From SARS-CoV-2 to Global Preparedness: A Graphical Interface for Standardised High-Throughput Bioinformatics Analysis in Pandemic Scenarios and Surveillance of Drug Resistance
Abstract
1. Introduction
2. Results
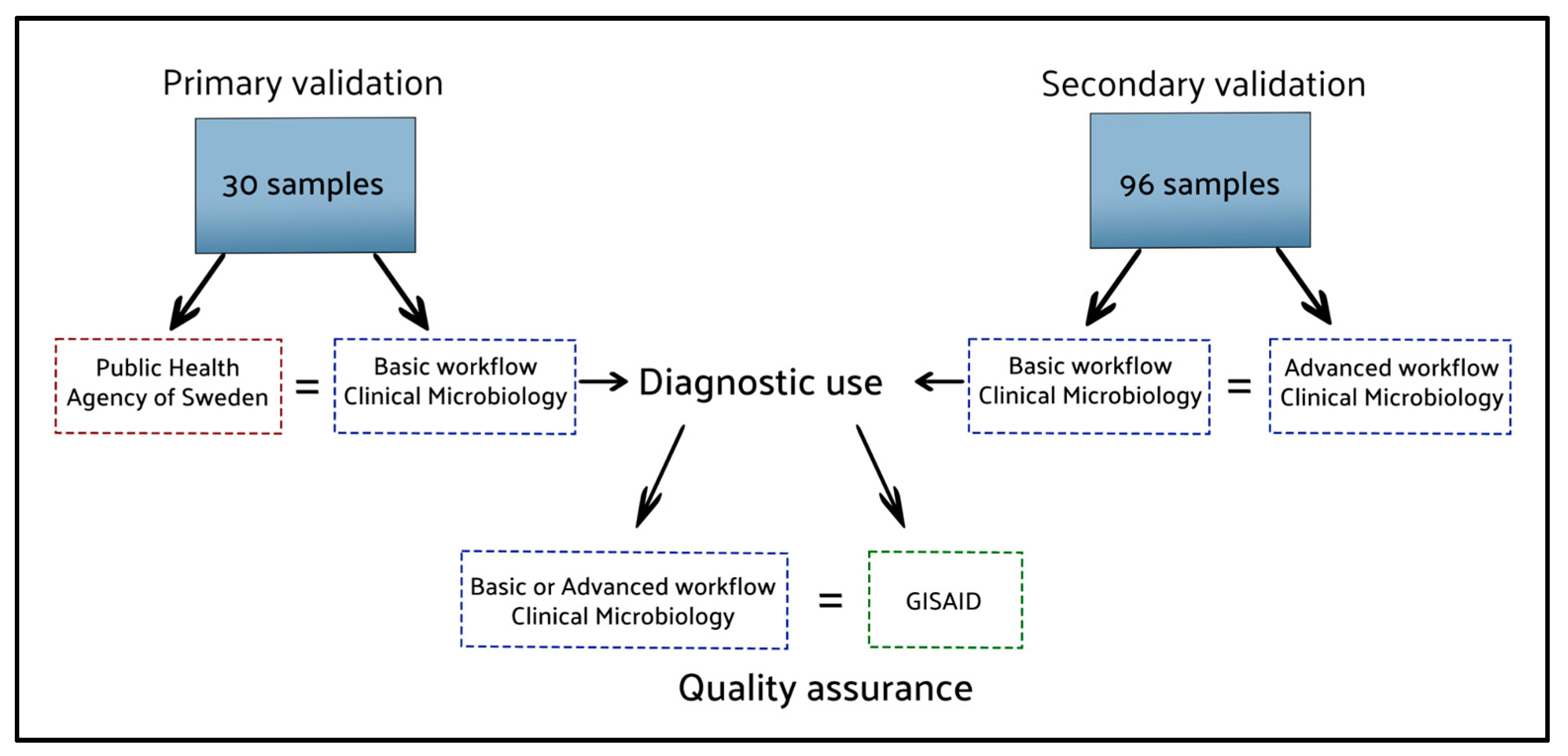
2.1. Workflow Validation
2.2. Test Data
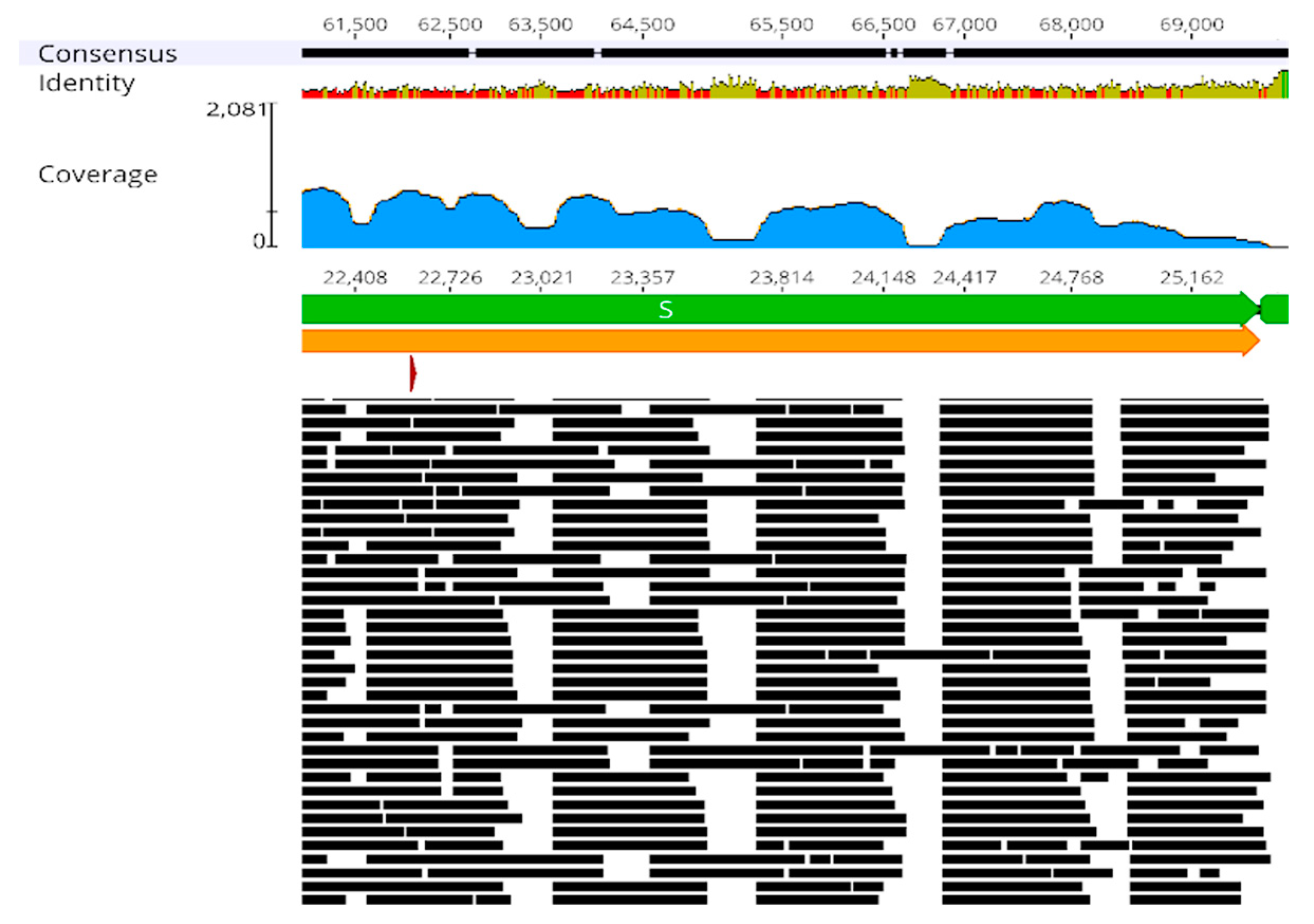
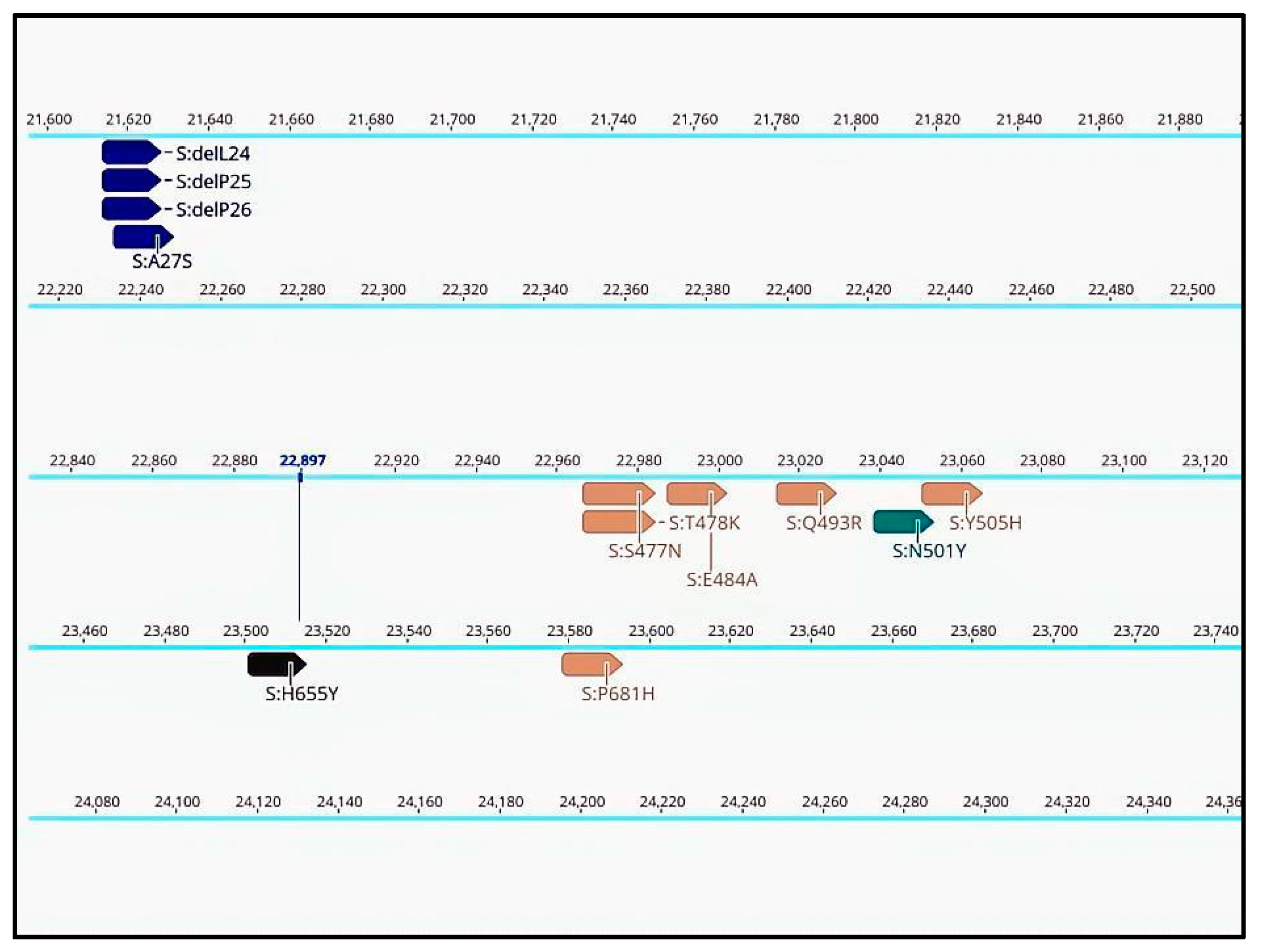
2.3. Visual Inspections in Geneious Prime
3. Discussion
4. Materials and Methods
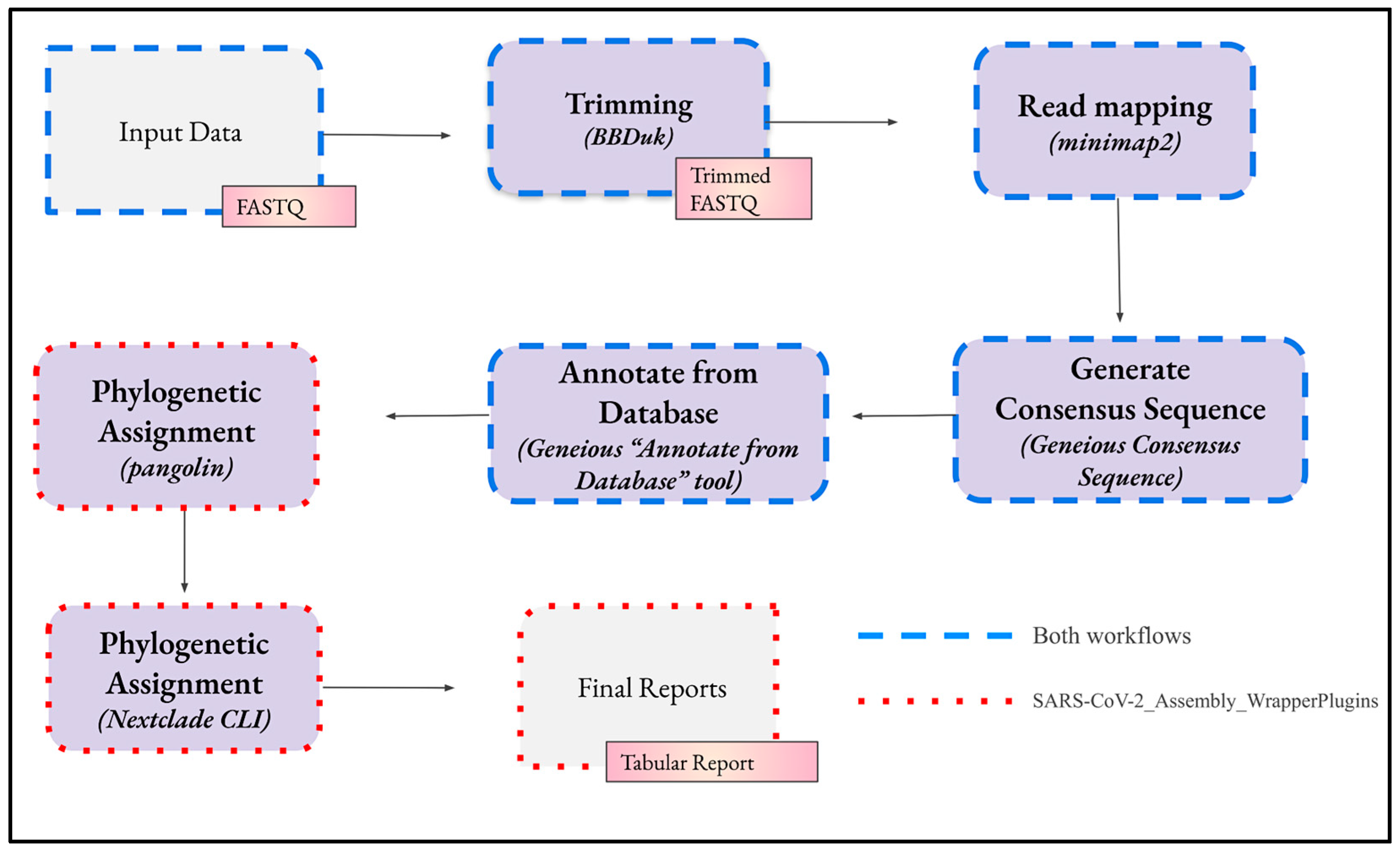
4.1. Geneious Workflows
4.1.1. Workflow Structure
- Mutations distinguishing unique variants, such as Alpha, Beta, Delta, and Omicron.
- Functional mutations in the coronavirus spike glycoprotein.
- Mutations providing antiviral resistance against Ritonavir-boosted nirmatrelvir (Paxlovid) or Remdesivir in the coronavirus non-structural protein (nsp) 5 and 12.
4.2. Plugins
4.3. Set-Up of the Workflows
4.3.1. Software
4.3.2. Files
4.3.3. Workflow Installation and Usage
4.4. Validation and Test Data
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Worldometer. COVID—Coronavirus Statistics—Worldometer. Available online: https://www.worldometers.info/coronavirus/ (accessed on 18 April 2024).
- Doleman, G.; De Leo, A.; Bloxsome, D. The impact of pandemics on healthcare providers’ workloads: A scoping review. J. Adv. Nurs. 2023, 79, 4434–4454. [Google Scholar] [CrossRef]
- Liu, R.; Han, H.; Liu, F.; Lv, Z.; Wu, K.; Liu, Y.; Feng, Y.; Zhu, C. Positive rate of RT-PCR detection of SARS-CoV-2 infection in 4880 cases from one hospital in Wuhan, China, from Jan to Feb 2020. Clin. Chim. Acta Int. J. Clin. Chem. 2020, 505, 172–175. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; Holmes, E.C.; Hill, V.; O’Toole, Á.; McCrone, J.T.; Ruis, C.; du Plessis, L.; Pybus, O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020, 5, 1403–1407. [Google Scholar] [CrossRef] [PubMed]
- Brinkmann, A.; Ulm, S.-L.; Uddin, S.; Förster, S.; Seifert, D.; Oehme, R.; Corty, M.; Schaade, L.; Michel, J.; Nitsche, A. AmpliCoV: Rapid Whole-Genome Sequencing Using Multiplex PCR Amplification and Real-Time Oxford Nanopore MinION Sequencing Enables Rapid Variant Identification of SARS-CoV-2. Front. Microbiol. 2021, 12, 651151. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Wu, K.; Chen, J.; Liu, H.; Huang, Y.; Zhang, Y.; Xiong, J.; Quan, W.; Wu, X.; Liang, Y.; et al. Rapid Acquisition of High-Quality SARS-CoV-2 Genome via Amplicon-Oxford Nanopore Sequencing. Virol. Sin. 2021, 36, 901–912. [Google Scholar] [CrossRef] [PubMed]
- Bull, R.A.; Adikari, T.N.; Ferguson, J.M.; Hammond, J.M.; Stevanovski, I.; Beukers, A.G.; Naing, Z.; Yeang, M.; Verich, A.; Gamaarachchi, H.; et al. Analytical validity of nanopore sequencing for rapid SARS-CoV-2 genome analysis. Nat. Commun. 2020, 11, 6272. [Google Scholar] [CrossRef] [PubMed]
- Sedlazeck, F.J.; Lee, H.; Darby, C.A.; Schatz, M.C. Piercing the dark matter: Bioinformatics of long-range sequencing and mapping. Nat. Rev. Genet. 2018, 19, 329–346. [Google Scholar] [CrossRef] [PubMed]
- Artic Network. Available online: https://artic.network/ncov-2019/ncov2019-bioinformatics-sop.html (accessed on 13 March 2023).
- Lang, J. NanoCoV19: An analytical pipeline for rapid detection of severe acute respiratory syndrome coronavirus 2. Front. Genet. 2022, 13, 1008792. [Google Scholar] [CrossRef] [PubMed]
- Brandt, C.; Krautwurst, S.; Spott, R.; Lohde, M.; Jundzill, M.; Marquet, M.; Hölzer, M. poreCov-An Easy to Use, Fast, and Robust Workflow for SARS-CoV-2 Genome Reconstruction via Nanopore Sequencing. Front. Genet. 2021, 12, 711437. [Google Scholar] [CrossRef] [PubMed]
- Mata Aroco, P. SARS-CoV-2 Whole Genome Sequencing Data Analysis for National Viral Monitoring in Public Health. Master’s Thesis, E.T.S. de Ingeniería Agronómica, Alimentaria y de Biosistemas (UPM), Madrid, Spain, 2023. Available online: https://oa.upm.es/75342/ (accessed on 30 November 2023).
- Geneious|Bioinformatics Software for Sequence Data Analysis. Geneious. Available online: https://www.geneious.com/ (accessed on 17 October 2023).
- O’Toole, Á.; Scher, E.; Underwood, A.; Jackson, B.; Hill, V.; McCrone, J.T.; Colquhoun, R.; Ruis, C.; Abu-Dahab, K.; Taylor, B.; et al. Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool. Virus Evol. 2021, 7, veab064. [Google Scholar] [CrossRef] [PubMed]
- Aksamentov, I.; Roemer, C.; Hodcroft, E.B.; Neher, R.A. Nextclade: Clade assignment, mutation calling and quality control for viral genomes. J. Open Source Softw. 2021, 6, 3773. [Google Scholar] [CrossRef]
- Martinell, M.; Andersson, T.; Mannsverk, S.S.; Bergholm, J.; Ellström, P.; Hill, A.; Lindh, J.; Kaden, R. In-Flight Transmission of a SARS-CoV-2 Lineage B.1.617.2 Harbouring the Rare S:E484Q Immune Escape Mutation. Viruses 2022, 14, 504. [Google Scholar] [CrossRef] [PubMed]
- Mannsverk, S.; Bergholm, J.; Palanisamy, N.; Ellström, P.; Kaden, R.; Lindh, J.; Lennerstrand, J. SARS-CoV-2 variants of concern and spike protein mutational dynamics in a Swedish cohort during 2021, studied by Nanopore sequencing. Virol. J. 2022, 19, 164. [Google Scholar] [CrossRef] [PubMed]
- Haars, J.; Palanisamy, N.; Wallin, F.; Mölling, P.; Lindh, J.; Sundqvist, M.; Ellström, P.; Kaden, R.; Lennerstrand, J. Prevalence of SARS-CoV-2 Omicron Sublineages and Spike Protein Mutations Conferring Resistance against Monoclonal Antibodies in a Swedish Cohort during 2022–2023. Microorganisms 2023, 11, 2417. [Google Scholar] [CrossRef] [PubMed]
- Freed, N.; Silander, O. SARS-CoV2 genome sequencing protocol (1200 bp amplicon ‘midnight’ primer set, using Nanopore Rapid kit) v5. In nCoV-2019 Sequencing Protocol v2 (GunIt); protocols.io: Berkeley, CA, USA, 2021. [Google Scholar] [CrossRef]
- Bushnell, B. BBMap. Available online: https://sourceforge.net/projects/bbmap/ (accessed on 2 February 2023).
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef]
- Sequencing of SARS-CoV-2—First Update. Available online: https://www.ecdc.europa.eu/en/publications-data/sequencing-sars-cov-2 (accessed on 4 October 2023).
- Shu, Y.; McCauley, J. GISAID: Global initiative on sharing all influenza data—From vision to reality. Eurosurveillance 2017, 22, 30494. [Google Scholar] [CrossRef] [PubMed]
- Wingett, S.W.; Andrews, S. FastQ Screen: A tool for multi-genome mapping and quality control. F1000Research 2018, 7, 1338. [Google Scholar] [CrossRef]
| Sample | Lineage (Pangolin Web Interface) | Lineage (Pangolin Plugin) | Lineage (Nextclade Web Interface) | Lineage (Nextclade Plugin) |
|---|---|---|---|---|
| 1 | B.1.1.7 | B.1.1.7 | B.1.1.7 | B.1.1.7 |
| 4 | AY.9.2 | AY.9.2 | AY.9.2 | AY.9.2 |
| 5 | AY.94 | AY.94 | AY.94 | AY.94 |
| 6 | AY.122 | AY.122 | AY.122 | AY.122 |
| 7 | BA.1 | BA.1 | BA.1 | BA.1 |
| 9 | BA.2 | BA.2 | BA.2 | BA.2 |
| 10 | BA.4.2 | BA.4.2 | BA.4.2 | BA.4.2 |
| 11 | BA.5 | BA.5 | BA.5.3 | BA.5.3 |
| 13 | BQ.1 | BQ.1 | BQ.1 | BQ.1 |
| 14 | BQ.1.1 | BQ.1.1 | BQ.1.1 | BQ.1.1 |
| 15 | XBB.1.5 | XBB.1.5 | XBB.1.5 | XBB.1.5 |
| 16 | XBB.1.9.2 | XBB.1.9.2 | XBB.1.9.2 | XBB.1.9.2 |
| 17 | CH.1.1.19 | CH.1.1.19 | CH.1.1.19 | CH.1.1.19 |
| 18 | EG.5.1.1 | EG.5.1.1 | EG.5.1.1 | EG.5.1.1 |
| File | Purpose |
|---|---|
| SARS-CoV-2_Assembly_Basic.geneiousWorkflow | The basic workflow without pango-lineage assignment. |
| SARS-CoV-2_Assembly_WrapperPlugins.geneiousWorkflow | The workflow that includes pango-lineage assignment. |
| SARS-CoV-2 reference sequence.geneious (NC_045512) | SARS-CoV2 Wuhan-Hu-1 reference sequence for alignment including gene annotations. |
| Annotation features (Dec 2023 version).geneious | Example database of common mutations. |
| Primer SARS-CoV-2 Artic.geneious | Artic primers that are to be trimmed from the data. |
| Primer SARS-CoV-2 Midnight1200.geneious | Midnight primers that are to be trimmed from the data. |
| Software | Workflow | Link |
|---|---|---|
| Geneious Prime (version 2021.1.1 or later) | Both | geneious.com (accessed on 17 October 2023) |
| Python 3 | “SARS-CoV-2_Assembly_WrapperPlugins” | python.org (accessed on 5 December 2023) |
| Docker Desktop (version 4.9.0 or later) | “SARS-CoV-2_Assembly_WrapperPlugins” | docker.com (accessed on 5 December 2023) |
| Nextclade CLI (version 2.14.0 or later) | “SARS-CoV-2_Assembly_WrapperPlugins” | nextstrain.org (accessed on 21 December 2023) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cumlin, T.; Karlsson, I.; Haars, J.; Rosengren, M.; Lennerstrand, J.; Pimushyna, M.; Feuk, L.; Ladenvall, C.; Kaden, R. From SARS-CoV-2 to Global Preparedness: A Graphical Interface for Standardised High-Throughput Bioinformatics Analysis in Pandemic Scenarios and Surveillance of Drug Resistance. Int. J. Mol. Sci. 2024, 25, 6645. https://doi.org/10.3390/ijms25126645
Cumlin T, Karlsson I, Haars J, Rosengren M, Lennerstrand J, Pimushyna M, Feuk L, Ladenvall C, Kaden R. From SARS-CoV-2 to Global Preparedness: A Graphical Interface for Standardised High-Throughput Bioinformatics Analysis in Pandemic Scenarios and Surveillance of Drug Resistance. International Journal of Molecular Sciences. 2024; 25(12):6645. https://doi.org/10.3390/ijms25126645
Chicago/Turabian StyleCumlin, Tomas, Ida Karlsson, Jonathan Haars, Maria Rosengren, Johan Lennerstrand, Maryna Pimushyna, Lars Feuk, Claes Ladenvall, and Rene Kaden. 2024. "From SARS-CoV-2 to Global Preparedness: A Graphical Interface for Standardised High-Throughput Bioinformatics Analysis in Pandemic Scenarios and Surveillance of Drug Resistance" International Journal of Molecular Sciences 25, no. 12: 6645. https://doi.org/10.3390/ijms25126645
APA StyleCumlin, T., Karlsson, I., Haars, J., Rosengren, M., Lennerstrand, J., Pimushyna, M., Feuk, L., Ladenvall, C., & Kaden, R. (2024). From SARS-CoV-2 to Global Preparedness: A Graphical Interface for Standardised High-Throughput Bioinformatics Analysis in Pandemic Scenarios and Surveillance of Drug Resistance. International Journal of Molecular Sciences, 25(12), 6645. https://doi.org/10.3390/ijms25126645






