Genetic Basis Identification of a NLR Gene, TaRGA5-like, That Confers Partial Powdery Mildew Resistance in Wheat SJ106
Abstract
:1. Introduction
2. Results
2.1. Genetic Analysis of Powdery Mildew Resistance in SJ106
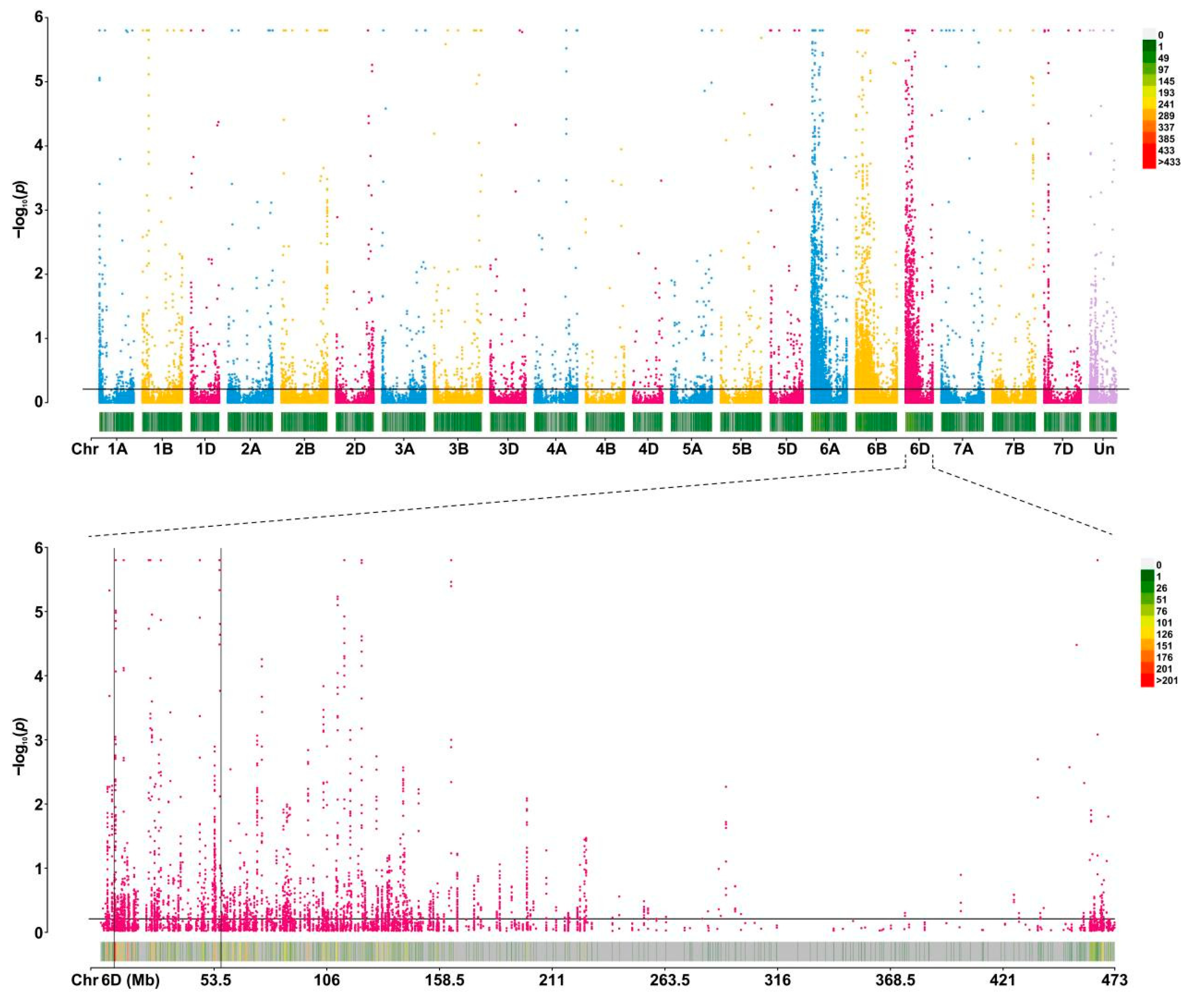
2.2. SNP Calling and Confirmation of Candidate Interval
2.3. Mapping of Powdery Mildew Resistance Gene
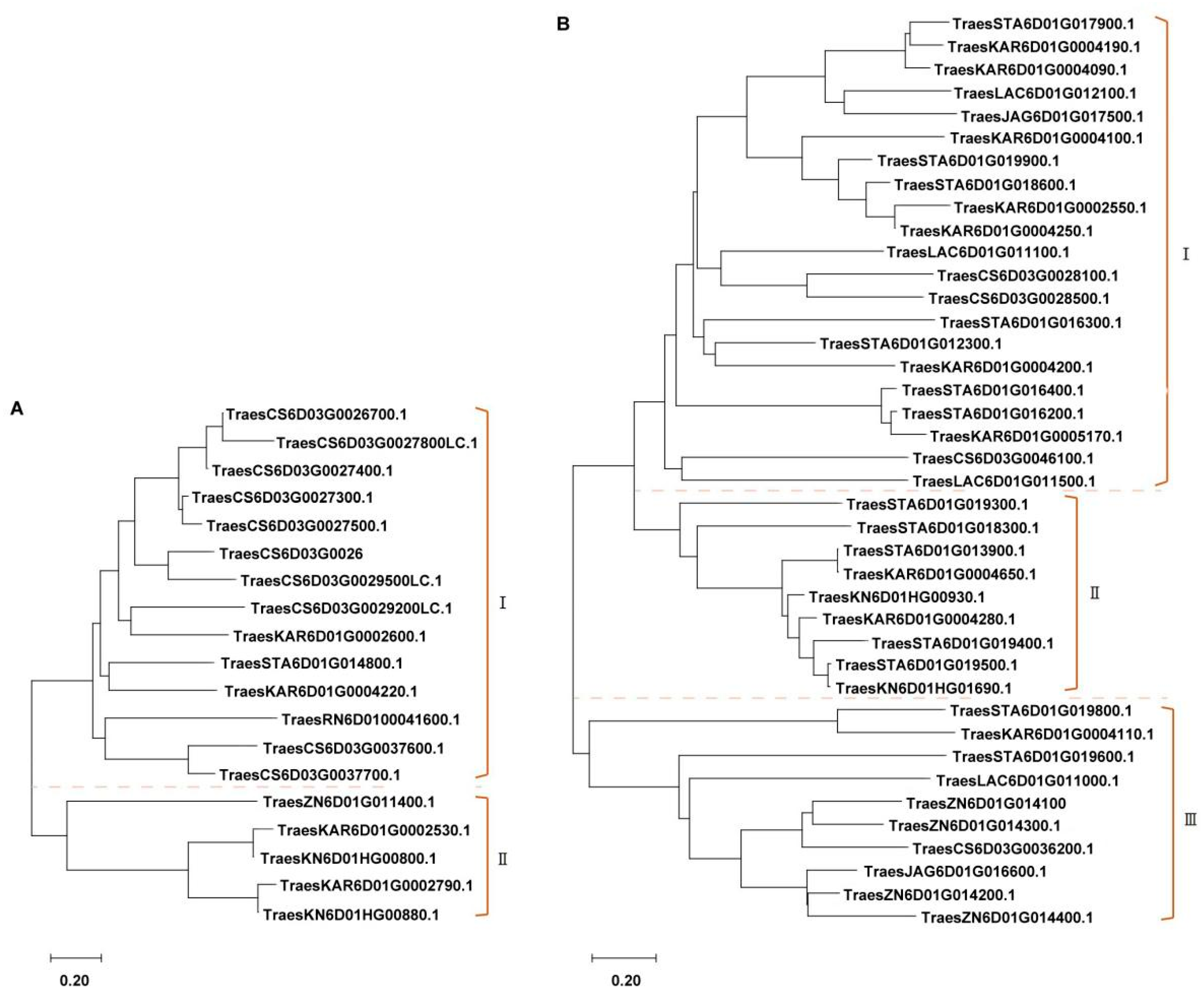
2.4. PmSJ106 Locus Region in Pan-Genome Is Enriched with Multiple NLR and Kinase Genes
2.5. Cloning of the Candidate Powdery Mildew Resistance Gene in SJ106
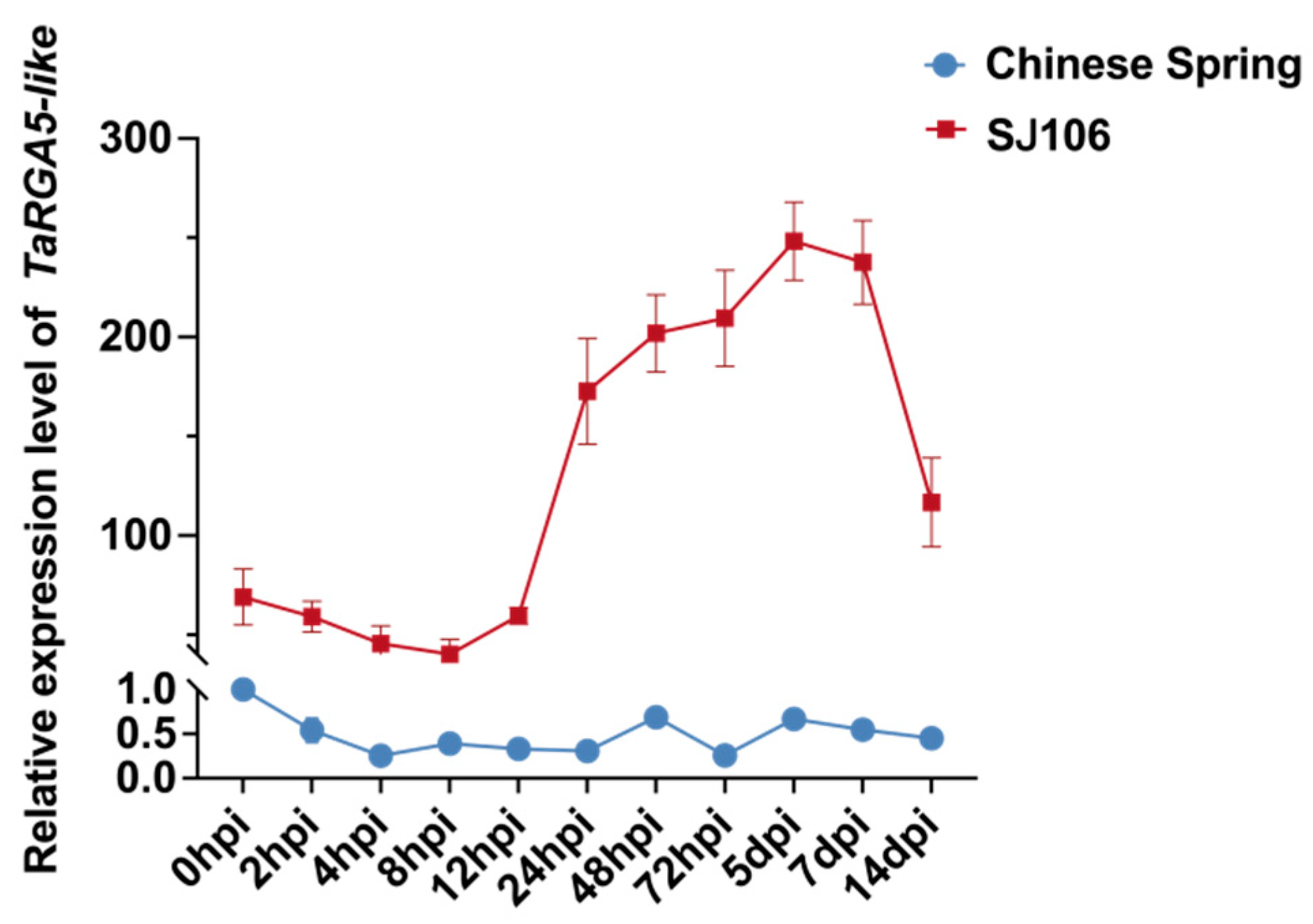
2.6. Expression Analysis of TaRGA5-like
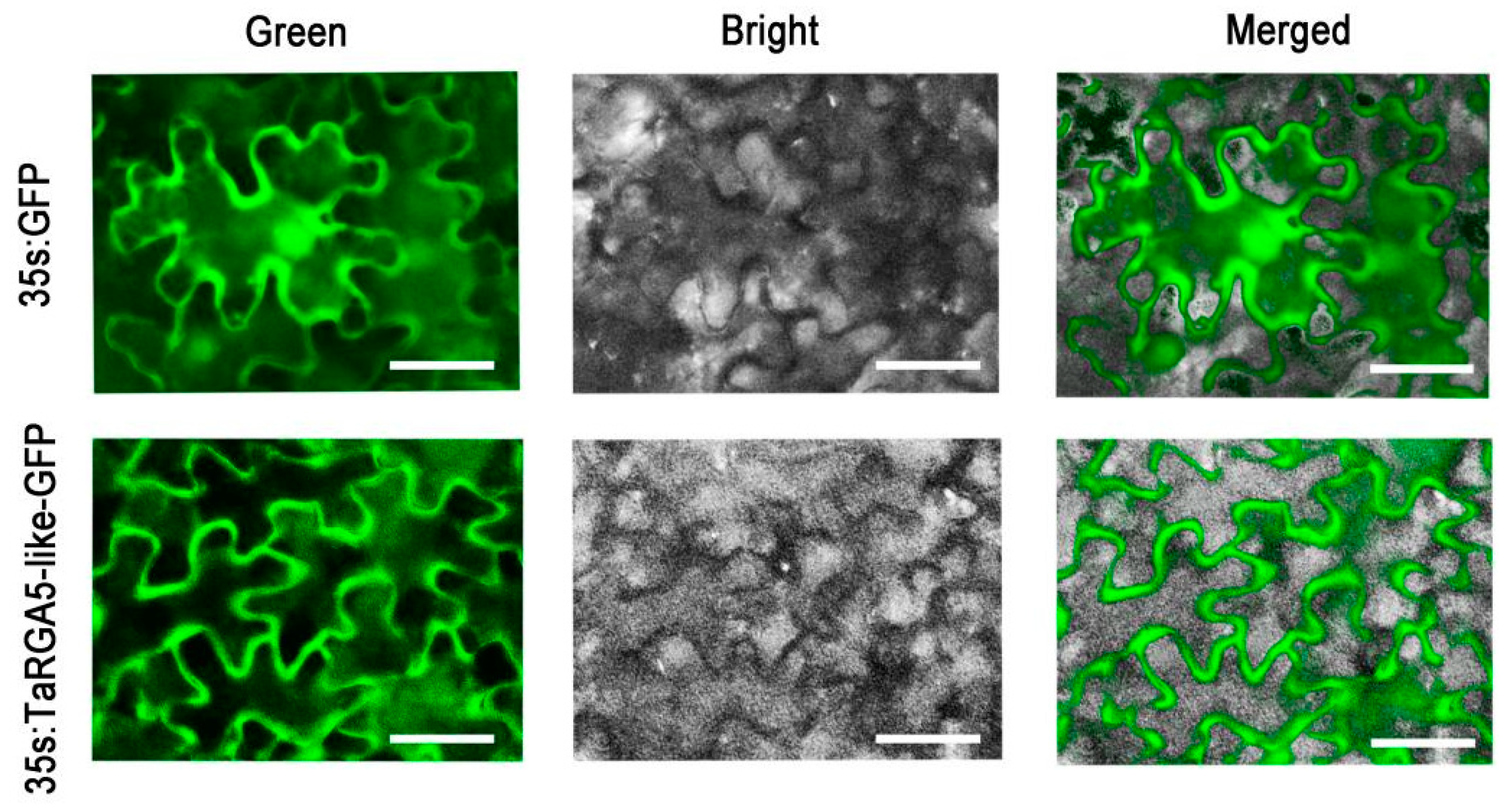
2.7. Subcellular Localization of TaRGA5-like
2.8. VIGS of of TaRGA5-like
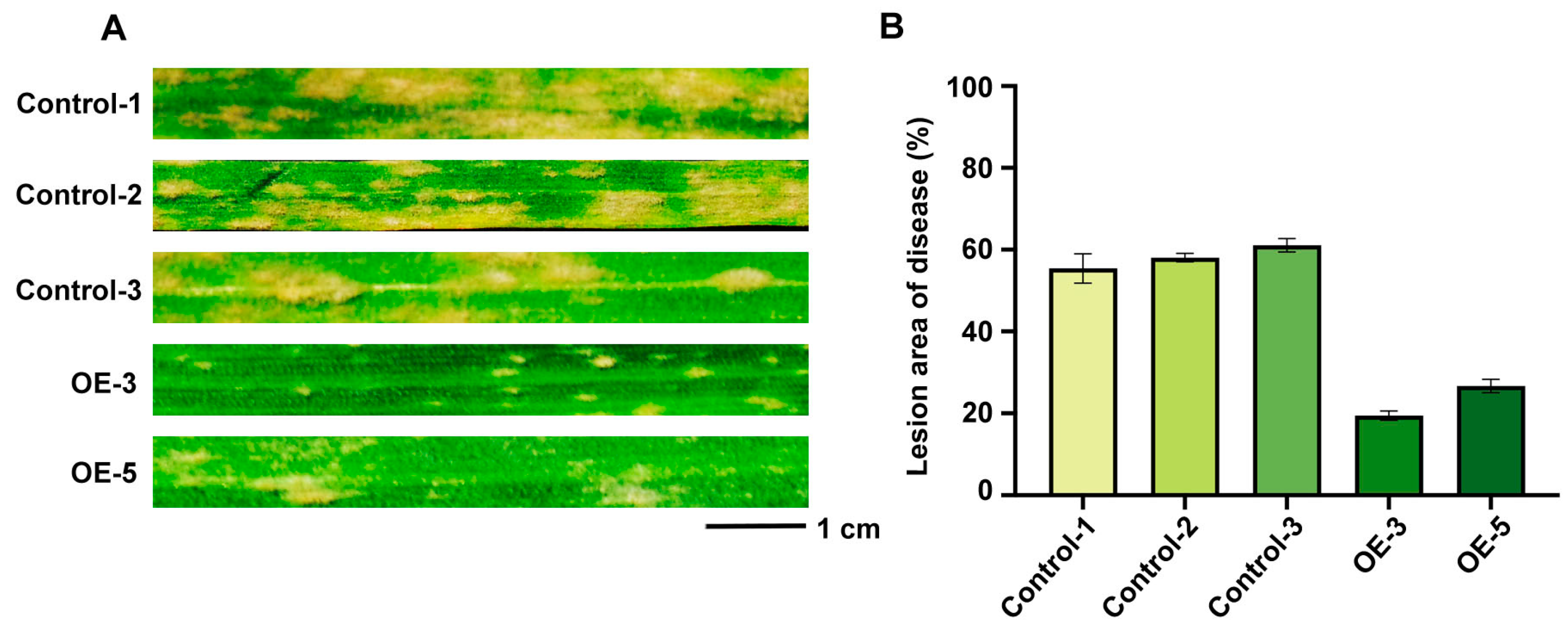
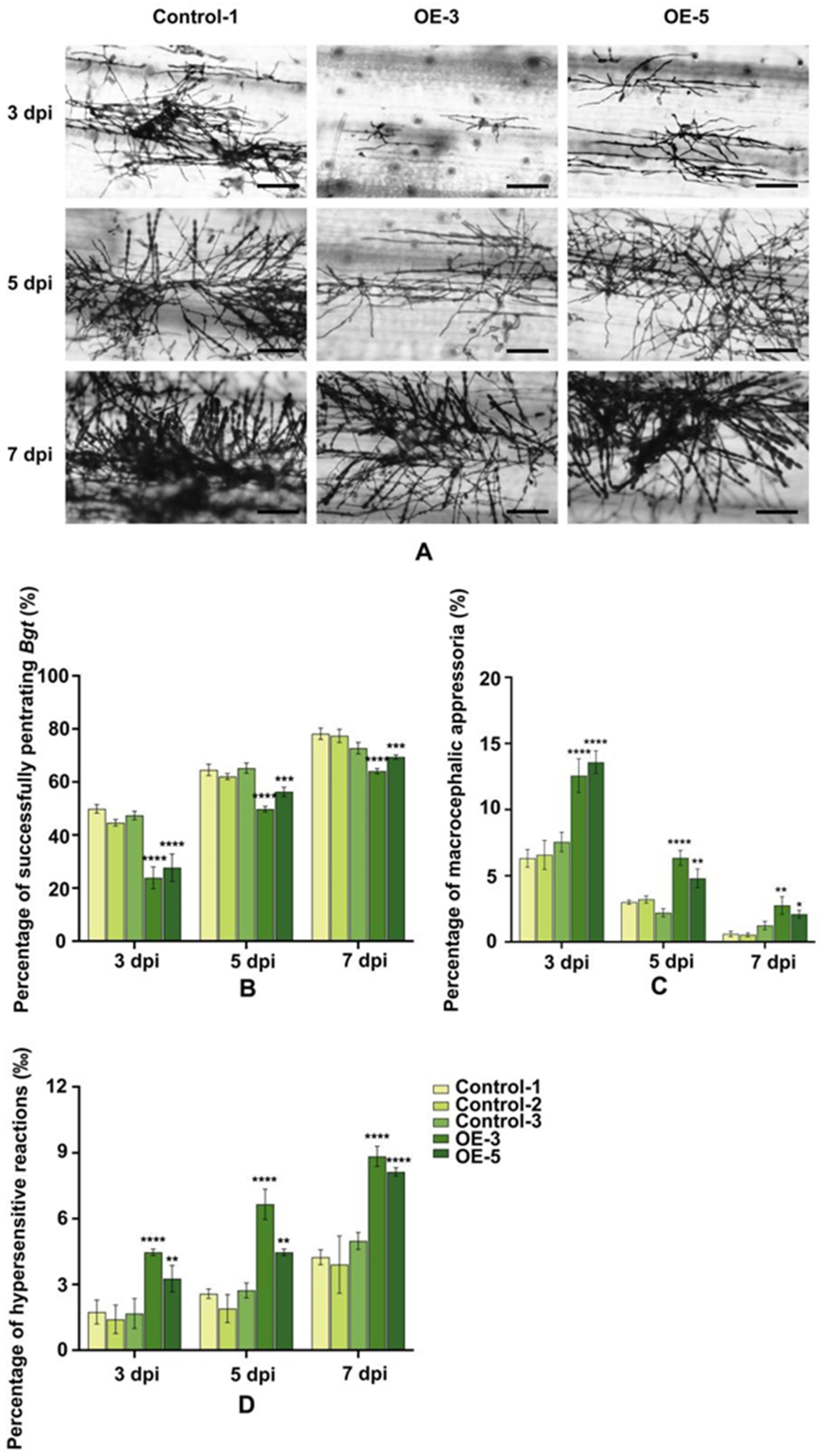
2.9. Over-Expression of of TaRGA5-like in Susceptible Wheat
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. BSR-Seq and RNA-Seq Analysis
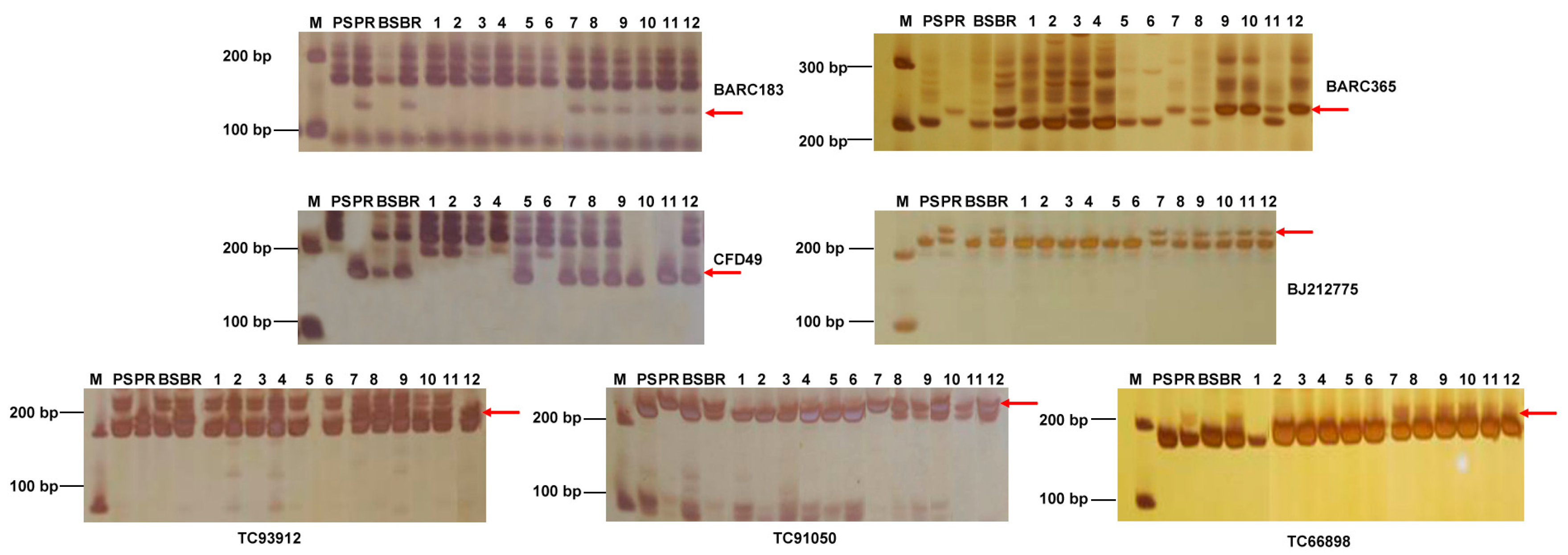
4.3. SSR Analysis
4.4. Candidate Gene Prediction and Functional Analysis
4.5. Isolation and Sequence Analysis of TaRGA5-like in SJ106
4.6. Expression Analysis of TaRGA5-like
4.7. Subcellular Localization of TaRGA5-like
4.8. Function Analysis of TaRGA5-like by VIGS
4.9. Function Analysis of TaRGA5-like by OE
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hurni, S.; Brunner, S.; Buchmann, G.; Herren, G.; Jordan, T.; Krukowski, P.; Wicker, T.; Yahiaoui, N.; Mago, R.; Keller, B. Rye Pm8 and wheat Pm3 are orthologous genes and show evolutionary conservation of resistance function against powdery mildew. Plant J. 2013, 76, 957–969. [Google Scholar] [CrossRef] [PubMed]
- Yahiaoui, N.; Srichumpa, P.; Dudler, R.; Keller, B. Genome analysis at different ploidy levels allows cloning of the powdery mildew resistance gene Pm3b from hexaploid wheat. Plant J. 2004, 37, 528–538. [Google Scholar] [CrossRef] [PubMed]
- Juroszek, P.; von Tiedemann, A. Climate change and potential future risks through wheat diseases: A review. Eur. J. Plant Pathol. 2013, 136, 21–33. [Google Scholar] [CrossRef]
- Sánchez-Martín, J.; Steuernagel, B.; Ghosh, S.; Herren, G.; Hurni, S.; Adamski, N.; Vrána, J.; Kubaláková, M.; Krattinger, S.G.; Wicker, T.; et al. Rapid gene isolation in barley and wheat by mutant chromosome sequencing. Genome Biol. 2016, 17, 221. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.P.; Hurni, S.; Ruinelli, M.; Brunner, S.; Sanchez-Martin, J.; Krukowski, P.; Peditto, D.; Buchmann, G.; Zbinden, H.; Keller, B. Evolutionary divergence of the rye Pm17 and Pm8 resistance genes reveals ancient diversity. Plant Mol. Biol. 2018, 98, 249–260. [Google Scholar] [CrossRef] [PubMed]
- Xing, L.; Hu, P.; Liu, J.; Witek, K.; Zhou, S.; Xu, J.; Zhou, W.; Gao, L.; Huang, Z.; Zhang, R.; et al. Pm21 from Haynaldia villosa encodes a CC-NBS- LRR protein conferring powdery mildew resistance in wheat. Mol. Plant 2018, 11, 874–878. [Google Scholar] [CrossRef] [PubMed]
- Zou, S.; Wang, H.; Li, Y.; Kong, Z.; Tang, D. The NB-LRR gene Pm60 confers powdery mildew resistance in wheat. New Phytol. 2018, 218, 298–309. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Dong, L.; Li, B.; Wang, Z.; Xie, J.; Qiu, D.; Li, Y.; Shi, W.; Yang, L.; Wu, Q.; et al. A CNL protein in wild emmer wheat confers powdery mildew resistance. New Phytol. 2020, 228, 1027–1037. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Guo, G.; Wang, Y.; Hu, T.; Wang, L.; Li, J.; Qiu, D.; Li, Y.; Wu, Q.; Lu, P.; et al. A rare single nucleotide variant in Pm5e confers powdery mildew resistance in common wheat. New Phytol. 2020, 228, 1011–1026. [Google Scholar] [CrossRef]
- Li, Y.; Wei, Z.-Z.; Sela, H.; Govta, L.; Klymiuk, V.; Roychowdhury, R.; Chawla, H.S.; Ens, J.; Wiebe, K.; Bocharova, V.; et al. Dissection of a rapidly evolving wheat resistance gene cluster by long-read genome sequencing accelerated the cloning of Pm69. Plant Commun. 2024, 5, 100646. [Google Scholar] [CrossRef]
- Wu, Q.; Chen, Y.; Li, B.; Li, J.; Zhang, P.; Xie, J.; Zhang, H.; Guo, G.; Lu, P.; Li, M.; et al. Functional characterization of powdery mildew resistance gene MlIW172, a new Pm60 allele and its allelic variation in wild emmer wheat. J. Genet. Genom. 2022, 49, 787–795. [Google Scholar] [CrossRef]
- Wu, Q.; Zhao, F.; Chen, Y.; Zhang, P.; Zhang, H.; Guo, G.; Xie, J.; Dong, L.; Lu, P.; Li, M.; et al. Bulked segregant CGT-Seq-facilitated map-based cloning of a powdery mildew resistance gene originating from wild emmer wheat (Triticum dicoccoides). Plant Biotechnol. J. 2021, 19, 1288–1290. [Google Scholar] [CrossRef]
- Hewitt, T.; Müller, M.C.; Molnár, I.; Mascher, M.; Holušová, K.; Šimková, H.; Kunz, L.; Zhang, J.; Li, J.; Bhatt, D.; et al. A highly differentiated region of wheat chromosome 7AL encodes a Pm1a immune receptor that recognizes its corresponding AvrPm1a effector from Blumeria graminis. New Phytol. 2021, 229, 2812–2826. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Liu, C.; Gong, S.; Chen, Z.; Chen, R.; Liu, T.; Liu, R.; Du, H.; Guo, R.; Li, G.; et al. Orthologous genes Pm12 and Pm21 from two wild relatives of wheat show evolutionary conservation but divergent powdery mildew resistance. Plant Commun. 2023, 4, 100472. [Google Scholar] [CrossRef] [PubMed]
- Lu, P.; Guo, L.; Wang, Z.; Li, B.; Li, J.; Li, Y.; Qiu, D.; Shi, W.; Yang, L.; Wang, N.; et al. A rare gain of function mutation in a wheat tandem kinase confers resistance to powdery mildew. Nat. Commun. 2020, 11, 680. [Google Scholar] [CrossRef]
- Gaurav, K.; Arora, S.; Silva, P.; Sánchez-Martín, J.; Horsnell, R.; Gao, L.; Brar, G.S.; Widrig, V.; John Raupp, W.; Singh, N.; et al. Population genomic analysis of Aegilops tauschii identifies targets for bread wheat improvement. Nat. Biotechnol. 2022, 40, 422–431. [Google Scholar] [CrossRef]
- Sánchez-Martín, J.; Widrig, V.; Herren, G.; Wicker, T.; Zbinden, H.; Gronnier, J.; Spörri, L.; Praz, C.R.; Heuberger, M.; Kolodziej, M.C.; et al. Wheat Pm4 resistance to powdery mildew is controlled by alternative splice variants encoding chimeric proteins. Nat. Plants 2021, 7, 327–341. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Men, W.; Ma, C.; Liu, Q.; Dong, Z.; Tian, X.; Wang, C.; Liu, C.; Gill, H.S.; Ma, P.; et al. Wheat powdery mildew resistance gene Pm13 encodes a mixed lineage kinase domain-like protein. Nat. Commun. 2024, 15, 2449. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhang, H.; Xiao, H.; Zhu, K.; Shi, W.; Zhang, D.; Wang, Y.; Yang, L.; Wu, Q.; Xie, J.; et al. A membrane associated tandem kinase from wild emmer wheat confers broad-spectrum resistance to powdery mildew. Nat. Commun. 2024, 15, 3124. [Google Scholar] [CrossRef]
- Krattinger, S.G.; Lagudah, E.S.; Spielmeyer, W.; Singh, R.P.; Huerta-Espino, J.; McFadden, H.; Bossolini, E.; Selter, L.L.; Keller, B. A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 2009, 323, 1360–1363. [Google Scholar] [CrossRef]
- Moore, J.W.; Herrera-Foessel, S.; Lan, C.; Schnippenkoetter, W.; Ayliffe, M.; Huerta-Espino, J.; Lillemo, M.; Viccars, L.; Milne, R.; Periyannan, S.; et al. A recently evolved hexose transporter variant confers resistance to multiple pathogens in wheat. Nat. Genet. 2015, 47, 1494–1498. [Google Scholar] [CrossRef] [PubMed]
- Dracatos, P.M.; Lu, J.; Sánchez-Martín, J.; Wulff, B.B.H. Resistance that stacks up: Engineering rust and mildew disease control in the cereal crops wheat and barley. Plant Biotechnol. J. 2023, 21, 1938–1951. [Google Scholar] [CrossRef]
- Wang, Z.L.; Li, L.H.; He, Z.H.; Duan, X.Y.; Zhou, Y.L.; Chen, X.M.; Lillemo, M.; Singh, R.P.; Wang, H.; Xia, X.C. Seedling and adult plant resistance to powdery mildew in Chinese bread wheat cultivars and lines. Plant Dis. 2005, 89, 457–463. [Google Scholar] [CrossRef] [PubMed]
- Zeng, F.; Yang, L.; Gong, S.; Shi, W.; Zhang, X.; Wang, H.; Xiang, L.; Xue, M.; Yu, D. Virulence and Diversity of Blumeria graminis f. sp. tritici Populations in China. J. Integr. Agric. 2014, 13, 2424–2437. [Google Scholar] [CrossRef]
- Xiao, J.; Liu, B.; Yao, Y.; Guo, Z.; Jia, H.; Kong, L.; Zhang, A.; Ma, W.; Ni, Z.; Xu, S.; et al. Wheat genomic study for genetic improvement of traits in China. Sci. China Life Sci. 2022, 65, 1718–1775. [Google Scholar] [CrossRef]
- He, H.; Ji, J.; Li, H.; Tong, J.; Feng, Y.; Wang, X.; Han, R.; Bie, T.; Liu, C.; Zhu, S. Genetic Diversity and Evolutionary Analyses Reveal the Powdery Mildew Resistance Gene Pm21 Undergoing Diversifying Selection. Front. Genet. 2020, 11, 489. [Google Scholar] [CrossRef]
- Wang, C.; Liang, D.; Wang, J.; Liu, D.; Gong, H.; Liu, J. Breeding and cultivation techniques of high-quality strong gluten spring wheat variety Jinqiang 9. Bull. Agric. Sci. Technol. 2016, 9, 221–222. (In Chinese) [Google Scholar]
- Wang, B.; Meng, T.; Xiao, B.; Yu, T.; Yue, T.; Jin, Y.; Ma, P. Fighting wheat powdery mildew: From genes to fields. Theor. Appl. Genet. 2023, 136, 196. [Google Scholar] [CrossRef]
- Rinaldo, A.; Gilbert, B.; Boni, R.; Krattinger, S.G.; Singh, D.; Park, R.F.; Lagudah, E.; Ayliffe, M. The Lr34 adult plant rust resistance gene provides seedling resistance in durum wheat without senescence. Plant Biotechnol. J. 2017, 15, 894–905. [Google Scholar] [CrossRef]
- Risk, J.M.; Selter, L.L.; Chauhan, H.; Krattinger, S.G.; Kumlehn, J.; Hensel, G.; Viccars, L.A.; Richardson, T.M.; Buesing, G.; Troller, A.; et al. The wheat Lr34 gene provides resistance against multiple fungal pathogens in barley. Plant Biotechnol. J. 2013, 11, 847–854. [Google Scholar] [CrossRef]
- Krattinger, S.G.; Sucher, J.; Selter, L.L.; Chauhan, H.; Zhou, B.; Tang, M.; Upadhyaya, N.M.; Mieulet, D.; Guiderdoni, E.; Weidenbach, D.; et al. The wheat durable, multipathogen resistance gene Lr34 confers partial blast resistance in rice. Plant Biotechnol. J. 2016, 14, 1261–1268. [Google Scholar] [CrossRef] [PubMed]
- Hatta, M.A.M.; Arora, S.; Ghosh, S.; Matny, O.; Smedley, M.A.; Yu, G.; Chakraborty, S.; Bhatt, D.; Xia, X.; Steuernagel, B.; et al. The wheat Sr22, Sr33, Sr35 and Sr45 genes confer resistance against stem rust in barley. Plant Biotechnol. J. 2021, 19, 273–284. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Kielsmeier-Cook, J.; Bajgain, P.; Zhang, X.; Anderson, J. Development of genotyping by sequencing (GBS)- and array-derived SNP markers for stem rust resistance gene Sr42. Mol. Breed. 2015, 35, 1–13. [Google Scholar] [CrossRef]
- Ma, S.; Wang, M.; Wu, J.; Guo, W.; Chen, Y.; Li, G.; Wang, Y.; Shi, W.; Xia, G.; Fu, D.; et al. WheatOmics: A platform combining multiple omics data to accelerate functional genomics studies in wheat. Mol. Plant 2021, 14, 1965–1968. [Google Scholar] [CrossRef] [PubMed]
- 35. Mackenzie, A.; Norman, M.; Gessese, M.; Chen, C.; Sørensen, C.; Hovmøller, M.; Ma, L.; Forrest, K.; Hickey, L.; Bariana, H.; et al. Wheat stripe rust resistance locus YR63 is a hot spot for evolution of defence genes – a pangenome discovery. BMC Plant Biol. 2023, 23, 590. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, X.; Yang, C.; Wang, G.; Fan, B.; Shang, Y.; Dang, C.; Xie, C.; Wang, Z. Genome-wide identification of TaCIPK gene family members in wheat and their roles in host response to Blumeria graminis f. sp. tritici infection. Int. J. Biol. Macromol. 2023, 248, 125691. [Google Scholar] [CrossRef]
- Liu, X.; Wang, J.; Fan, B.; Shang, Y.; Sun, Y.; Dang, C.; Xie, C.; Wang, Z.; Peng, Y. A COI1 gene in wheat contributes to the early defence response against wheat powdery mildew. J. Phytopathol. 2018, 166, 116–122. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, X.; Yang, C.; Wu, S.; Dong, H.; Wang, G.; Han, X.; Fan, B.; Shang, Y.; Dang, C.; Xie, C.; et al. Genetic Basis Identification of a NLR Gene, TaRGA5-like, That Confers Partial Powdery Mildew Resistance in Wheat SJ106. Int. J. Mol. Sci. 2024, 25, 6603. https://doi.org/10.3390/ijms25126603
Liu X, Yang C, Wu S, Dong H, Wang G, Han X, Fan B, Shang Y, Dang C, Xie C, et al. Genetic Basis Identification of a NLR Gene, TaRGA5-like, That Confers Partial Powdery Mildew Resistance in Wheat SJ106. International Journal of Molecular Sciences. 2024; 25(12):6603. https://doi.org/10.3390/ijms25126603
Chicago/Turabian StyleLiu, Xiaoying, Chenxiao Yang, Siqi Wu, Huixuan Dong, Guangyu Wang, Xinyue Han, Baoli Fan, Yuntao Shang, Chen Dang, Chaojie Xie, and et al. 2024. "Genetic Basis Identification of a NLR Gene, TaRGA5-like, That Confers Partial Powdery Mildew Resistance in Wheat SJ106" International Journal of Molecular Sciences 25, no. 12: 6603. https://doi.org/10.3390/ijms25126603





