta, C.; Feinberg, A.P.; Hansen, K.D.; Irizarry, R.A. Minfi: A flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 2014, 30, 1363–1369. [Google Scholar] [CrossRef]
- Du, P.; Zhang, X.; Huang, C.C.; Jafari, N.; Kibbe, W.A.; Hou, L.; Lin, S.M. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinform. 2010, 11, 587. [Google Scholar] [CrossRef]
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberl, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0: Fundamental Algorithms for Scientific Computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef]
- Seabold, S.; Perktold, J. Statsmodels: Econometric and statistical modeling with python. In Proceedings of the 9th Python in Science Conference, Austin, TX, USA, 28–30 June 2010. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Griesel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- McInnes, L.; Healy, J.; Saul, N.; Grossberger, L. UMAP: Uniform Manifold Approximation and Projection. J. Open Source Softw. 2018, 3, 861. [Google Scholar] [CrossRef]
- Hunter, J. Matplotlib: A 2D graphics environment. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]
- Waskom, M.L. seaborn: Statistical data visualization. J. Open Source Softw. 2021, 6, 3021. [Google Scholar] [CrossRef]
- Pruitt, K.; Tatusova, T.; Maglott, D.R. NCBI Reference Sequence (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucl. Acids Res. 2005, 35, D61–D65. [Google Scholar] [CrossRef]


| Regions | % | % | ||
|---|---|---|---|---|
| TSS200 | 1237 | 12.37 | 50,040 | 13.51 |
| TSS1500 | 1643 | 16.43 | 64,419 | 17.39 |
| 5UTR | 1482 | 14.82 | 50,649 | 13.67 |
| Body | 3805 | 38.05 | 134,277 | 36.25 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Burgermeister, S.; Stoykova, S.; Krebs, F.S.; Zoete, V.; Mbefo, M.; Egervari, K.; Reinhard, A.; Bisig, B.; Hewer, E. Methylation-Based Characterization of a New IDH2 Mutation in Sinonasal Undifferentiated Carcinoma. Int. J. Mol. Sci. 2024, 25, 6518. https://doi.org/10.3390/ijms25126518
Burgermeister S, Stoykova S, Krebs FS, Zoete V, Mbefo M, Egervari K, Reinhard A, Bisig B, Hewer E. Methylation-Based Characterization of a New IDH2 Mutation in Sinonasal Undifferentiated Carcinoma. International Journal of Molecular Sciences. 2024; 25(12):6518. https://doi.org/10.3390/ijms25126518
Chicago/Turabian StyleBurgermeister, Simon, Simona Stoykova, Fanny S. Krebs, Vincent Zoete, Martial Mbefo, Kristof Egervari, Antoine Reinhard, Bettina Bisig, and Ekkehard Hewer. 2024. "Methylation-Based Characterization of a New IDH2 Mutation in Sinonasal Undifferentiated Carcinoma" International Journal of Molecular Sciences 25, no. 12: 6518. https://doi.org/10.3390/ijms25126518
APA StyleBurgermeister, S., Stoykova, S., Krebs, F. S., Zoete, V., Mbefo, M., Egervari, K., Reinhard, A., Bisig, B., & Hewer, E. (2024). Methylation-Based Characterization of a New IDH2 Mutation in Sinonasal Undifferentiated Carcinoma. International Journal of Molecular Sciences, 25(12), 6518. https://doi.org/10.3390/ijms25126518







