Adipocyte Mitochondria: Deciphering Energetic Functions across Fat Depots in Obesity and Type 2 Diabetes
Abstract
1. Introduction
2. White Adipocyte Function
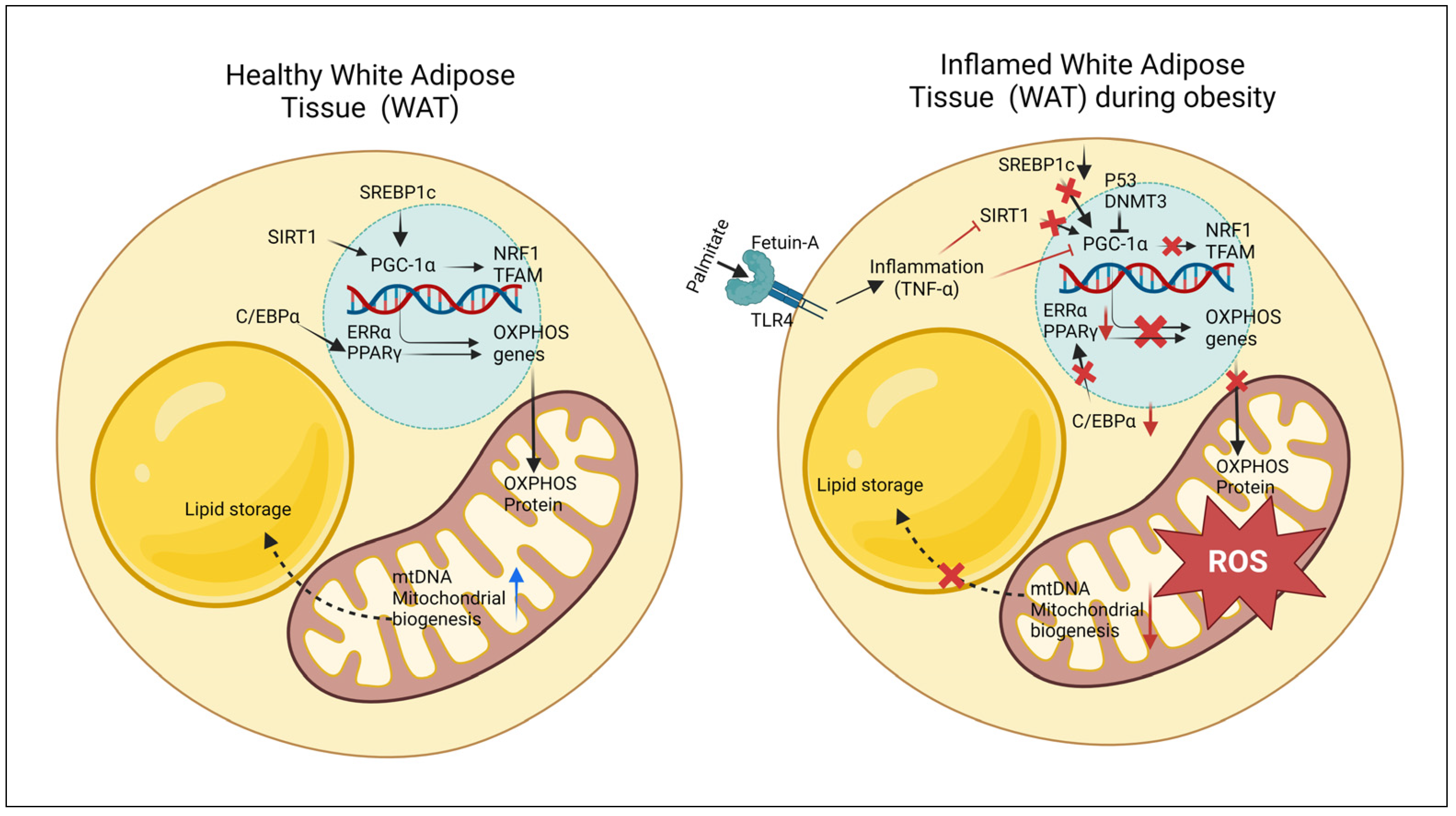
3. Mitochondrial Dysfunction in White Adipocytes in Obesity
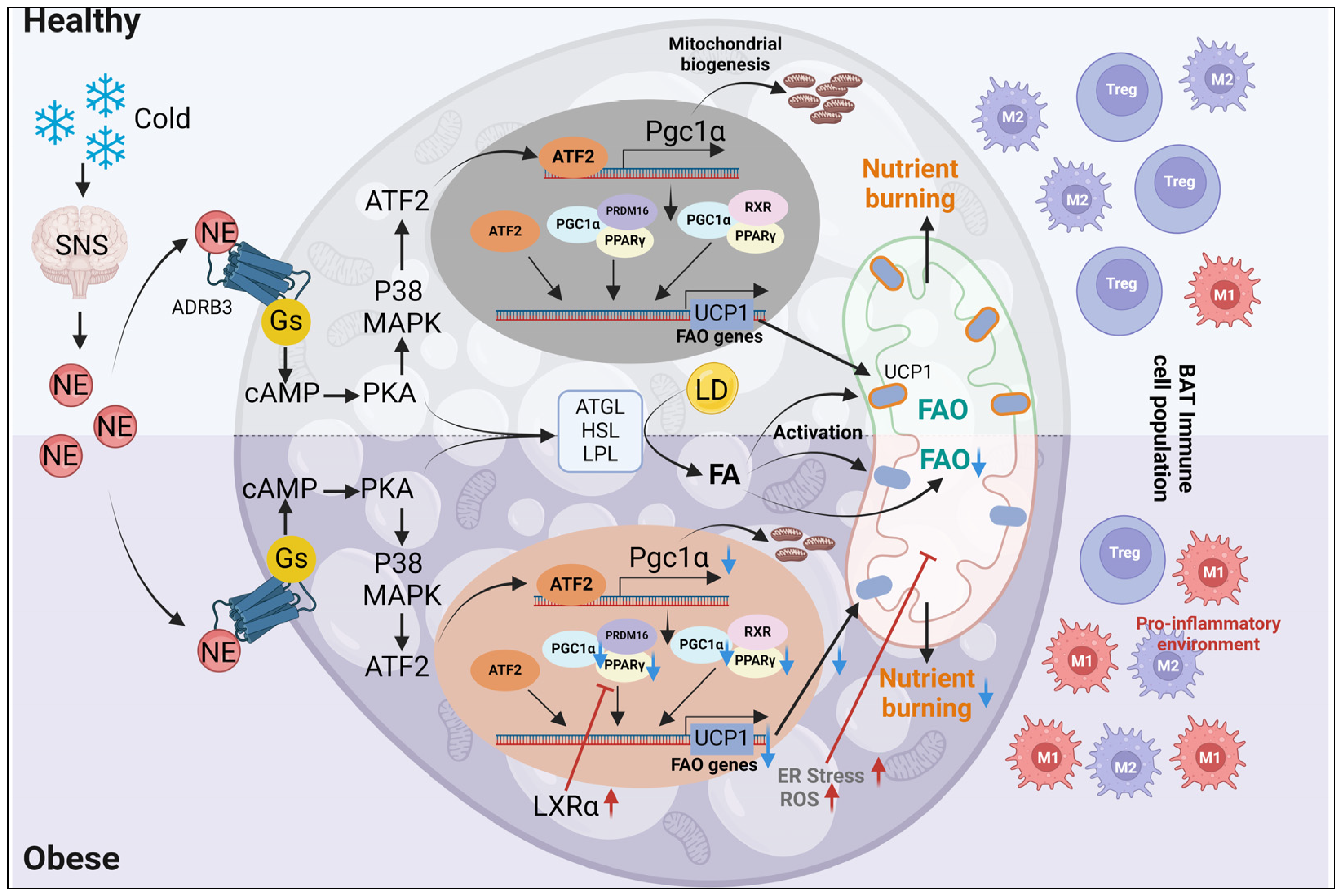
4. Obesity-Induced Changes in Mitochondrial Function in Brown Adipocytes
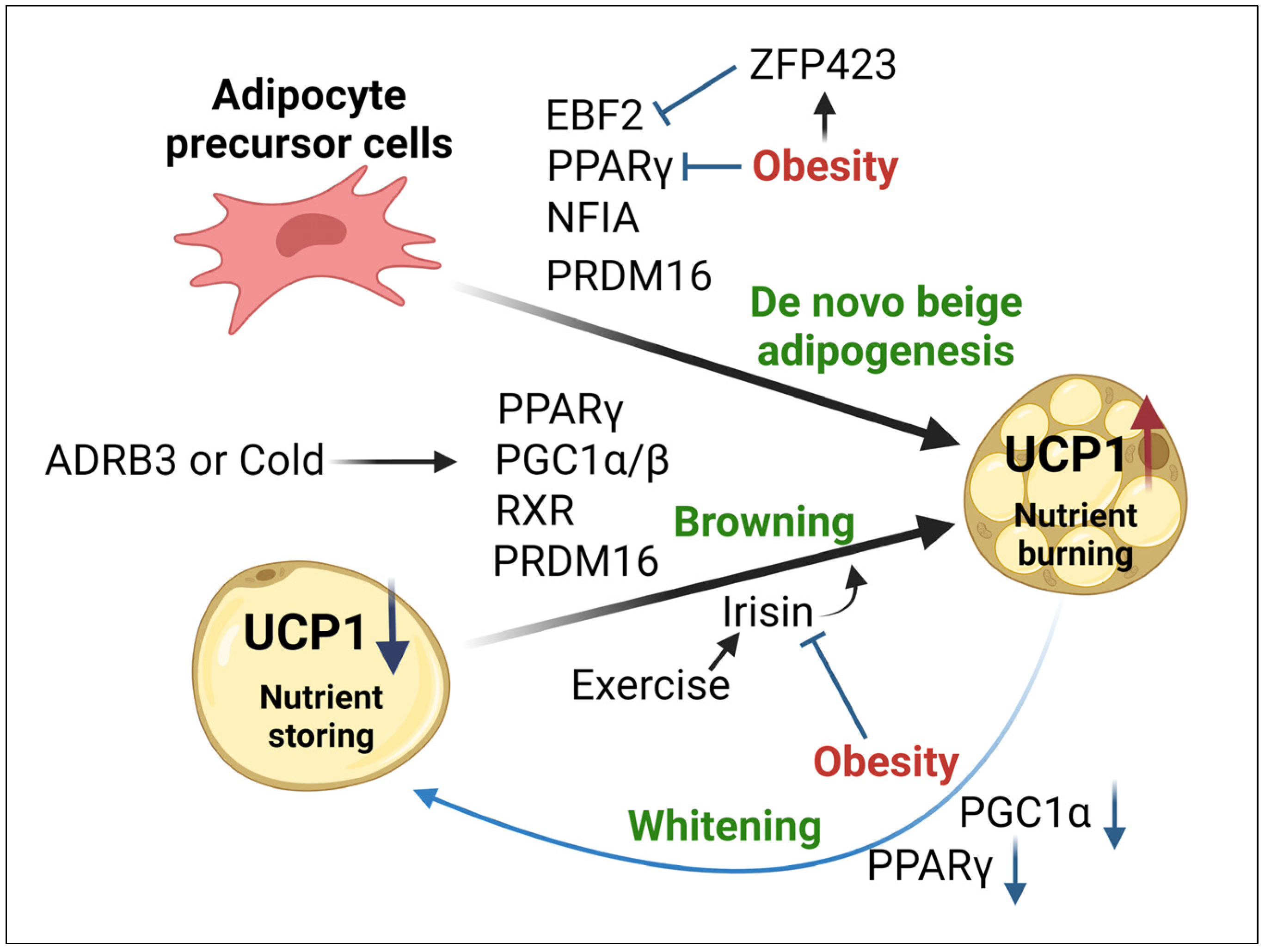
5. Mitochondrial Impairments in Beige Adipocytes during Obesity
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Carobbio, S.; Guénantin, A.C.; Samuelson, I.; Bahri, M.; Vidal-Puig, A. Brown and beige fat: From molecules to physiology and pathophysiology. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2019, 1864, 37–50. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Gurmaches, J.; Guertin, D.A. Adipocytes arise from multiple lineages that are heterogeneously and dynamically distributed. Nat. Commun. 2014, 5, 4099. [Google Scholar] [CrossRef] [PubMed]
- Van Nguyen, T.T.; Vu, V.V.; Pham, P.V. Transcriptional Factors of Thermogenic Adipocyte Development and Generation of Brown and Beige Adipocytes From Stem Cells. Stem Cell Rev. Rep. 2020, 16, 876–892. [Google Scholar] [CrossRef] [PubMed]
- Ziqubu, K.; Dludla, P.V.; Mthembu, S.X.H.; Nkambule, B.B.; Mabhida, S.E.; Jack, B.U.; Nyambuya, T.M.; Mazibuko-Mbeje, S.E. An insight into brown/beige adipose tissue whitening, a metabolic complication of obesity with the multifactorial origin. Front. Endocrinol. 2023, 14, 1114767. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Bostrom, P.; Sparks, L.M.; Ye, L.; Choi, J.H.; Giang, A.H.; Khandekar, M.; Virtanen, K.A.; Nuutila, P.; Schaart, G.; et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell 2012, 150, 366–376. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, M.M. Subcutaneous and visceral adipose tissue: Structural and functional differences. Obes. Rev. 2010, 11, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Kwok, K.H.; Lam, K.S.; Xu, A. Heterogeneity of white adipose tissue: Molecular basis and clinical implications. Exp. Mol. Med. 2016, 48, e215. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Hernandez, A.; Beneit, N.; Diaz-Castroverde, S.; Escribano, O. Differential Role of Adipose Tissues in Obesity and Related Metabolic and Vascular Complications. Int. J. Endocrinol. 2016, 2016, 1216783. [Google Scholar] [CrossRef] [PubMed]
- Goossens, G.H. The Metabolic Phenotype in Obesity: Fat Mass, Body Fat Distribution, and Adipose Tissue Function. Obes. Facts 2017, 10, 207–215. [Google Scholar] [CrossRef]
- Kusminski, C.M.; Scherer, P.E. Mitochondrial dysfunction in white adipose tissue. Trends Endocrinol. Metab. 2012, 23, 435–443. [Google Scholar] [CrossRef]
- Lee, J.H.; Park, A.; Oh, K.J.; Lee, S.C.; Kim, W.K.; Bae, K.H. The Role of Adipose Tissue Mitochondria: Regulation of Mitochondrial Function for the Treatment of Metabolic Diseases. Int. J. Mol. Sci. 2019, 20, 4924. [Google Scholar] [CrossRef] [PubMed]
- Bhatti, J.S.; Bhatti, G.K.; Reddy, P.H. Mitochondrial dysfunction and oxidative stress in metabolic disorders—A step towards mitochondria based therapeutic strategies. Bba-Mol. Basis Dis. 2017, 1863, 1066–1077. [Google Scholar] [CrossRef] [PubMed]
- Prasun, P. Mitochondrial dysfunction in metabolic syndrome. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165838. [Google Scholar] [CrossRef] [PubMed]
- Kajimura, S.; Spiegelman, B.M.; Seale, P. Brown and Beige Fat: Physiological Roles beyond Heat Generation. Cell Metab. 2015, 22, 546–559. [Google Scholar] [CrossRef] [PubMed]
- Park, H.; He, A.; Lodhi, I.J. Lipid Regulators of Thermogenic Fat Activation. Trends Endocrinol. Metab. 2019, 30, 710–723. [Google Scholar] [CrossRef] [PubMed]
- Lynes, M.D.; Kodani, S.D.; Tseng, Y.H. Lipokines and Thermogenesis. Endocrinology 2019, 160, 2314–2325. [Google Scholar] [CrossRef] [PubMed]
- Longo, M.; Zatterale, F.; Naderi, J.; Parrillo, L.; Formisano, P.; Raciti, G.A.; Beguinot, F.; Miele, C. Adipose Tissue Dysfunction as Determinant of Obesity-Associated Metabolic Complications. Int. J. Mol. Sci. 2019, 20, 2358. [Google Scholar] [CrossRef] [PubMed]
- Wikstrom, J.D.; Mahdaviani, K.; Liesa, M.; Sereda, S.B.; Si, Y.; Las, G.; Twig, G.; Petrovic, N.; Zingaretti, C.; Graham, A.; et al. Hormone-induced mitochondrial fission is utilized by brown adipocytes as an amplification pathway for energy expenditure. EMBO J. 2014, 33, 418–436. [Google Scholar] [CrossRef] [PubMed]
- Cannon, B.; Nedergaard, J. Brown adipose tissue: Function and physiological significance. Physiol. Rev. 2004, 84, 277–359. [Google Scholar] [CrossRef]
- Rosen, E.D.; Spiegelman, B.M. Adipocytes as regulators of energy balance and glucose homeostasis. Nature 2006, 444, 847–853. [Google Scholar] [CrossRef]
- Curtis, J.M.; Grimsrud, P.A.; Wright, W.S.; Xu, X.; Foncea, R.E.; Graham, D.W.; Brestoff, J.R.; Wiczer, B.M.; Ilkayeva, O.; Cianflone, K.; et al. Downregulation of Adipose Glutathione S-Transferase A4 Leads to Increased Protein Carbonylation, Oxidative Stress, and Mitochondrial Dysfunction. Diabetes 2010, 59, 1132–1142. [Google Scholar] [CrossRef] [PubMed]
- Lolmede, K.; Duffaut, C.; Zakaroff-Girard, A.; Bouloumie, A. Immune cells in adipose tissue: Key players in metabolic disorders. Diabetes Metab. 2011, 37, 283–290. [Google Scholar] [CrossRef]
- Sun, K.; Kusminski, C.M.; Scherer, P.E. Adipose tissue remodeling and obesity. J. Clin. Invest. 2011, 121, 2094–2101. [Google Scholar] [CrossRef]
- Wronska, A.; Kmiec, Z. Structural and biochemical characteristics of various white adipose tissue depots. Acta Physiol. (Oxf.) 2012, 205, 194–208. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y. Angiogenesis and vascular functions in modulation of obesity, adipose metabolism, and insulin sensitivity. Cell Metab. 2013, 18, 478–489. [Google Scholar] [CrossRef] [PubMed]
- Sun, K.; Tordjman, J.; Clement, K.; Scherer, P.E. Fibrosis and adipose tissue dysfunction. Cell Metab. 2013, 18, 470–477. [Google Scholar] [CrossRef]
- Wang, Q.A.; Tao, C.; Gupta, R.K.; Scherer, P.E. Tracking adipogenesis during white adipose tissue development, expansion and regeneration. Nat. Med. 2013, 19, 1338–1344. [Google Scholar] [CrossRef]
- Pellegrinelli, V.; Carobbio, S.; Vidal-Puig, A. Adipose tissue plasticity: How fat depots respond differently to pathophysiological cues. Diabetologia 2016, 59, 1075–1088. [Google Scholar] [CrossRef]
- Carrière, A.; Fernandez, Y.; Rigoulet, M.; Pénicaud, L.; Casteilla, L. Inhibition of preadipocyte proliferation by mitochondrial reactive oxygen species. Febs Lett. 2003, 550, 163–167. [Google Scholar] [CrossRef]
- Kajimura, S.; Seale, P.; Tomaru, T.; Erdjument-Bromage, H.; Cooper, M.P.; Ruas, J.L.; Chin, S.; Tempst, P.; Lazar, M.A.; Spiegelman, B.M. Regulation of the brown and white fat gene programs through a PRDM16/CtBP transcriptional complex. Genes Dev. 2008, 22, 1397–1409. [Google Scholar] [CrossRef]
- Seale, P.; Kajimura, S.; Yang, W.; Chin, S.; Rohas, L.M.; Uldry, M.; Tavernier, G.; Langin, D.; Spiegelman, B.M. Transcriptional control of brown fat determination by PRDM16. Cell Metab. 2007, 6, 38–54. [Google Scholar] [CrossRef]
- Ohno, H.; Shinoda, K.; Spiegelman, B.M.; Kajimura, S. PPARgamma agonists induce a white-to-brown fat conversion through stabilization of PRDM16 protein. Cell Metab. 2012, 15, 395–404. [Google Scholar] [CrossRef] [PubMed]
- Harms, M.; Seale, P. Brown and beige fat: Development, function and therapeutic potential. Nat. Med. 2013, 19, 1252–1263. [Google Scholar] [CrossRef]
- Bae, J.Y.; Chen, J.G.; Zhao, L. Chronic activation of pattern recognition receptors suppresses brown adipogenesis of multipotent mesodermal stem cells and brown pre-adipocytes. Biochem. Cell Biol. 2015, 93, 251–261. [Google Scholar] [CrossRef]
- Castro, J.P.; Grune, T.; Speckmann, B. The two faces of reactive oxygen species (ROS) in adipocyte function and dysfunction. Biol. Chem. 2016, 397, 709–724. [Google Scholar] [CrossRef]
- Jones, D.P.; Sies, H. The Redox Code. Antioxid. Redox Signal 2015, 23, 734–746. [Google Scholar] [CrossRef] [PubMed]
- Mracek, T.; Cannon, B.; Houstek, J. IL-1 and LPS but not IL-6 inhibit differentiation and downregulate PPAR gamma in brown adipocytes. Cytokine 2004, 26, 9–15. [Google Scholar] [CrossRef]
- Sanchez-Infantes, D.; Cereijo, R.; Peyrou, M.; Piquer-Garcia, I.; Stephens, J.M.; Villarroya, F. Oncostatin m impairs brown adipose tissue thermogenic function and the browning of subcutaneous white adipose tissue. Obesity (Silver Spring) 2017, 25, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Zoller, V.; Funcke, J.B.; Keuper, M.; Abd El Hay, M.; Debatin, K.M.; Wabitsch, M.; Fischer-Posovszky, P. TRAIL (TNF-related apoptosis-inducing ligand) inhibits human adipocyte differentiation via caspase-mediated downregulation of adipogenic transcription factors. Cell Death Dis. 2016, 7, e2412. [Google Scholar] [CrossRef][Green Version]
- Xia, W.; Veeragandham, P.; Cao, Y.; Xu, Y.; Rhyne, T.E.; Qian, J.; Hung, C.W.; Zhao, P.; Jones, Y.; Gao, H.; et al. Obesity causes mitochondrial fragmentation and dysfunction in white adipocytes due to RalA activation. Nat. Metab. 2024, 6, 273–289. [Google Scholar] [CrossRef]
- Xiong, X.Q.; Chen, D.; Sun, H.J.; Ding, L.; Wang, J.J.; Chen, Q.; Li, Y.H.; Zhou, Y.B.; Han, Y.; Zhang, F.; et al. FNDC5 overexpression and irisin ameliorate glucose/lipid metabolic derangements and enhance lipolysis in obesity. Biochim. Biophys. Acta 2015, 1852, 1867–1875. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, R.; Meng, Y.; Li, S.; Donelan, W.; Zhao, Y.; Qi, L.; Zhang, M.; Wang, X.; Cui, T.; et al. Irisin stimulates browning of white adipocytes through mitogen-activated protein kinase p38 MAP kinase and ERK MAP kinase signaling. Diabetes 2014, 63, 514–525. [Google Scholar] [CrossRef] [PubMed]
- Pereira, R.O.; Marti, A.; Olvera, A.C.; Tadinada, S.M.; Bjorkman, S.H.; Weatherford, E.T.; Morgan, D.A.; Westphal, M.; Patel, P.H.; Kirby, A.K.; et al. OPA1 deletion in brown adipose tissue improves thermoregulation and systemic metabolism via FGF21. eLife 2021, 10, e66519. [Google Scholar] [CrossRef] [PubMed]
- Yao, C.H.; Wang, R.; Wang, Y.; Kung, C.P.; Weber, J.D.; Patti, G.J. Mitochondrial fusion supports increased oxidative phosphorylation during cell proliferation. eLife 2019, 8, e41351. [Google Scholar] [CrossRef]
- Krisko, T.I.; Nicholls, H.T.; Bare, C.J.; Holman, C.D.; Putzel, G.G.; Jansen, R.S.; Sun, N.; Rhee, K.Y.; Banks, A.S.; Cohen, D.E. Dissociation of Adaptive Thermogenesis from Glucose Homeostasis in Microbiome-Deficient Mice. Cell Metab. 2020, 31, 592–604.e9. [Google Scholar] [CrossRef] [PubMed]
- Choo, H.J.; Kim, J.H.; Kwon, O.B.; Lee, C.S.; Mun, J.Y.; Han, S.S.; Yoon, Y.S.; Yoon, G.; Choi, K.M.; Ko, Y.G. Mitochondria are impaired in the adipocytes of type 2 diabetic mice. Diabetologia 2006, 49, 784–791. [Google Scholar] [CrossRef] [PubMed]
- Fruhbeck, G. Overview of adipose tissue and its role in obesity and metabolic disorders. Methods Mol. Biol. 2008, 456, 1–22. [Google Scholar] [PubMed]
- Salvador, J.; Silva, C.; Pujante, P.; Fruhbeck, G. Abdominal obesity: An indicator of cardiometabolic risk. Endocrinol. Nutr. 2008, 55, 420–432. [Google Scholar] [CrossRef] [PubMed]
- Gabriely, I.; Ma, X.H.; Yang, X.M.; Atzmon, G.; Rajala, M.W.; Berg, A.H.; Scherer, P.; Rossetti, L.; Barzilai, N. Removal of visceral fat prevents insulin resistance and glucose intolerance of aging: An adipokine-mediated process? Diabetes 2002, 51, 2951–2958. [Google Scholar] [CrossRef]
- Indulekha, K.; Anjana, R.M.; Surendar, J.; Mohan, V. Association of visceral and subcutaneous fat with glucose intolerance, insulin resistance, adipocytokines and inflammatory markers in Asian Indians (CURES-113). Clin. Biochem. 2011, 44, 281–287. [Google Scholar] [CrossRef]
- Preis, S.R.; Massaro, J.M.; Robins, S.J.; Hoffmann, U.; Vasan, R.S.; Irlbeck, T.; Meigs, J.B.; Sutherland, P.; D’Agostino, R.B., Sr.; O’Donnell, C.J.; et al. Abdominal subcutaneous and visceral adipose tissue and insulin resistance in the Framingham heart study. Obesity (Silver Spring) 2010, 18, 2191–2198. [Google Scholar] [CrossRef]
- Zhang, P.; He, Y.; Wu, S.; Li, X.; Lin, X.; Gan, M.; Chen, L.; Zhao, Y.; Niu, L.; Zhang, S.; et al. Factors Associated with White Fat Browning: New Regulators of Lipid Metabolism. Int. J. Mol. Sci. 2022, 23, 7641. [Google Scholar] [CrossRef]
- Kloting, N.; Fasshauer, M.; Dietrich, A.; Kovacs, P.; Schon, M.R.; Kern, M.; Stumvoll, M.; Bluher, M. Insulin-sensitive obesity. Am. J. Physiol. Endocrinol. Metab. 2010, 299, E506–E515. [Google Scholar] [CrossRef]
- Deveaud, C.; Beauvoit, B.; Salin, B.; Schaeffer, J.; Rigoulet, M. Regional differences in oxidative capacity of rat white adipose tissue are linked to the mitochondrial content of mature adipocytes. Mol. Cell. Biochem. 2004, 267, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Kraunsoe, R.; Boushel, R.; Hansen, C.N.; Schjerling, P.; Qvortrup, K.; Stockel, M.; Mikines, K.J.; Dela, F. Mitochondrial respiration in subcutaneous and visceral adipose tissue from patients with morbid obesity. J. Physiol. 2010, 588 Pt 12, 2023–2032. [Google Scholar] [CrossRef]
- Asterholm, I.W.; Mundy, D.I.; Weng, J.; Anderson, R.G.W.; Scherer, P.E. Altered Mitochondrial Function and Metabolic Inflexibility Associated with Loss of Caveolin-1. Cell Metab. 2012, 15, 171–185. [Google Scholar]
- Wilson-Fritch, L.; Nicoloro, S.; Chouinard, M.; Lazar, M.A.; Chui, P.C.; Leszyk, J.; Straubhaar, J.; Czech, M.P.; Corvera, S. Mitochondrial remodeling in adipose tissue associated with obesity and treatment with rosiglitazone. J. Clin. Investig. 2004, 114, 1281–1289. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, M.; Deguchi, Y.; Nozaki, Y.; Higami, Y. Contribution of PGC-1alpha to Obesity- and Caloric Restriction-Related Physiological Changes in White Adipose Tissue. Int. J. Mol. Sci. 2021, 22, 6025. [Google Scholar] [CrossRef] [PubMed]
- Gleyzer, N.; Vercauteren, K.; Scarpulla, R.C. Control of Mitochondrial Transcription Specificity Factors (TFB1M and TFB2M) by Nuclear Respiratory Factors (NRF-1 and NRF-2) and PGC-1 Family Coactivators. Mol. Cell. Biol. 2005, 25, 1354–1366. [Google Scholar] [CrossRef]
- Vina, J.; Gomez-Cabrera, M.C.; Borras, C.; Froio, T.; Sanchis-Gomar, F.; Martinez-Bello, V.E.; Pallardo, F.V. Mitochondrial biogenesis in exercise and in ageing. Adv. Drug Deliv. Rev. 2009, 61, 1369–1374. [Google Scholar] [CrossRef]
- Heinonen, S.; Buzkova, J.; Muniandy, M.; Kaksonen, R.; Ollikainen, M.; Ismail, K.; Hakkarainen, A.; Lundbom, J.; Lundbom, N.; Vuolteenaho, K.; et al. Impaired Mitochondrial Biogenesis in Adipose Tissue in Acquired Obesity. Diabetes 2015, 64, 3135–3145. [Google Scholar] [CrossRef] [PubMed]
- Heinonen, S.; Muniandy, M.; Buzkova, J.; Mardinoglu, A.; Rodríguez, A.; Frühbeck, G.; Hakkarainen, A.; Lundbom, J.; Lundbom, N.; Kaprio, J. Mitochondria-related transcriptional signature is downregulated in adipocytes in obesity: A study of young healthy MZ twins. Diabetologia 2017, 60, 169–181. [Google Scholar] [CrossRef] [PubMed]
- Kleiner, S.; Mepani, R.J.; Laznik, D.; Ye, L.; Jurczak, M.J.; Jornayvaz, F.R.; Estall, J.L.; Bhowmick, D.C.; Shulman, G.I.; Spiegelman, B.M. Development of insulin resistance in mice lacking PGC-1α in adipose tissues. Proc. Natl. Acad. Sci. USA 2012, 109, 9635–9640. [Google Scholar] [CrossRef] [PubMed]
- Devarakonda, S.; Gupta, K.; Chalmers, M.J.; Hunt, J.F.; Griffin, P.R.; Van Duyne, G.D.; Spiegelman, B.M. Disorder-to-order transition underlies the structural basis for the assembly of a transcriptionally active PGC-1α/ERRγ complex. Proc. Natl. Acad. Sci. USA 2011, 108, 18678–18683. [Google Scholar] [CrossRef] [PubMed]
- Valerio, A.; Cardile, A.; Cozzi, V.; Bracale, R.; Tedesco, L.; Pisconti, A.; Palomba, L.; Cantoni, O.; Clementi, E.; Moncada, S. TNF-α downregulates eNOS expression and mitochondrial biogenesis in fat and muscle of obese rodents. J. Clin. Investig. 2006, 116, 2791–2798. [Google Scholar] [CrossRef] [PubMed]
- Dasgupta, S.; Bhattacharya, S.; Biswas, A.; Majumdar, S.S.; Mukhopadhyay, S.; Ray, S.; Bhattacharya, S. NF-kappaB mediates lipid-induced fetuin-A expression in hepatocytes that impairs adipocyte function effecting insulin resistance. Biochem. J. 2010, 429, 451–462. [Google Scholar] [CrossRef]
- Pal, D.; Dasgupta, S.; Kundu, R.; Maitra, S.; Das, G.; Mukhopadhyay, S.; Ray, S.; Majumdar, S.S.; Bhattacharya, S. Fetuin-A acts as an endogenous ligand of TLR4 to promote lipid-induced insulin resistance. Nat. Med. 2012, 18, 1279–1285. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, M.; Mukherjee, S.; Chatterjee, S.K.; Chattopadhyay, D.; Das, S.; Majumdar, S.S.; Mukhopadhyay, S.; Mukherjee, S.; Bhattarcharya, S. Impairment of energy sensors, SIRT1 and AMPK, in lipid induced inflamed adipocyte is regulated by Fetuin A. Cell. Signal. 2018, 42, 67–76. [Google Scholar] [CrossRef]
- Das, S.; Chattopadhyay, D.; Chatterjee, S.K.; Mondal, S.A.; Majumdar, S.S.; Mukhopadhyay, S.; Saha, N.; Velayutham, R.; Bhattacharya, S.; Mukherjee, S. Increase in PPARgamma inhibitory phosphorylation by Fetuin-A through the activation of Ras-MEK-ERK pathway causes insulin resistance. Biochim. Biophys. Acta Mol. Basis Dis. 2021, 1867, 166050. [Google Scholar] [CrossRef]
- Gao, C.L.; Zhu, C.; Zhao, Y.P.; Chen, X.H.; Ji, C.B.; Zhang, C.M.; Zhu, J.G.; Xia, Z.K.; Tong, M.L.; Guo, X.R. Mitochondrial dysfunction is induced by high levels of glucose and free fatty acids in 3T3-L1 adipocytes. Mol. Cell Endocrinol. 2010, 320, 25–33. [Google Scholar] [CrossRef]
- Wenz, T. Regulation of mitochondrial biogenesis and PGC-1alpha under cellular stress. Mitochondrion 2013, 13, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Brookheart, R.T.; Michel, C.I.; Schaffer, J.E. As a Matter of Fat. Cell Metab. 2009, 10, 9–12. [Google Scholar] [CrossRef] [PubMed]
- Mori, M.; Nakagami, H.; Rodriguez-Araujo, G.; Nimura, K.; Kaneda, Y. Essential role for miR-196a in brown adipogenesis of white fat progenitor cells. PLoS Biol. 2012, 10, e1001314. [Google Scholar] [CrossRef] [PubMed]
- Gaudet, A.D.; Fonken, L.K.; Gushchina, L.V.; Aubrecht, T.G.; Maurya, S.K.; Periasamy, M.; Nelson, R.J.; Popovich, P.G. miR-155 Deletion in Female Mice Prevents Diet-Induced Obesity. Sci. Rep. 2016, 6, 22862. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Tang, M.; Xiao, T.; Liu, H.; Liu, W.; Li, G.; Zhang, F.; Xiao, Y.; Zhou, Z.; Liu, F.; et al. Obesity-Associated miR-199a/214 Cluster Inhibits Adipose Browning via PRDM16-PGC-1alpha Transcriptional Network. Diabetes 2018, 67, 2585–2600. [Google Scholar] [CrossRef] [PubMed]
- Valera-Alberni, M.; Canto, C. Mitochondrial stress management: A dynamic journey. Cell Stress 2018, 2, 253–274. [Google Scholar] [CrossRef] [PubMed]
- Murphy, M.P. How mitochondria produce reactive oxygen species. Biochem. J. 2009, 417, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Alcalá, M.; Sánchez-Vera, I.; Sevillano, J.; Herrero, L.; Serra, D.; Ramos, M.P.; Viana, M. Vitamin E reduces adipose tissue fibrosis, inflammation, and oxidative stress and improves metabolic profile in obesity. Obesity 2015, 23, 1598–1606. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, S.; Fujita, T.; Shimabukuro, M.; Iwaki, M.; Yamada, Y.; Nakajima, Y.; Nakayama, O.; Makishima, M.; Matsuda, M.; Shimomura, I. Increased oxidative stress in obesity and its impact on metabolic syndrome. J. Clin. Investig. 2004, 114, 1752–1761. [Google Scholar] [CrossRef]
- Jankovic, A.; Korac, A.; Srdic-Galic, B.; Buzadzic, B.; Otasevic, V.; Stancic, A.; Vucetic, M.; Markelic, M.; Velickovic, K.; Golic, I.; et al. Differences in the redox status of human visceral and subcutaneous adipose tissues-relationships to obesity and metabolic risk. Metabolism 2014, 63, 661–671. [Google Scholar] [CrossRef]
- Isakson, P.; Hammarstedt, A.; Gustafson, B.; Smith, U. Impaired preadipocyte differentiation in human abdominal obesity: Role of Wnt, tumor necrosis factor-alpha, and inflammation. Diabetes 2009, 58, 1550–1557. [Google Scholar] [CrossRef] [PubMed]
- Wilson-Fritch, L.; Burkart, A.; Bell, G.; Mendelson, K.; Leszyk, J.; Nicoloro, S.; Czech, M.; Corvera, S. Mitochondrial biogenesis and remodeling during adipogenesis and in response to the insulin sensitizer rosiglitazone. Mol. Cell. Biol. 2003, 23, 1085–1094. [Google Scholar] [CrossRef] [PubMed]
- Kaaman, M.; Sparks, L.M.; van Harmelen, V.; Smith, S.R.; Sjolin, E.; Dahlman, I.; Arner, P. Strong association between mitochondrial DNA copy number and lipogenesis in human white adipose tissue. Diabetologia 2007, 50, 2526–2533. [Google Scholar] [CrossRef] [PubMed]
- Rylova, S.N.; Albertioni, F.; Flygh, G.; Eriksson, S. Activity profiles of deoxynucleoside kinases and 5′-nucleotidases in cultured adipocytes and myoblastic cells: Insights into mitochondrial toxicity of nucleoside analogs. Biochem. Pharmacol. 2005, 69, 951–960. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Burkart, A.; Nicoloro, S.M.; Czech, M.P.; Straubhaar, J.; Corvera, S. Paradoxical effect of mitochondrial respiratory chain impairment on insulin signaling and glucose transport in adipose cells. J. Biol. Chem. 2008, 283, 30658–30667. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.H.; Wang, C.C.; Wei, Y.H. Mitochondrial dysfunction in insulin insensitivity: Implication of mitochondrial role in type 2 diabetes. Ann. N. Y Acad. Sci. 2010, 1201, 157–165. [Google Scholar] [CrossRef]
- Vankoningsloo, S.; De Pauw, A.; Houbion, A.; Tejerina, S.; Demazy, C.; de Longueville, F.; Bertholet, V.; Renard, P.; Remacle, J.; Holvoet, P.; et al. CREB activation induced by mitochondrial dysfunction triggers triglyceride accumulation in 3T3-L1 preadipocytes. J. Cell Sci. 2006, 119 Pt 7, 1266–1282. [Google Scholar] [CrossRef]
- Li, Y.; Fromme, T. Uncoupling Protein 1 Does Not Produce Heat without Activation. Int. J. Mol. Sci. 2022, 23, 2406. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, R.; Diwoky, C.; Schoiswohl, G.; Feiler, U.; Wongsiriroj, N.; Abdellatif, M.; Kolb, D.; Hoeks, J.; Kershaw, E.E.; Sedej, S.; et al. Cold-Induced Thermogenesis Depends on ATGL-Mediated Lipolysis in Cardiac Muscle, but Not Brown Adipose Tissue. Cell Metab. 2017, 26, 753–763. [Google Scholar] [CrossRef]
- Shin, H.; Ma, Y.; Chanturiya, T.; Cao, Q.; Wang, Y.; Kadegowda, A.K.G.; Jackson, R.; Rumore, D.; Xue, B.; Shi, H.; et al. Lipolysis in Brown Adipocytes Is Not Essential for Cold-Induced Thermogenesis in Mice. Cell Metab. 2017, 26, 764–777.e5. [Google Scholar] [CrossRef]
- Cinti, S. The adipose organ. Prostaglandins Leukot. Essent. Fatty Acids 2005, 73, 9–15. [Google Scholar] [CrossRef]
- Cypess, A.M.; Kahn, C.R. The role and importance of brown adipose tissue in energy homeostasis. Curr. Opin. Pediatr. 2010, 22, 478–484. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, K.; Yamada, T. UCP1 Dependent and Independent Thermogenesis in Brown and Beige Adipocytes. Front. Endocrinol. 2020, 11, 498. [Google Scholar] [CrossRef] [PubMed]
- Vernochet, C.; Damilano, F.; Mourier, A.; Bezy, O.; Mori, M.A.; Smyth, G.; Rosenzweig, A.; Larsson, N.G.; Kahn, C.R. Adipose tissue mitochondrial dysfunction triggers a lipodystrophic syndrome with insulin resistance, hepatosteatosis, and cardiovascular complications. FASEB J. 2014, 28, 4408–4419. [Google Scholar] [CrossRef] [PubMed]
- Orava, J.; Nuutila, P.; Noponen, T.; Parkkola, R.; Viljanen, T.; Enerback, S.; Rissanen, A.; Pietilainen, K.H.; Virtanen, K.A. Blunted metabolic responses to cold and insulin stimulation in brown adipose tissue of obese humans. Obesity (Silver Spring) 2013, 21, 2279–2287. [Google Scholar] [CrossRef]
- Cannon, B.; Jacobsson, A.; Rehnmark, S.; Nedergaard, J. Signal transduction in brown adipose tissue recruitment: Noradrenaline and beyond. Int. J. Obes. Relat. Metab. Disord. 1996, 20 (Suppl. 3), S36–S42. [Google Scholar]
- Sharma, B.K.; Patil, M.; Satyanarayana, A. Negative regulators of brown adipose tissue (BAT)-mediated thermogenesis. J. Cell Physiol. 2014, 229, 1901–1907. [Google Scholar] [CrossRef]
- Laurencikiene, J.; Ryden, M. Liver X receptors and fat cell metabolism. Int. J. Obes. (Lond.) 2012, 36, 1494–1502. [Google Scholar] [CrossRef]
- Wong, K.E.; Szeto, F.L.; Zhang, W.; Ye, H.; Kong, J.; Zhang, Z.; Sun, X.J.; Li, Y.C. Involvement of the vitamin D receptor in energy metabolism: Regulation of uncoupling proteins. Am. J. Physiol. Endocrinol. Metab. 2009, 296, E820–E828. [Google Scholar] [CrossRef]
- Narvaez, C.J.; Matthews, D.; Broun, E.; Chan, M.; Welsh, J. Lean phenotype and resistance to diet-induced obesity in vitamin D receptor knockout mice correlates with induction of uncoupling protein-1 in white adipose tissue. Endocrinology 2009, 150, 651–661. [Google Scholar] [CrossRef]
- Wong, K.E.; Kong, J.; Zhang, W.; Szeto, F.L.; Ye, H.; Deb, D.K.; Brady, M.J.; Li, Y.C. Targeted expression of human vitamin D receptor in adipocytes decreases energy expenditure and induces obesity in mice. J. Biol. Chem. 2011, 286, 33804–33810. [Google Scholar] [CrossRef] [PubMed]
- van den Berg, S.M.; van Dam, A.D.; Rensen, P.C.; de Winther, M.P.; Lutgens, E. Immune Modulation of Brown(ing) Adipose Tissue in Obesity. Endocr. Rev. 2017, 38, 46–68. [Google Scholar] [CrossRef]
- Nguyen, K.D.; Qiu, Y.; Cui, X.; Goh, Y.P.; Mwangi, J.; David, T.; Mukundan, L.; Brombacher, F.; Locksley, R.M.; Chawla, A. Alternatively activated macrophages produce catecholamines to sustain adaptive thermogenesis. Nature 2011, 480, 104–108. [Google Scholar] [CrossRef]
- Lee, Y.H.; Petkova, A.P.; Granneman, J.G. Identification of an adipogenic niche for adipose tissue remodeling and restoration. Cell Metab. 2013, 18, 355–367. [Google Scholar] [CrossRef]
- Shan, B.; Wang, X.; Wu, Y.; Xu, C.; Xia, Z.; Dai, J.; Shao, M.; Zhao, F.; He, S.; Yang, L.; et al. The metabolic ER stress sensor IRE1alpha suppresses alternative activation of macrophages and impairs energy expenditure in obesity. Nat. Immunol. 2017, 18, 519–529. [Google Scholar] [CrossRef] [PubMed]
- Medrikova, D.; Sijmonsma, T.P.; Sowodniok, K.; Richards, D.M.; Delacher, M.; Sticht, C.; Gretz, N.; Schafmeier, T.; Feuerer, M.; Herzig, S. Brown adipose tissue harbors a distinct sub-population of regulatory T cells. PLoS ONE 2015, 10, e0118534. [Google Scholar] [CrossRef]
- Alcala, M.; Calderon-Dominguez, M.; Bustos, E.; Ramos, P.; Casals, N.; Serra, D.; Viana, M.; Herrero, L. Increased inflammation, oxidative stress and mitochondrial respiration in brown adipose tissue from obese mice. Sci. Rep. 2017, 7, 16082. [Google Scholar] [CrossRef]
- Fitzgibbons, T.P.; Kogan, S.; Aouadi, M.; Hendricks, G.M.; Straubhaar, J.; Czech, M.P. Similarity of mouse perivascular and brown adipose tissues and their resistance to diet-induced inflammation. Am. J. Physiol. Heart Circ. Physiol. 2011, 301, H1425–H1437. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, T.; Nitta, T.; Maruno, K.; Yeh, Y.S.; Kuwata, H.; Tomita, K.; Goto, T.; Takahashi, N.; Kawada, T. Macrophage infiltration into obese adipose tissues suppresses the induction of UCP1 level in mice. Am. J. Physiol. Endocrinol. Metab. 2016, 310, E676–E687. [Google Scholar] [CrossRef]
- Alcala, M.; Calderon-Dominguez, M.; Serra, D.; Herrero, L.; Viana, M. Mechanisms of Impaired Brown Adipose Tissue Recruitment in Obesity. Front. Physiol. 2019, 10, 94. [Google Scholar] [CrossRef]
- Bae, J.; Ricciardi, C.J.; Esposito, D.; Komarnytsky, S.; Hu, P.; Curry, B.J.; Brown, P.L.; Gao, Z.G.; Biggerstaff, J.P.; Chen, J.G.; et al. Activation of pattern recognition receptors in brown adipocytes induces inflammation and suppresses uncoupling protein 1 expression and mitochondrial respiration. Am. J. Physiol-Cell Physiol. 2014, 306, C918–C930. [Google Scholar] [CrossRef] [PubMed]
- Ron, D.; Brasier, A.R.; McGehee, R.E., Jr.; Habener, J.F. Tumor necrosis factor-induced reversal of adipocytic phenotype of 3T3-L1 cells is preceded by a loss of nuclear CCAAT/enhancer binding protein (C/EBP). J. Clin. Investig. 1992, 89, 223–233. [Google Scholar] [CrossRef]
- Miranda, S.; Gonzalez-Rodriguez, A.; Revuelta-Cervantes, J.; Rondinone, C.M.; Valverde, A.M. Beneficial effects of PTP1B deficiency on brown adipocyte differentiation and protection against apoptosis induced by pro- and anti-inflammatory stimuli. Cell. Signal. 2010, 22, 645–659. [Google Scholar] [CrossRef]
- Valladares, A.; Alvarez, A.M.; Ventura, J.J.; Roncero, C.; Benito, M.; Porras, A. p38 mitogen-activated protein kinase mediates tumor necrosis factor-alpha-induced apoptosis in rat fetal brown adipocytes. Endocrinology 2000, 141, 4383–4395. [Google Scholar] [CrossRef]
- Liu, Z.; Gu, H.; Gan, L.; Xu, Y.; Feng, F.; Saeed, M.; Sun, C. Reducing Smad3/ATF4 was essential for Sirt1 inhibiting ER stress-induced apoptosis in mice brown adipose tissue. Oncotarget 2017, 8, 9267–9279. [Google Scholar] [CrossRef]
- Lim, S.; Honek, J.; Xue, Y.; Seki, T.; Cao, Z.; Andersson, P.; Yang, X.; Hosaka, K.; Cao, Y. Cold-induced activation of brown adipose tissue and adipose angiogenesis in mice. Nat. Protoc. 2012, 7, 606–615. [Google Scholar] [CrossRef]
- Xiao, C.; Goldgof, M.; Gavrilova, O.; Reitman, M.L. Anti-obesity and metabolic efficacy of the beta3-adrenergic agonist, CL316243, in mice at thermoneutrality compared to 22 degrees C. Obesity (Silver Spring) 2015, 23, 1450–1459. [Google Scholar] [CrossRef] [PubMed]
- Shao, M.; Wang, Q.A.; Song, A.; Vishvanath, L.; Busbuso, N.C.; Scherer, P.E.; Gupta, R.K. Cellular Origins of Beige Fat Cells Revisited. Diabetes 2019, 68, 1874–1885. [Google Scholar] [CrossRef] [PubMed]
- Shapira, S.N.; Seale, P. Transcriptional Control of Brown and Beige Fat Development and Function. Obesity (Silver Spring) 2019, 27, 13–21. [Google Scholar] [CrossRef]
- Seale, P.; Conroe, H.M.; Estall, J.; Kajimura, S.; Frontini, A.; Ishibashi, J.; Cohen, P.; Cinti, S.; Spiegelman, B.M. Prdm16 determines the thermogenic program of subcutaneous white adipose tissue in mice. J. Clin. Investig. 2011, 121, 96–105. [Google Scholar] [CrossRef]
- Perez-Sotelo, D.; Roca-Rivada, A.; Baamonde, I.; Baltar, J.; Castro, A.I.; Dominguez, E.; Collado, M.; Casanueva, F.F.; Pardo, M. Lack of Adipocyte-Fndc5/Irisin Expression and Secretion Reduces Thermogenesis and Enhances Adipogenesis. Sci. Rep. 2017, 7, 16289. [Google Scholar] [CrossRef] [PubMed]
- Cairo, M.; Villarroya, J. The role of autophagy in brown and beige adipose tissue plasticity. J. Physiol. Biochem. 2020, 76, 213–226. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, I.; Aprahamian, T.; Kikuchi, R.; Shimizu, A.; Papanicolaou, K.N.; MacLauchlan, S.; Maruyama, S.; Walsh, K. Vascular rarefaction mediates whitening of brown fat in obesity. J. Clin. Investig. 2014, 124, 2099–2112. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Li, L.; Li, M.; Lam, S.M.; Wang, G.; Wu, Y.; Zhang, H.; Niu, C.; Zhang, X.; Liu, X.; et al. Microbiota Depletion Impairs Thermogenesis of Brown Adipose Tissue and Browning of White Adipose Tissue. Cell Rep. 2019, 26, 2720–2737.e5. [Google Scholar] [CrossRef] [PubMed]
- Ng, R.; Hussain, N.A.; Zhang, Q.; Chang, C.; Li, H.; Fu, Y.; Cao, L.; Han, W.; Stunkel, W.; Xu, F. miRNA-32 Drives Brown Fat Thermogenesis and Trans-activates Subcutaneous White Fat Browning in Mice. Cell Rep. 2017, 19, 1229–1246. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Guan, M.; Townsend, K.L.; Huang, T.L.; An, D.; Yan, X.; Xue, R.; Schulz, T.J.; Winnay, J.; Mori, M.; et al. MicroRNA-455 regulates brown adipogenesis via a novel HIF1an-AMPK-PGC1alpha signaling network. EMBO Rep. 2015, 16, 1378–1393. [Google Scholar] [CrossRef] [PubMed]
- Tiraby, C.; Tavernier, G.; Lefort, C.; Larrouy, D.; Bouillaud, F.; Ricquier, D.; Langin, D. Acquirement of brown fat cell features by human white adipocytes. J. Biol. Chem. 2003, 278, 33370–33376. [Google Scholar] [CrossRef] [PubMed]
- Semple, R.; Crowley, V.; Sewter, C.; Laudes, M.; Christodoulides, C.; Considine, R.; Vidal-Puig, A.; O’rahilly, S. Expression of the thermogenic nuclear hormone receptor coactivator PGC-1 α is reduced in the adipose tissue of morbidly obese subjects. Int. J. Obes. 2004, 28, 176–179. [Google Scholar] [CrossRef] [PubMed]
- Kubo, N.; Kawahara, M.; Okamatsu-Ogura, Y.; Miyazaki, Y.; Otsuka, R.; Fukuchi, K. Evaluation of Glucose Uptake and Uncoupling Protein 1 Activity in Adipose Tissue of Diabetic Mice upon beta-Adrenergic Stimulation. Mol. Imaging Biol. 2019, 21, 249–256. [Google Scholar] [CrossRef]
- Singh, A.K.; Aryal, B.; Chaube, B.; Rotllan, N.; Varela, L.; Horvath, T.L.; Suarez, Y.; Fernandez-Hernando, C. Brown adipose tissue derived ANGPTL4 controls glucose and lipid metabolism and regulates thermogenesis. Mol. Metab. 2018, 11, 59–69. [Google Scholar] [CrossRef]
- Sidossis, L.S.; Porter, C.; Saraf, M.K.; Borsheim, E.; Radhakrishnan, R.S.; Chao, T.; Ali, A.; Chondronikola, M.; Mlcak, R.; Finnerty, C.C.; et al. Browning of Subcutaneous White Adipose Tissue in Humans after Severe Adrenergic Stress. Cell Metab. 2015, 22, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Kepple, J.D.; Liu, Y.; Kim, T.; Cero, C.; Johnson, J.W.; Rowe, G.C.; Cypess, A.M.; Habegger, K.M.; Young, M.; Hunter, C.S. The transcriptional co-regulator LDB1 is required for brown adipose function. Mol. Metab. 2021, 53, 101284. [Google Scholar] [CrossRef] [PubMed]
- Cypess, A.M.; Weiner, L.S.; Roberts-Toler, C.; Franquet Elia, E.; Kessler, S.H.; Kahn, P.A.; English, J.; Chatman, K.; Trauger, S.A.; Doria, A.; et al. Activation of human brown adipose tissue by a beta3-adrenergic receptor agonist. Cell Metab. 2015, 21, 33–38. [Google Scholar] [CrossRef]
- Yao, L.; Cui, X.; Chen, Q.; Yang, X.; Fang, F.; Zhang, J.; Liu, G.; Jin, W.; Chang, Y. Cold-Inducible SIRT6 Regulates Thermogenesis of Brown and Beige Fat. Cell Rep. 2017, 20, 641–654. [Google Scholar] [CrossRef]
- de Jong, J.M.A.; Wouters, R.T.F.; Boulet, N.; Cannon, B.; Nedergaard, J.; Petrovic, N. The beta(3)-adrenergic receptor is dispensable for browning of adipose tissues. Am. J. Physiol. Endocrinol. Metab. 2017, 312, E508–E518. [Google Scholar] [CrossRef]
- Rodriguez, A.; Becerril, S.; Ezquerro, S.; Mendez-Gimenez, L.; Fruhbeck, G. Crosstalk between adipokines and myokines in fat browning. Acta Physiol. 2017, 219, 362–381. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.; Swarbrick, M.M.; Greenfield, J.R. The sum of all browning in FGF21 therapeutics. Cell Metab. 2015, 21, 795–796. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Struik, D.; Dommerholt, M.B.; Jonker, J.W. Fibroblast growth factors in control of lipid metabolism: From biological function to clinical application. Curr. Opin. Lipidol. 2019, 30, 235–243. [Google Scholar] [CrossRef]
- Kharitonenkov, A.; Shiyanova, T.L.; Koester, A.; Ford, A.M.; Micanovic, R.; Galbreath, E.J.; Sandusky, G.E.; Hammond, L.J.; Moyers, J.S.; Owens, R.A.; et al. FGF-21 as a novel metabolic regulator. J. Clin. Investig. 2005, 115, 1627–1635. [Google Scholar] [CrossRef]
- Kuzawa, C.W. Adipose tissue in human infancy and childhood: An evolutionary perspective. Am. J. Phys. Anthropol. 1998, 107 (Suppl. 27), 177–209. [Google Scholar] [CrossRef]
- Yu, H.; Dilbaz, S.; Cossmann, J.; Hoang, A.C.; Diedrich, V.; Herwig, A.; Harauma, A.; Hoshi, Y.; Moriguchi, T.; Landgraf, K.; et al. Breast milk alkylglycerols sustain beige adipocytes through adipose tissue macrophages. J. Clin. Investig. 2019, 129, 2485–2499. [Google Scholar] [CrossRef] [PubMed]
- Xue, R.; Lynes, M.D.; Dreyfuss, J.M.; Shamsi, F.; Schulz, T.J.; Zhang, H.; Huang, T.L.; Townsend, K.L.; Li, Y.; Takahashi, H.; et al. Clonal analyses and gene profiling identify genetic biomarkers of the thermogenic potential of human brown and white preadipocytes. Nat. Med. 2015, 21, 760–768. [Google Scholar] [CrossRef]
- Bostrom, P.; Wu, J.; Jedrychowski, M.P.; Korde, A.; Ye, L.; Lo, J.C.; Rasbach, K.A.; Bostrom, E.A.; Choi, J.H.; Long, J.Z.; et al. A PGC1-alpha-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature 2012, 481, 463–468. [Google Scholar] [CrossRef]
- Wu, R.; Park, J.; Qian, Y.; Shi, Z.; Hu, R.; Yuan, Y.; Xiong, S.; Wang, Z.; Yan, G.; Ong, S.G.; et al. Genetically prolonged beige fat in male mice confers long-lasting metabolic health. Nat. Commun. 2023, 14, 2731. [Google Scholar] [CrossRef] [PubMed]
- Varshney, R.; Das, S.; Trahan, G.D.; Farriester, J.W.; Mullen, G.P.; Kyere-Davies, G.; Presby, D.M.; Houck, J.A.; Webb, P.G.; Dzieciatkowska, M.; et al. Neonatal intake of Omega-3 fatty acids enhances lipid oxidation in adipocyte precursors. iScience 2023, 26, 105750. [Google Scholar] [CrossRef] [PubMed]
- Cawthorn, W.P.; Scheller, E.L.; MacDougald, O.A. Adipose tissue stem cells meet preadipocyte commitment: Going back to the future. J. Lipid Res. 2012, 53, 227–246. [Google Scholar] [CrossRef]
- Exley, M.A.; Hand, L.; O’Shea, D.; Lynch, L. Interplay between the immune system and adipose tissue in obesity. J. Endocrinol. 2014, 223, R41–R48. [Google Scholar] [CrossRef]
- Saely, C.H.; Geiger, K.; Drexel, H. Brown versus white adipose tissue: A mini-review. Gerontology 2012, 58, 15–23. [Google Scholar] [CrossRef]
- Karise, I.; Bargut, T.C.; Del Sol, M.; Aguila, M.B.; Mandarim-de-Lacerda, C.A. Metformin enhances mitochondrial biogenesis and thermogenesis in brown adipocytes of mice. Biomed. Pharmacother. 2019, 111, 1156–1165. [Google Scholar] [CrossRef]
- Zheng, Y.; Yang, N.; Pang, Y.; Gong, Y.; Yang, H.; Ding, W.; Yang, H. Mitochondria-associated regulation in adipose tissues and potential reagents for obesity intervention. Front. Endocrinol. 2023, 14, 1132342. [Google Scholar] [CrossRef]
- Xu, Z.; Huo, J.; Ding, X.; Yang, M.; Li, L.; Dai, J.; Hosoe, K.; Kubo, H.; Mori, M.; Higuchi, K.; et al. Coenzyme Q10 Improves Lipid Metabolism and Ameliorates Obesity by Regulating CaMKII-Mediated PDE4 Inhibition. Sci. Rep. 2017, 7, 8253. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Galilea, M.; Perez-Matute, P.; Prieto-Hontoria, P.L.; Houssier, M.; Burrell, M.A.; Langin, D.; Martinez, J.A.; Moreno-Aliaga, M.J. alpha-Lipoic acid treatment increases mitochondrial biogenesis and promotes beige adipose features in subcutaneous adipocytes from overweight/obese subjects. Biochim. Biophys. Acta 2015, 1851, 273–281. [Google Scholar] [CrossRef] [PubMed]
- Chacinska, M.; Zabielski, P.; Ksiazek, M.; Szalaj, P.; Jarzabek, K.; Kojta, I.; Chabowski, A.; Blachnio-Zabielska, A.U. The Impact of OMEGA-3 Fatty Acids Supplementation on Insulin Resistance and Content of Adipocytokines and Biologically Active Lipids in Adipose Tissue of High-Fat Diet Fed Rats. Nutrients 2019, 11, 835. [Google Scholar] [CrossRef] [PubMed]
- Bogacka, I.; Xie, H.; Bray, G.A.; Smith, S.R. Pioglitazone induces mitochondrial biogenesis in human subcutaneous adipose tissue in vivo. Diabetes 2005, 54, 1392–1399. [Google Scholar] [CrossRef] [PubMed]
- Boutant, M.; Kulkarni, S.S.; Joffraud, M.; Ratajczak, J.; Valera-Alberni, M.; Combe, R.; Zorzano, A.; Canto, C. Mfn2 is critical for brown adipose tissue thermogenic function. EMBO J. 2017, 36, 1543–1558. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yang, L.; Mao, Z.; Pan, X.; Zhao, Y.; Gu, X.; Eckel-Mahan, K.; Zuo, Z.; Tong, Q.; Hartig, S.M.; et al. Novel role of dynamin-related-protein 1 in dynamics of ER-lipid droplets in adipose tissue. FASEB J. 2020, 34, 8265–8282. [Google Scholar] [CrossRef] [PubMed]
- Mendham, A.E.; Larsen, S.; George, C.; Adams, K.; Hauksson, J.; Olsson, T.; Fortuin-de Smidt, M.C.; Nono Nankam, P.A.; Hakim, O.; Goff, L.M.; et al. Exercise training results in depot-specific adaptations to adipose tissue mitochondrial function. Sci. Rep. 2020, 10, 3785. [Google Scholar] [CrossRef] [PubMed]
- Demine, S.; Renard, P.; Arnould, T. Mitochondrial Uncoupling: A Key Controller of Biological Processes in Physiology and Diseases. Cells 2019, 8, 795. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.Y.; Brychta, R.J.; Israni, N.S.; Jiang, A.; Lea, H.J.; Lentz, T.N.; Pierce, A.E.; Cypess, A.M. Activating Human Adipose Tissue with the beta3-Adrenergic Agonist Mirabegron. Methods Mol. Biol. 2022, 2448, 83–96. [Google Scholar]
- Sun, X.; Sui, W.; Mu, Z.; Xie, S.; Deng, J.; Li, S.; Seki, T.; Wu, J.; Jing, X.; He, X.; et al. Mirabegron displays anticancer effects by globally browning adipose tissues. Nat. Commun. 2023, 14, 7610. [Google Scholar] [CrossRef]
- Cypess, A.M. Reassessing Human Adipose Tissue. N. Engl. J. Med. 2022, 386, 768–779. [Google Scholar] [CrossRef] [PubMed]
- Cypess, A.M. Does activating brown fat contribute to important metabolic benefits in humans? Yes! J. Clin. Investig. 2023, 133, e175282. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.K. Human brown fat and metabolic disease: A heated debate. J. Clin. Investig. 2023, 133, e176678. [Google Scholar] [CrossRef] [PubMed]
| White Adipocyte | Brown Adipocyte | Beige Adipocyte | |
|---|---|---|---|
| Function | Energy storage, Endocrine signaling | Thermogenesis, Endocrine signaling | Adaptive thermogenesis, Endocrine signaling |
| Mitochondrial density | Low | High | Medium |
| Origin | Myf5- progenitors | Myf5+ progenitors | Myf5- progenitors |
| Regulators of fate determination | BMP-4, BMP-10, FGF10, PPARγ, C/EBPs | BMP-7, PRDM16, PPARγ, C/EBPs, PGC1α | BMP-7, PRDM16, PPARγ, C/EBPs, PGC1α |
| UCP1 expression | Low | High | High |
| Lipid droplet | Large unilocular | Small multilocular | Medium multilocular |
| Markers | Leptin, Hoxc8, Hoxc9 | UCP1, Zic1, Lhx8 | UCP1, Cd137, Tmem26 |
| Morphology |  |  |  |
| Functional Implications | Model or Clinical Condition | Citation |
|---|---|---|
| White adipocytes | ||
| Mitochondria-generated ROS send physiological “distress” signals in WAT during caloric excess | GSTA4-silenced 3T3-L1 adipocytes and GSTA4-null mice | [21] |
| Recruitment of inflammatory cells into WAT | B6.V-Lepob/OlaHsd mice, CD11c-DTR mice | [22] |
| WAT stored fuels as triacylglycerols, until whole-body energy demand signals for lipid release | ob/ob mice | [23] |
| Asymmetry in substrate availability causes mitochondrial malfunction | db/db mice, ob/ob mice | [10] |
| WAT is highly vascularized and innervated for its endocrine signaling functions | Type 2 Diabetic patients | [24] |
| WAT can remodel vasculature and extracellular matrix (ECM) to allow tissue expansion, oxygenation, and mobilization of nutrients | ob/ob mice, mice fed a high-calorie diet; Type 2 Diabetic patients | [25,26] |
| WAT expansion occurs through increased adipocyte size (hypertrophy) and/or number (hyperplasia) | Cross of adiponectinPrtTA (adnP-rtTA) transgenic mice with TRE-cre and Rosa26-loxP-stoploxP-lacZ transgenic mice | [27] |
| Browning of adipocytes blocks lipid spillover | Engrailed-1 (En1)-CreERT-inducible mice crossed with Rosa-floxed Stop-LacZ mouse, Sox10-Cre/Rosa26-YFP model, A-Zip mice | [28] |
| Adaptive thermogenesis of BAT and browning of WAT maintain metabolic homeostasis in obese diabetic mice | 3T3L1 cells, db/db mice, ob/ob mice, Glut4-knockout mice, Pgc1α-knockout mice, Mfn2-knockout mice | [11] |
| Brown adipocytes | ||
| ROS prevent establishment of healthy BAT by inhibiting preadipocyte proliferation and differentiation | High-Fat Diet (HFD)-fed mice, Obese patients | [29] |
| PRDM16 protein stabilization by Thiazolidinediones (TZDs) promotes white-to-brown adipose tissue conversion | 3T3-F442A cells, CtBP-1+/− CtBP-2+/− and CtBP-1−/− CtBP-2−/− MEFs, 3T3-L1 cells | [30] |
| Loss of PRDM16 in brown adipose precursors results in the loss of brown adipocyte features | Primary brown pre-adipocytes and myoblasts from P0–P4 Swiss-Webster mice, Cross of Myf5Cre/1 mice18 with rosa26R3 (R26R3)-YFP mice19, Prdm16-knockout mice | [31] |
| PGC1α stimulates FNDC5 to promote irisin release, which directly stimulates “browning” of white adipocytes | C57BL/6J mice, Obese BALB/c mice | [5,32] |
| PRDM16 controls thermogenic programming and preserves the fate of brown adipocytes | Myf5Cre, Rosa26Cre mice, Prdm16flox and Prdm3flox mice | [33] |
| Brown preadipocyte development and adipogenesis are suppressed in an NF-κB-dependent manner | C3H10T1/2 cells | [34] |
| Diminished or excessive ROS affects BAT recruitment | 3T3-L1 cells, adipocytes isolated from lean and obese patients | [35,36] |
| Brown adipocyte differentiation is inhibited by pro-inflammatory cytokines such as TNF-α, IL-1, LPS, and Oncostatin M, released by T cells and macrophages | Stromal vascular cells from C57BL/6J mice, High-fat diet (HFD)-fed C57BL/6J mice, BAT preadipocytes from C57BL/6J mice, Primary human stromal vascular cells | [37,38,39] |
| Persistent activation of RalA suppresses energy expenditure in obese adipose tissue | Ralaf/f (n = 8) and RalaAKO fed with HFD | [40] |
| Beige adipocytes | ||
| Impaired Ca2+ homeostasis, decreased ATP synthesis, increased ROS generation, changed mitochondrial enzyme activity, and anomalies in systemic energy negatively impact mitochondrial function in BeAT | Adipocytes from C57BL/6J mice, 3T3-L1 cells, Adipose tissue from obese patients | [41,42] |
| When stimulated by cold, mitochondria in brown and beige adipocytes adopt distinctive morphologies and inter-organelle interactions | C57BL/6J mice | [18] |
| Non-shivering thermogenesis is mostly found in brown and beige adipocytes | β(3)-adrenoceptor-knockout mice, human obese patients, 3T3L1 cells | [14] |
| Loss of mitochondrial dynamics due to Mfn2 or Opa1 knockdown induces buildup of intracellular triacylglycerols in adipocytes | Adipose tissue from obese patients, HFD mice | [43,44] |
| Impairment of phosphorylation of DRP1 at serine 600 by protein kinase A (PKA) impairs adipocyte function | Adipocytes from C57BL/6J mice, GF gnotobiotic mice | [45] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Das, S.; Mukhuty, A.; Mullen, G.P.; Rudolph, M.C. Adipocyte Mitochondria: Deciphering Energetic Functions across Fat Depots in Obesity and Type 2 Diabetes. Int. J. Mol. Sci. 2024, 25, 6681. https://doi.org/10.3390/ijms25126681
Das S, Mukhuty A, Mullen GP, Rudolph MC. Adipocyte Mitochondria: Deciphering Energetic Functions across Fat Depots in Obesity and Type 2 Diabetes. International Journal of Molecular Sciences. 2024; 25(12):6681. https://doi.org/10.3390/ijms25126681
Chicago/Turabian StyleDas, Snehasis, Alpana Mukhuty, Gregory P. Mullen, and Michael C. Rudolph. 2024. "Adipocyte Mitochondria: Deciphering Energetic Functions across Fat Depots in Obesity and Type 2 Diabetes" International Journal of Molecular Sciences 25, no. 12: 6681. https://doi.org/10.3390/ijms25126681
APA StyleDas, S., Mukhuty, A., Mullen, G. P., & Rudolph, M. C. (2024). Adipocyte Mitochondria: Deciphering Energetic Functions across Fat Depots in Obesity and Type 2 Diabetes. International Journal of Molecular Sciences, 25(12), 6681. https://doi.org/10.3390/ijms25126681






