Phytaspase Does Not Require Proteolytic Activity for Its Stress-Induced Internalization
Abstract
1. Introduction
2. Results
2.1. Setup of the Phytaspase Transport Assay
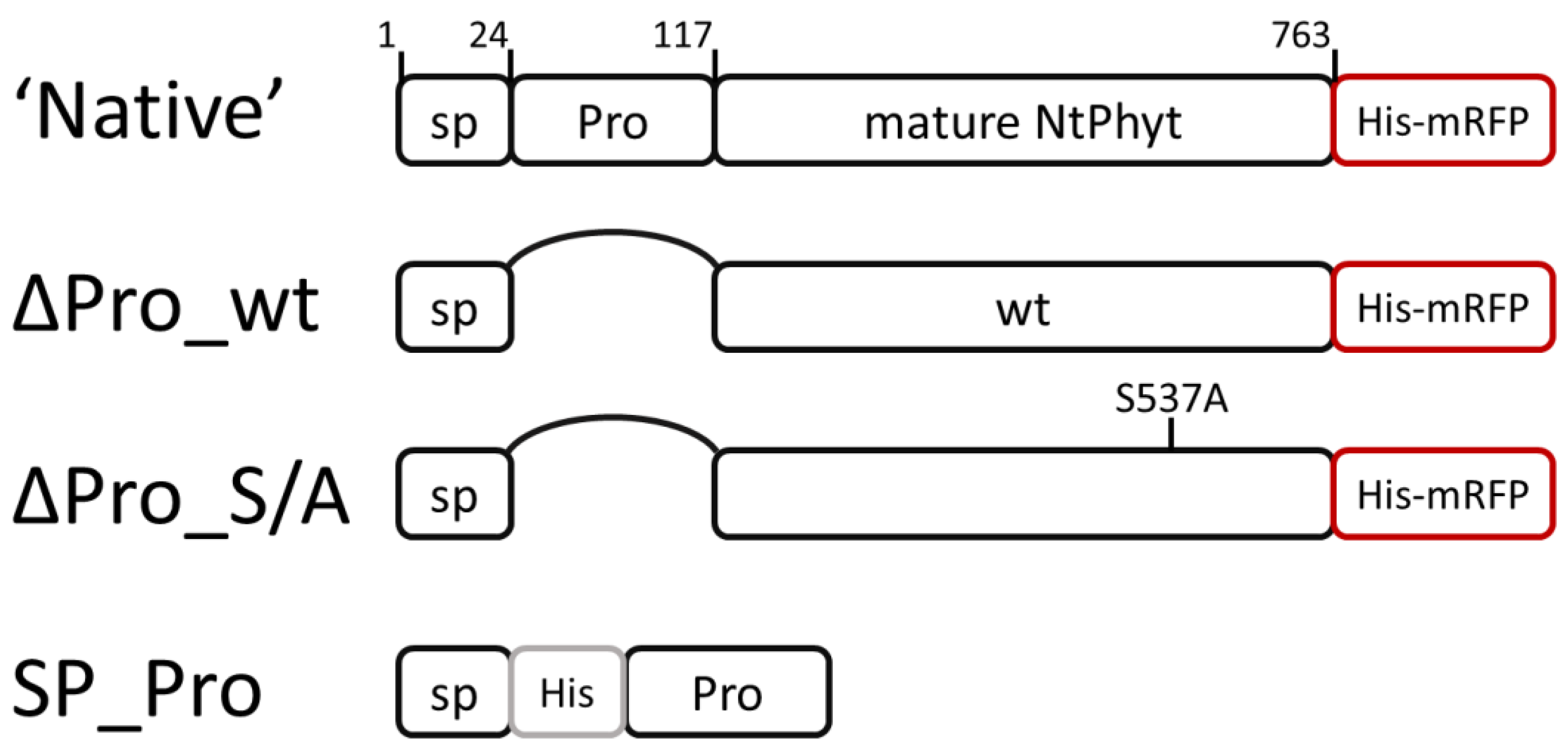
2.2. Elimination of the Prodomain from the NtPhyt Precursor Compromises Proteolytic Activity and Secretion of NtPhyt
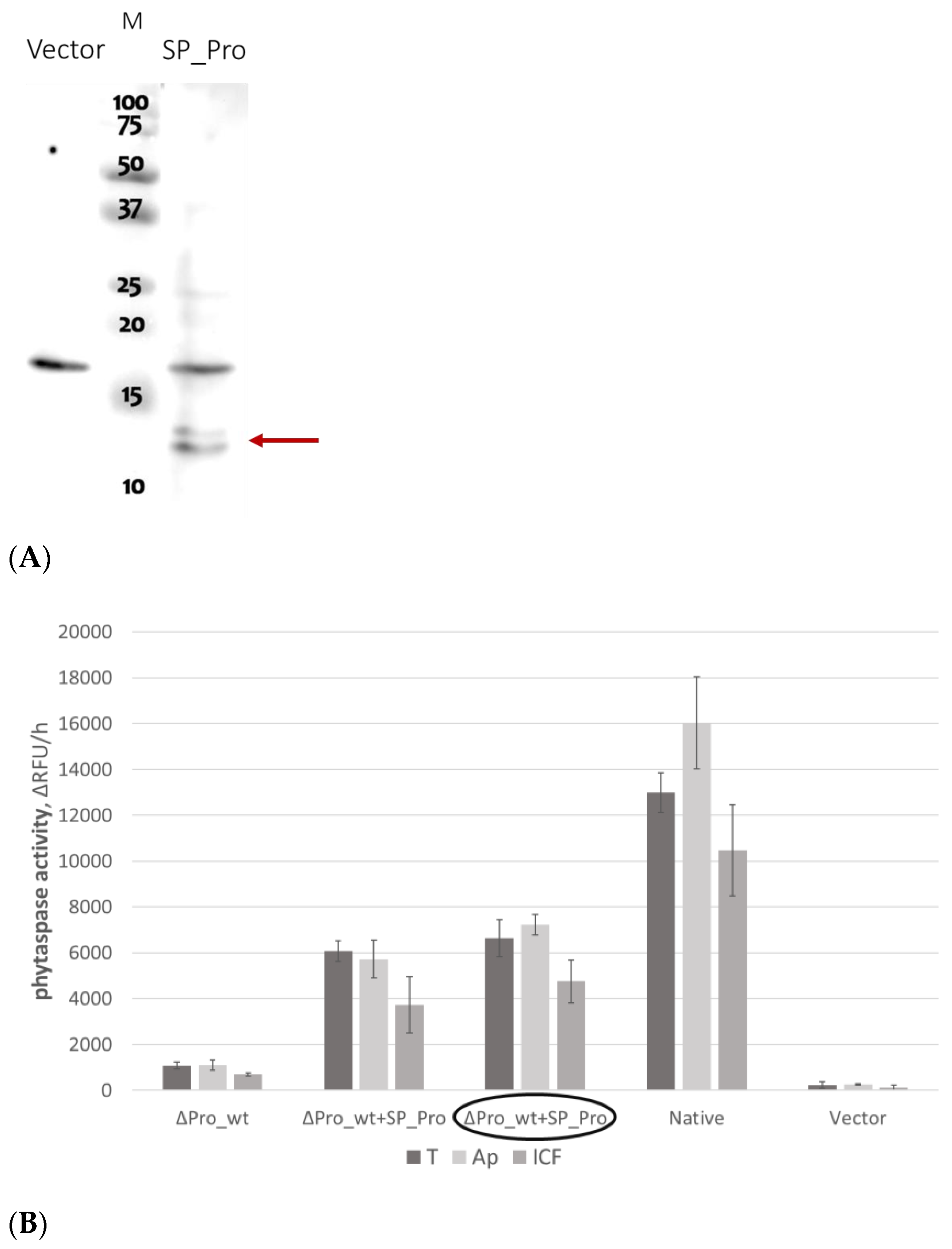
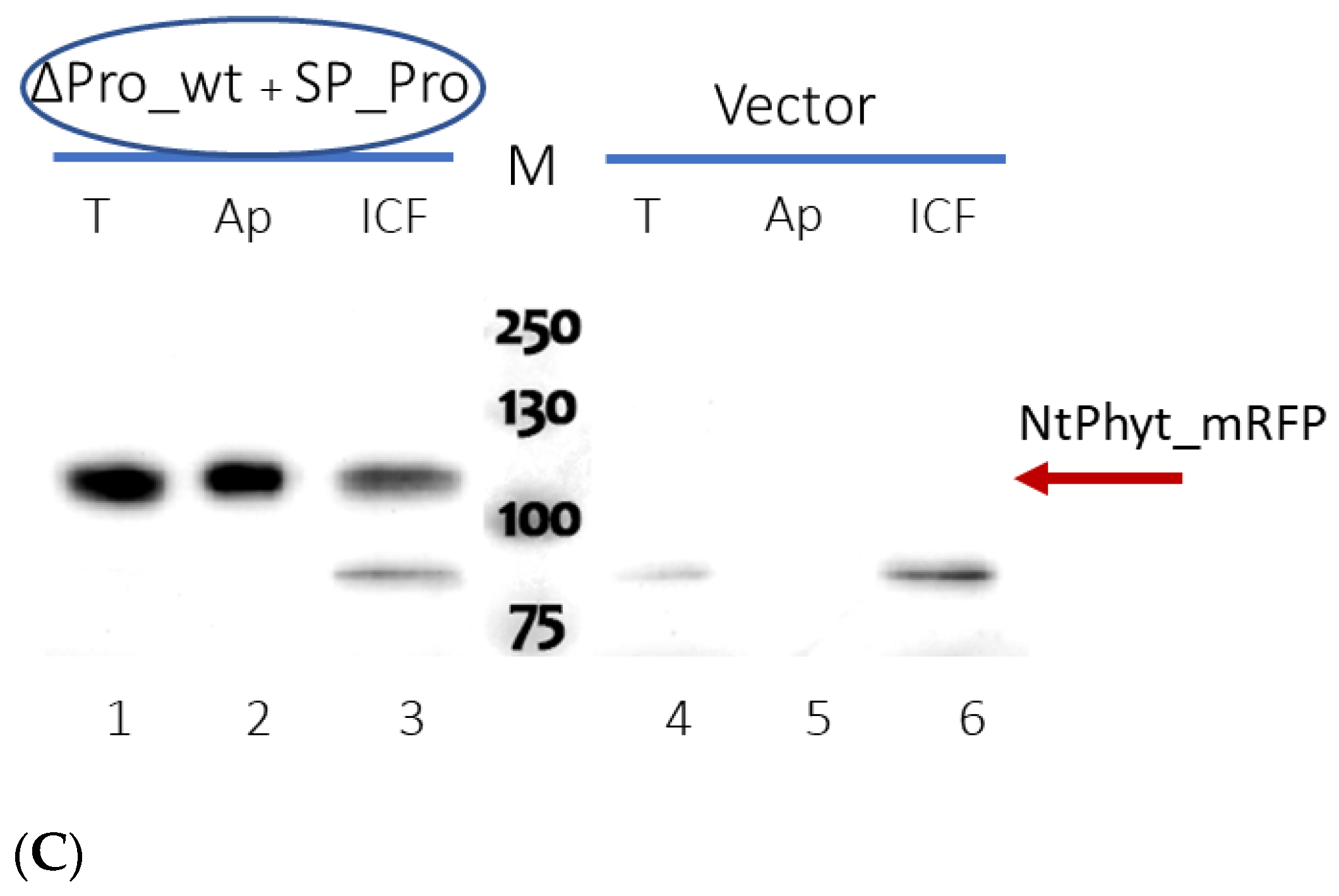
2.3. Rescuing the Prodomain-Less NtPhyt
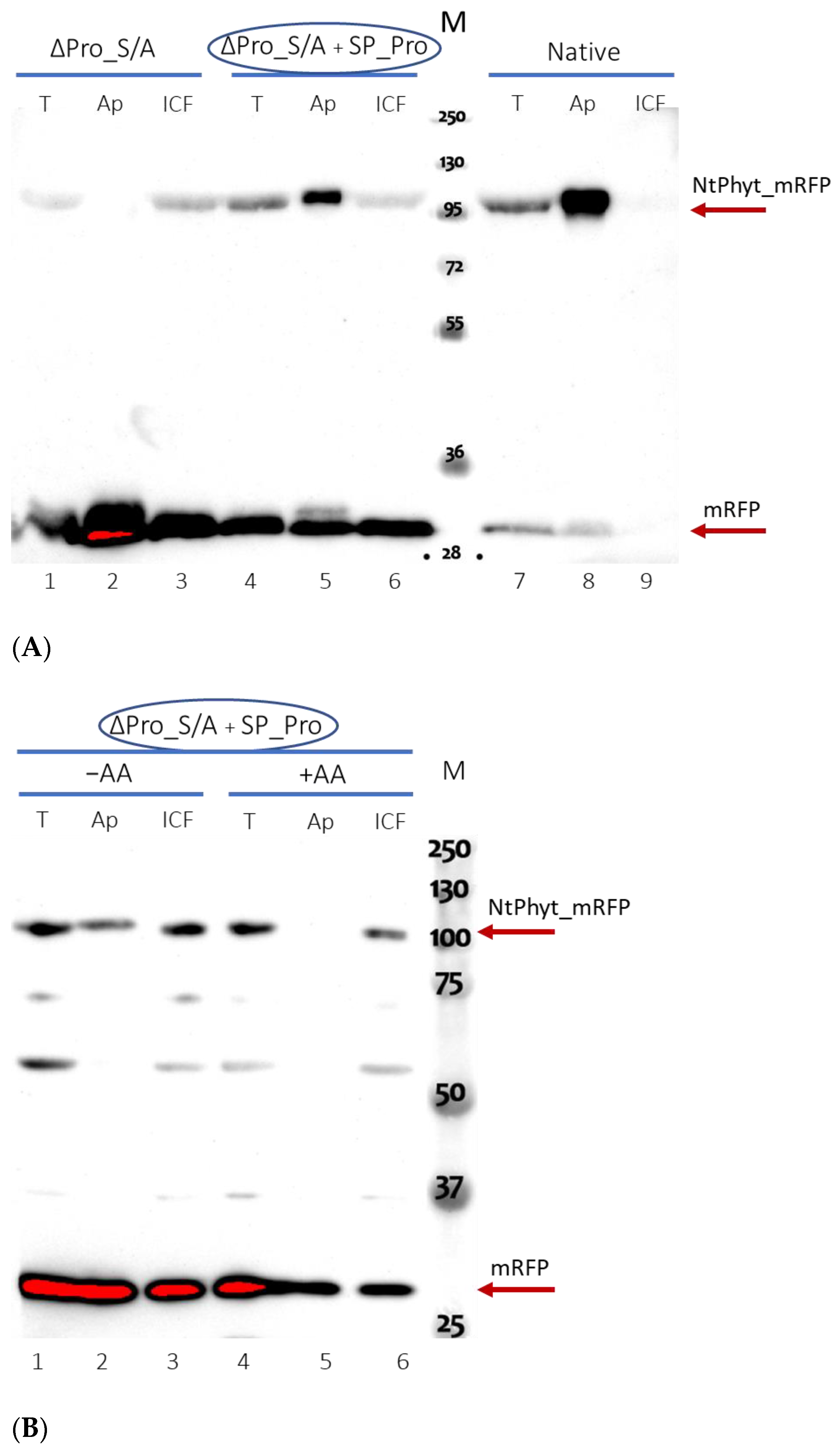
2.4. Catalytically Inactive NtPhyt Mutant Is Competent for Stress-Induced Internalization
3. Discussion
4. Materials and Methods
4.1. Plant Growth Conditions
4.2. Plasmid Construction
4.3. Transient Expression in N. benthamiana and Protein Fractionation
4.4. Phytaspase Activity Determination
4.5. Western Blot Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schaller, A.; Stintzi, A.; Rivas, S.; Serrano, I.; Chichkova, N.V.; Vartapetian, A.B.; Martínez, D.; Guiamét, J.J.; Sueldo, D.J.; van der Hoorn, R.A.L.; et al. From structure to function—A family portrait of plant subtilases. New Phytol. 2018, 218, 901–915. [Google Scholar] [CrossRef]
- Schaller, A.; Stintzi, A.; Graff, L. Subtilases—Versatile tools for protein turnover, plant development, and interactions with the environment. Physiol. Plant. 2012, 145, 52–66. [Google Scholar] [CrossRef]
- Trusova, S.V.; Golyshev, S.A.; Chichkova, N.V.; Vartapetian, A.B. Sometimes they come back: Endocytosis provides localization dynamics of a subtilase in cells committed to cell death. J. Exp. Bot. 2019, 70, 2003–2007. [Google Scholar] [CrossRef]
- Chichkova, N.V.; Shaw, J.; Galiullina, R.A.; Drury, G.E.; Tuzhikov, A.I.; Kim, S.H.; Kalkum, M.; Hong, T.B.; Gorshkova, E.N.; Torrance, L.; et al. Phytaspase, a relocalisable cell death promoting plant protease with caspase specificity. EMBO J. 2010, 29, 1149–1161. [Google Scholar] [CrossRef]
- Galiullina, R.A.; Kasperkiewicz, P.; Chichkova, N.V.; Szalek, A.; Serebryakova, M.V.; Poreba, M.; Drag, M.; Vartapetian, A.B. Substrate specificity and possible heterologous targets of phytaspase, a plant cell death protease. J. Biol. Chem. 2015, 290, 24806–24815. [Google Scholar] [CrossRef]
- Chichkova, N.V.; Tuzhikov, A.I.; Taliansky, M.; Vartapetian, A.B. Plant phytaspases and animal caspases: Structurally unrelated death proteases with a common role and specificity. Physiol. Plant. 2012, 145, 77–84. [Google Scholar] [CrossRef]
- Thornberry, N.A.; Rano, T.A.; Peterson, E.P.; Rasper, D.M.; Timkey, T.; Garcia-Calvo, M.; Houtzager, V.M.; Nordstrom, P.A.; Roy, S.; Vaillancourt, J.P.; et al. A combinatorial approach defines specificities of members of the caspase family and granzyme B. Functional relationships established for key mediators of apoptosis. J. Biol. Chem. 1997, 272, 17907–17911. [Google Scholar] [CrossRef]
- Talanian, R.V.; Quinlan, C.; Trautz, S.; Hackett, M.C.; Mankovich, J.A.; Banach, D.; Ghayur, T.; Brady, K.D.; Wong, W.W. Substrate specificities of caspase family proteases. J. Biol. Chem. 1997, 272, 9677–9682. [Google Scholar] [CrossRef]
- Timmer, J.C.; Salvesen, G.S. Caspase substrates. Cell Death Differ. 2007, 14, 66–72. [Google Scholar] [CrossRef]
- Chichkova, N.V.; Kim, S.H.; Titova, E.S.; Kalkum, M.; Morozov, V.S.; Rubtsov, Y.P.; Kalinina, N.O.; Taliansky, M.E.; Vartapetian, A.B. A plant caspase-like protease activated during the hypersensitive response. Plant Cell 2004, 16, 157–171. [Google Scholar] [CrossRef]
- Vartapetian, A.B.; Tuzhikov, A.I.; Chichkova, N.V.; Taliansky, M.; Wolpert, T.J. A plant alternative to animal caspases: Subtilisin-like proteases. Cell Death Differ. 2011, 18, 1289–1297. [Google Scholar] [CrossRef]
- Reichardt, S.; Repper, D.; Tuzhikov, A.I.; Galiullina, R.A.; Planas-Marquès, M.; Chichkova, N.V.; Vartapetian, A.B.; Stintzi, A.; Schaller, A. The tomato subtilase family includes several cell death-related proteinases with caspase specificity. Sci. Rep. 2018, 8, 10531. [Google Scholar] [CrossRef]
- Trusova, S.V.; Teplova, A.D.; Golyshev, S.A.; Galiullina, R.A.; Morozova, E.A.; Chichkova, N.V.; Vartapetian, A.B. Clathrin-mediated endocytosis delivers proteolytically active phytaspases into plant cells. Front. Plant Sci. 2019, 10, 873. [Google Scholar] [CrossRef]
- Cedzich, A.; Huttenlocher, F.; Kuhn, B.M.; Pfannstiel, J.; Gabler, L.; Stintzi, A.; Schaller, A. The protease-associated domain and C-terminal extension are required for zymogen processing, sorting within the secretory pathway, and activity of tomato subtilase 3 (SlSBT3). J. Biol. Chem. 2009, 284, 14068–14078. [Google Scholar] [CrossRef]
- Zhu, X.L.; Ohta, Y.; Jordan, F.; Inouye, M. Pro-sequence of subtilisin can guide the refolding of denatured subtilisin in an intermolecular process. Nature 1989, 339, 483–484. [Google Scholar] [CrossRef]
- Meyer, M.; Leptihn, S.; Welz, M.; Schaller, A. Functional characterization of propeptides in plant subtilases as intramolecular chaperones and inhibitors of the mature protease. J. Biol. Chem. 2016, 291, 19449–19461. [Google Scholar] [CrossRef]
- Zhang, W.; Planas-Marquès, M.; Mazier, M.; Šimkovicová, M.; Rocafort, M.; Mantz, M.; Huesgen, P.F.; Takken, F.L.W.; Stintzi, A.; Schaller, A.; et al. The tomato P69 subtilase family is involved in resistance to bacterial wilt. Plant J. 2024, 118, 388–404. [Google Scholar] [CrossRef]
- Nakagawa, M.; Ueyama, M.; Tsuruta, H.; Uno, T.; Kanamaru, K.; Mikami, B.; Yamagata, H. Functional analysis of the cucumisin propeptide as a potent inhibitor of its mature enzyme. J. Biol. Chem. 2010, 285, 29797–29807. [Google Scholar] [CrossRef]
- Eder, J.; Fersht, A.R. Pro-sequence-assisted protein folding. Mol. Microbiol. 1995, 16, 609–614. [Google Scholar] [CrossRef]
- Soh, U.J.; Dores, M.R.; Chen, B.; Trejo, J. Signal transduction by protease-activated receptors. Br. J. Pharmacol. 2010, 160, 191–203. [Google Scholar] [CrossRef]
- Vu, T.K.; Hung, D.T.; Wheaton, V.I.; Coughlin, S.R. Molecular cloning of a functional thrombin receptor reveals a novel proteolytic mechanism of receptor activation. Cell 1991, 64, 1057–1068. [Google Scholar] [CrossRef]
- Rasmussen, U.B.; Vouret-Craviari, V.; Jallat, S.; Schlesinger, Y.; Pagès, G.; Pavirani, A.; Lecocq, J.P.; Pouysségur, J.; Van Obberghen-Schilling, E. cDNA cloning and expression of a hamster alpha-thrombin receptor coupled to Ca2+ mobilization. FEBS Lett. 1991, 288, 123–128. [Google Scholar] [CrossRef]
- Peach, C.J.; Edgington-Mitchell, L.E.; Bunnett, N.W.; Schmidt, B.L. Protease-activated receptors in health and disease. Physiol. Rev. 2023, 103, 717–785. [Google Scholar] [CrossRef]
- Liu, X.; Yu, J.; Song, S.; Yue, X.; Li, Q. Protease-activated receptor-1 (PAR-1): A promising molecular target for cancer. Oncotarget 2017, 8, 107334–107345. [Google Scholar] [CrossRef]
- Flaumenhaft, R.; De Ceunynck, K. Targeting PAR1: Now What? Trends Pharmacol. Sci. 2017, 38, 701–716. [Google Scholar] [CrossRef]
- Seidah, N.G.; Mayer, G.; Zaid, A.; Rousselet, E.; Nassoury, N.; Poirier, S.; Essalmani, R.; Prat, A. The activation and physiological functions of the proprotein convertases. Int. J. Biochem. Cell Biol. 2008, 40, 1111–1125. [Google Scholar] [CrossRef]
- Remacle, A.G.; Shiryaev, S.A.; Oh, E.S.; Cieplak, P.; Srinivasan, A.; Wei, G.; Liddington, R.C.; Ratnikov, B.I.; Parent, A.; Desjardins, R.; et al. Substrate cleavage analysis of furin and related proprotein convertases. A comparative study. J. Biol. Chem. 2008, 283, 20897–20906. [Google Scholar] [CrossRef]
- Naureckiene, S.; Ma, L.; Sreekumar, K.; Purandare, U.; Lo, C.F.; Huang, Y.; Chiang, L.W.; Grenier, J.M.; Ozenberger, B.A.; Jacobsen, J.S.; et al. Functional characterization of Narc 1, a novel proteinase related to proteinase K. Arch. Biochem. Biophys. 2003, 420, 55–67. [Google Scholar] [CrossRef]
- Benjannet, S.; Rhainds, D.; Essalmani, R.; Mayne, J.; Wickham, L.; Jin, W.; Asselin, M.C.; Hamelin, J.; Varret, M.; Allard, D.; et al. NARC-1/PCSK9 and its natural mutants: Zymogen cleavage and effects on the low density lipoprotein (LDL) receptor and LDL cholesterol. J. Biol. Chem. 2004, 279, 48865–48875. [Google Scholar] [CrossRef]
- Lagace, T.A.; Curtis, D.E.; Garuti, R.; McNutt, M.C.; Park, S.W.; Prather, H.B.; Anderson, N.N.; Ho, Y.K.; Hammer, R.E.; Horton, J.D. Secreted PCSK9 decreases the number of LDL receptors in hepatocytes and in livers of parabiotic mice. J. Clin. Investig. 2006, 116, 2995–3005. [Google Scholar] [CrossRef]
- Zhang, D.W.; Lagace, T.A.; Garuti, R.; Zhao, Z.; McDonald, M.; Horton, J.D.; Cohen, J.C.; Hobbs, H.H. Binding of proprotein convertase subtilisin/kexin type 9 to epidermal growth factor-like repeat A of low density lipoprotein receptor decreases receptor recycling and increases degradation. J. Biol. Chem. 2007, 282, 18602–18612. [Google Scholar] [CrossRef]
- Cameron, J.; Holla, Ø.L.; Ranheim, T.; Kulseth, M.A.; Berge, K.E.; Leren, T.P. Effect of mutations in the PCSK9 gene on the cell surface LDL receptors. Hum. Mol. Genet. 2006, 15, 1551–1558. [Google Scholar] [CrossRef]
- Nassoury, N.; Blasiole, D.A.; Tebon Oler, A.; Benjannet, S.; Hamelin, J.; Poupon, V.; McPherson, P.S.; Attie, A.D.; Prat, A.; Seidah, N.G. The cellular trafficking of the secretory proprotein convertase PCSK9 and its dependence on the LDLR. Traffic 2007, 8, 718–732. [Google Scholar] [CrossRef]
- Durairaj, A.; Sabates, A.; Nieves, J.; Moraes, B.; Baum, S. Proprotein Convertase Subtilisin/Kexin Type 9 (PCSK9) and Its Inhibitors: A Review of Physiology, Biology, and Clinical Data. Curr. Treat. Options Cardiovasc. Med. 2017, 19, 58. [Google Scholar] [CrossRef]
- Mikaeeli, S.; Ben Djoudi Ouadda, A.; Evagelidis, A.; Essalmani, R.; Ramos, O.H.P.; Fruchart-Gaillard, C.; Seidah, N.G. Insights into PCSK9-LDLR Regulation and Trafficking via the Differential Functions of MHC-I Proteins HFE and HLA-C. Cells 2024, 13, 857. [Google Scholar] [CrossRef]
- McNutt, M.C.; Lagace, T.A.; Horton, J.D. Catalytic activity is not required for secreted PCSK9 to reduce low density lipoprotein receptors in HepG2 cells. J. Biol. Chem. 2007, 282, 20799–20803. [Google Scholar] [CrossRef]
- Li, J.; Tumanut, C.; Gavigan, J.A.; Huang, W.J.; Hampton, E.N.; Tumanut, R.; Suen, K.F.; Trauger, J.W.; Spraggon, G.; Lesley, S.A.; et al. Secreted PCSK9 promotes LDL receptor degradation independently of proteolytic activity. Biochem. J. 2007, 406, 203–207. [Google Scholar] [CrossRef]
- Teplova, A.D.; Pigidanov, A.A.; Serebryakova, M.V.; Golyshev, S.A.; Galiullina, R.A.; Chichkova, N.V.; Vartapetian, A.B. Phytaspase is capable of detaching the endoplasmic reticulum retrieval signal from tobacco calreticulin-3. Int. J. Mol. Sci. 2023, 24, 16527. [Google Scholar] [CrossRef]
- Lee, J.; Lee, H.J.; Shin, M.K.; Ryu, W.S. Versatile PCR-mediated insertion or deletion mutagenesis. Biotechniques 2004, 36, 398–400. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Torosian, T.A.; Barsukova, A.I.; Chichkova, N.V.; Vartapetian, A.B. Phytaspase Does Not Require Proteolytic Activity for Its Stress-Induced Internalization. Int. J. Mol. Sci. 2024, 25, 6729. https://doi.org/10.3390/ijms25126729
Torosian TA, Barsukova AI, Chichkova NV, Vartapetian AB. Phytaspase Does Not Require Proteolytic Activity for Its Stress-Induced Internalization. International Journal of Molecular Sciences. 2024; 25(12):6729. https://doi.org/10.3390/ijms25126729
Chicago/Turabian StyleTorosian, Tatevik A., Anastasia I. Barsukova, Nina V. Chichkova, and Andrey B. Vartapetian. 2024. "Phytaspase Does Not Require Proteolytic Activity for Its Stress-Induced Internalization" International Journal of Molecular Sciences 25, no. 12: 6729. https://doi.org/10.3390/ijms25126729
APA StyleTorosian, T. A., Barsukova, A. I., Chichkova, N. V., & Vartapetian, A. B. (2024). Phytaspase Does Not Require Proteolytic Activity for Its Stress-Induced Internalization. International Journal of Molecular Sciences, 25(12), 6729. https://doi.org/10.3390/ijms25126729




