Extracellular Microenvironment Alterations in Ductal Carcinoma In Situ and Invasive Breast Cancer Pathologies by Multiplexed Spatial Proteomics
Abstract
1. Introduction
2. Results
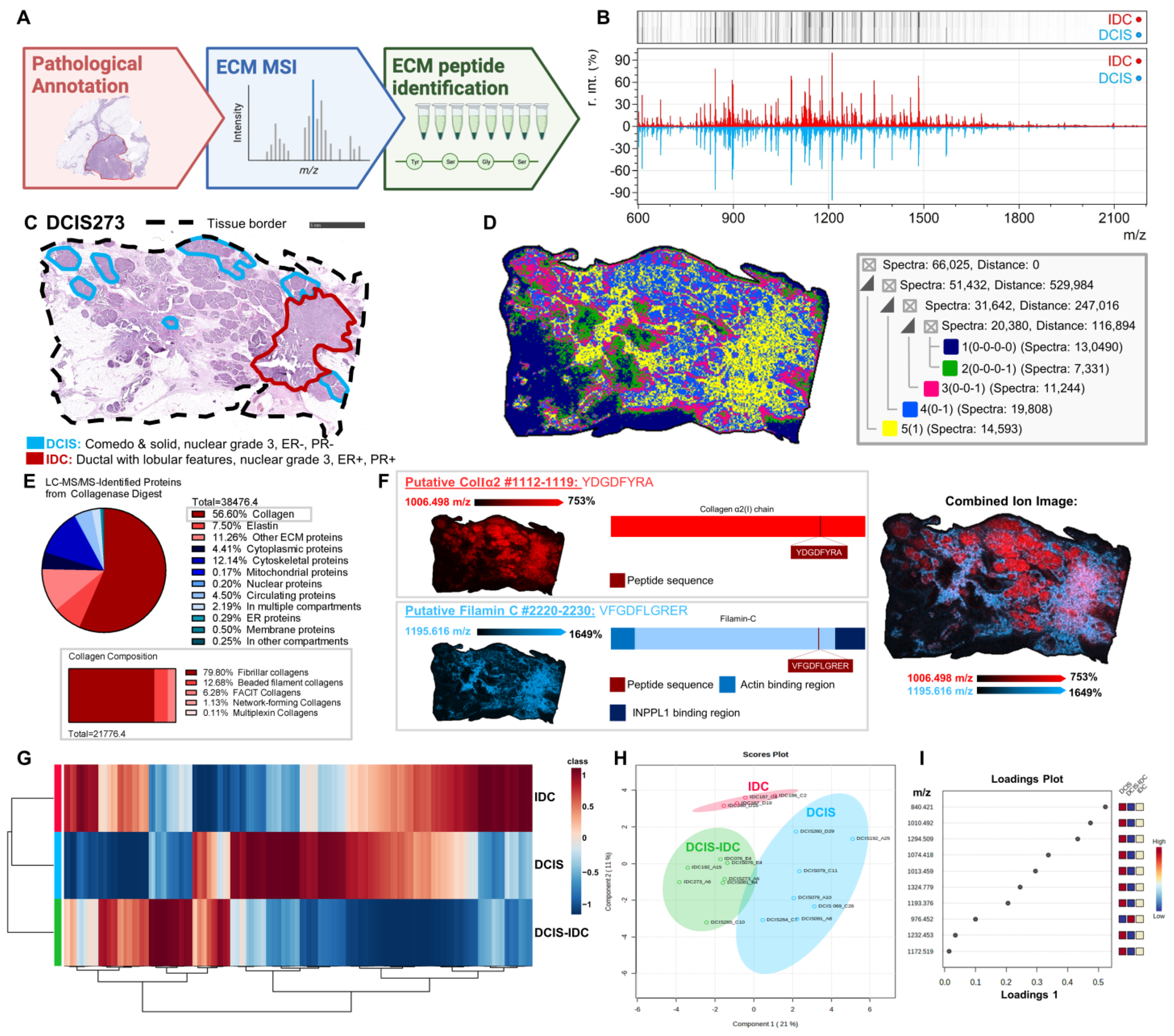
2.1. Study Overview
2.2. Spatial Mapping of the Extracellular Proteome Defines DCIS Histopathology
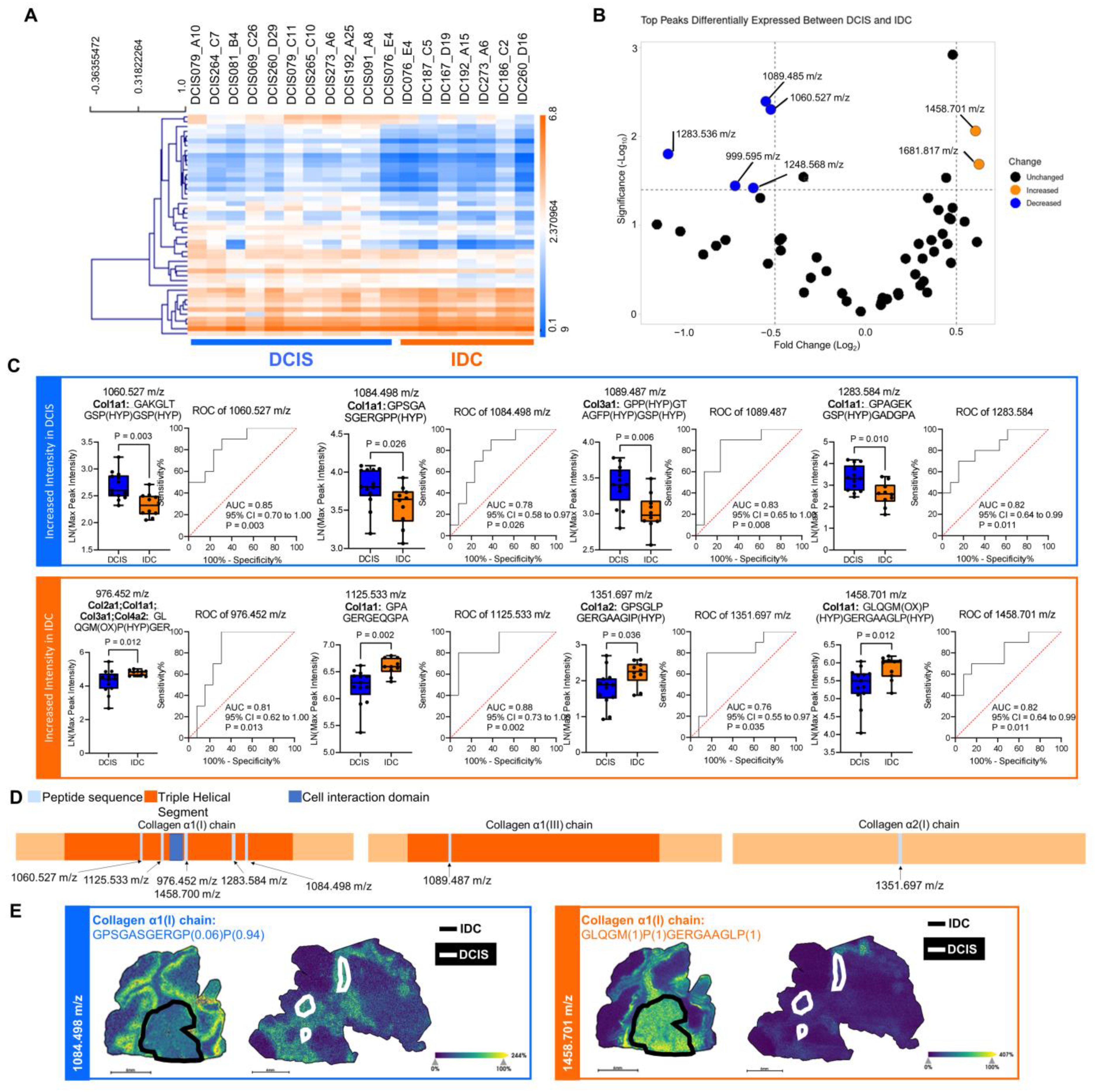
2.3. Fibrillar Collagen Domains Define Pathological Regions of DCIS and IDC
2.4. Extracellular Microenvironment Contributes to Intra-Tumoral Heterogeneity
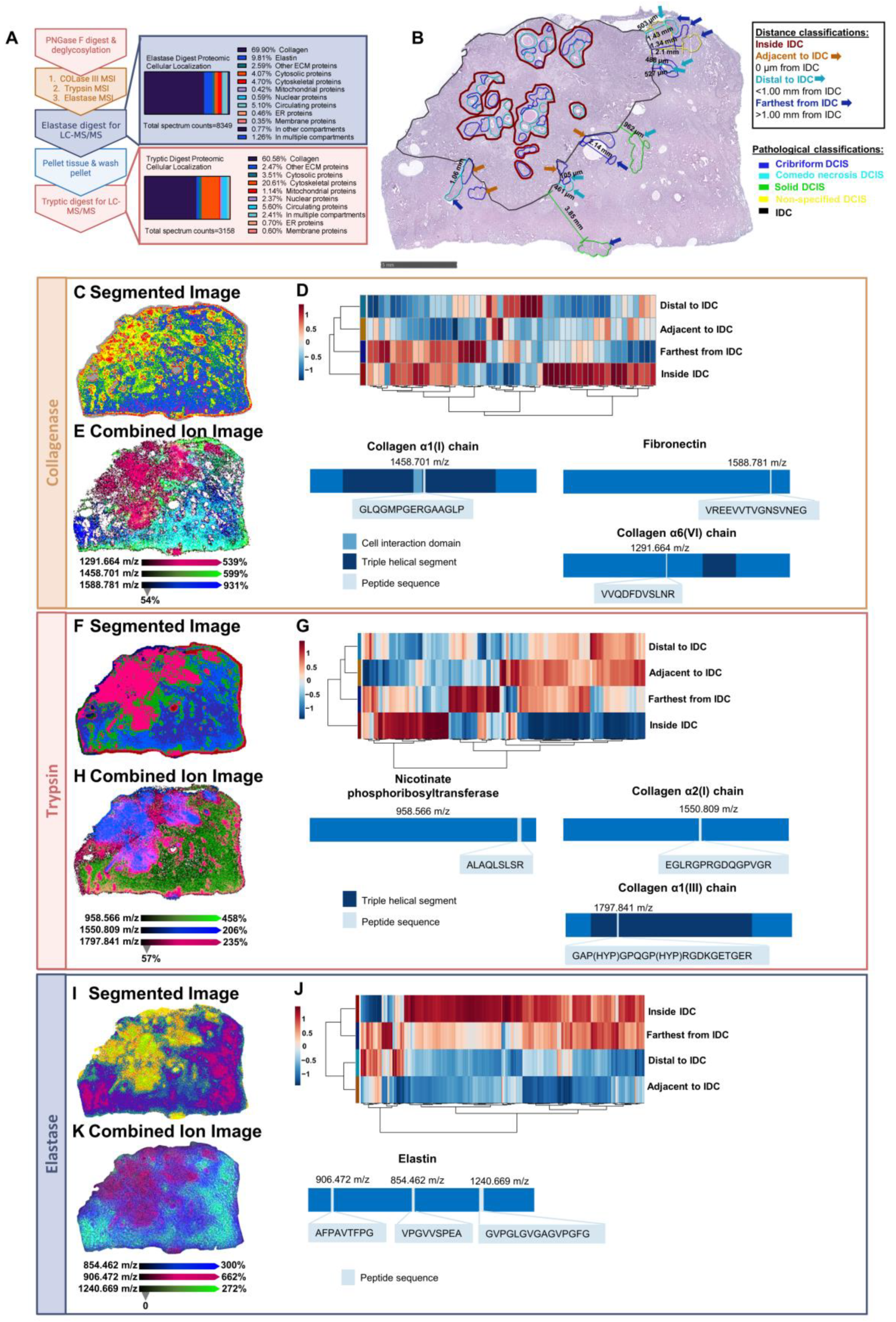
2.5. Distinct Tryptic Peptide Profiles Define Pathological Regions
2.6. Serial Enzymatic Digest Reveals Pathology-Specific Proteomes and Proteomic Field Cancerization
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Patient Cohort
4.3. Histological Staining
4.4. Matrix-Associated Laser Desorption/Ionization–Mass Spectrometry Imaging (MALDI-MSI) FFPE Tissue Preparation
4.5. MALDI-MSI
4.6. Sample Preparation for LC-MS/MS Proteomics
4.7. LC-MS/MS Peptide Sequencing
4.8. Proteomic Analysis
4.9. Statistics
4.10. Gene Ontology (GO) Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| DCIS | Ductal carcinoma in situ |
| HYP | Hydroxylated proline |
| IBC | Invasive breast cancer |
| IDC | Invasive ductal carcinoma |
| LC-MS/MS | Liquid chromatography tandem mass spectrometry |
| MALDI-MSI | Matrix-Assisted Laser Desorption/Ionization–Mass Spectrometry Imaging |
| MALDI-QTOF | Matrix-Assisted Laser Desorption/Ionization–Quadruple Time-of-Flight |
| OX | Oxidation |
| PPM | Parts per million |
| sPLS-DA | Sparse Partial Least Squares Discriminant Analysis |
References
- Feinberg, J.; Wetstone, R.; Greenstein, D.; Borgen, P. Is DCIS Overrated? Springer International Publishing: Berlin/Heidelberg, Germany, 2018; pp. 53–72. [Google Scholar] [CrossRef]
- Sørum, R.; Hofvind, S.; Skaane, P.; Haldorsen, T. Trends in incidence of ductal carcinoma in situ: The effect of a population-based screening programme. Breast 2010, 19, 499–505. [Google Scholar] [CrossRef]
- Solin, L.J.; Gray, R.; Hughes, L.L.; Wood, W.C.; Lowen, M.A.; Badve, S.S.; Baehner, F.L.; Ingle, J.N.; Perez, E.A.; Recht, A.; et al. Surgical Excision Without Radiation for Ductal Carcinoma in Situ of the Breast: 12-Year Results From the ECOG-ACRIN E5194 Study. J. Clin. Oncol. 2015, 33, 3938–3944. [Google Scholar] [CrossRef]
- Bartlett, J.M.; Nofech-Moses, S.; Rakovitch, E. Ductal carcinoma in situ of the breast: Can biomarkers improve current management? Clin. Chem. 2014, 60, 60–67. [Google Scholar] [CrossRef][Green Version]
- Leonard, G.D.; Swain, S.M. Ductal carcinoma in situ, complexities and challenges. J. Natl. Cancer Inst. 2004, 96, 906–920. [Google Scholar] [CrossRef]
- Allison, K.H.; Hammond, M.E.H.; Dowsett, M.; McKernin, S.E.; Carey, L.A.; Fitzgibbons, P.L.; Hayes, D.F.; Lakhani, S.R.; Chavez-MacGregor, M.; Perlmutter, J.; et al. Estrogen and progesterone receptor testing in breast cancer: ASCO/CAP guideline update. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2020, 38, 1346–1366. [Google Scholar] [CrossRef]
- Punglia, R.S.; Jiang, W.; Lipsitz, S.R.; Hughes, M.E.; Schnitt, S.J.; Hassett, M.J.; Nekhlyudov, L.; Achacoso, N.; Edge, S.; Javid, S.H. Clinical risk score to predict likelihood of recurrence after ductal carcinoma in situ treated with breast-conserving surgery. Breast Cancer Res. Treat. 2018, 167, 751–759. [Google Scholar] [CrossRef]
- Risom, T.; Glass, D.R.; Averbukh, I.; Liu, C.C.; Baranski, A.; Kagel, A.; McCaffrey, E.F.; Greenwald, N.F.; Rivero-Gutiérrez, B.; Strand, S.H.; et al. Transition to invasive breast cancer is associated with progressive changes in the structure and composition of tumor stroma. Cell 2022, 185, 299–310.e18. [Google Scholar] [CrossRef]
- Strand, S.H.; Rivero-Gutierrez, B.; Houlahan, K.E.; Seoane, J.A.; King, L.M.; Risom, T.; Simpson, L.A.; Vennam, S.; Khan, A.; Cisneros, L.; et al. Molecular classification and biomarkers of clinical outcome in breast ductal carcinoma in situ: Analysis of TBCRC 038 and RAHBT cohorts. Cancer Cell 2022, 40, 1521–1536.e7. [Google Scholar] [CrossRef]
- Acerbi, I.; Cassereau, L.; Dean, I.; Shi, Q.; Au, A.; Park, C.; Chen, Y.Y.; Liphardt, J.; Hwang, E.S.; Weaver, V.M. Human breast cancer invasion and aggression correlates with ECM stiffening and immune cell infiltration. Integr. Biol. 2015, 7, 1120–1134. [Google Scholar] [CrossRef]
- Toss, M.S.; Miligy, I.M.; Gorringe, K.L.; Aleskandarany, M.A.; Alkawaz, A.; Mittal, K.; Aneja, R.; Ellis, I.O.; Green, A.R.; Rakha, E.A. Collagen (XI) alpha-1 chain is an independent prognostic factor in breast ductal carcinoma in situ. Mod. Pathol. 2019, 32, 1460–1472. [Google Scholar] [CrossRef]
- Papanicolaou, M.; Parker, A.L.; Yam, M.; Filipe, E.C.; Wu, S.Z.; Chitty, J.L.; Wyllie, K.; Tran, E.; Mok, E.; Nadalini, A.; et al. Temporal profiling of the breast tumour microenvironment reveals collagen XII as a driver of metastasis. Nat. Commun. 2022, 13, 4587. [Google Scholar] [CrossRef]
- Wottawa, M.; Leisering, P.; Ahlen, M.V.; Schnelle, M.; Vogel, S.; Malz, C.; Bordoli, M.R.; Camenisch, G.; Hesse, A.; Napp, J.; et al. Knockdown of prolyl-4-hydroxylase domain 2 inhibits tumor growth of human breast cancer MDA-MB-231 cells by affecting TGF-β1 processing. Int. J. Cancer 2013, 132, 2787–2798. [Google Scholar] [CrossRef]
- Xiong, G.; Deng, L.; Zhu, J.; Rychahou, P.G.; Xu, R. Prolyl-4-hydroxylase α subunit 2 promotes breast cancer progression and metastasis by regulating collagen deposition. BMC Cancer 2014, 14, 1. [Google Scholar] [CrossRef]
- Angel, P.M.; Comte-Walters, S.; Ball, L.E.; Talbot, K.; Mehta, A.; Brockbank, K.G.M.; Drake, R.R. Mapping Extracellular Matrix Proteins in Formalin-Fixed, Paraffin-Embedded Tissues by MALDI Imaging Mass Spectrometry. J. Proteome Res. 2018, 17, 635–646. [Google Scholar] [CrossRef]
- Angel, P.M.; Bruner, E.; Bethard, J.; Clift, C.L.; Ball, L.; Drake, R.R.; Feghali-Bostwick, C. Extracellular matrix alterations in low-grade lung adenocarcinoma compared with normal lung tissue by imaging mass spectrometry. J. Mass. Spectrom. 2020, 55, e4450. [Google Scholar] [CrossRef]
- Angel, P.M.; Spruill, L.; Jefferson, M.; Bethard, J.R.; Ball, L.E.; Hughes-Halbert, C.; Drake, R.R. Zonal regulation of collagen-type proteins and posttranslational modifications in prostatic benign and cancer tissues by imaging mass spectrometry. Prostate 2020, 80, 1071–1086. [Google Scholar] [CrossRef]
- Clift, C.L.; Drake, R.R.; Mehta, A.; Angel, P.M. Multiplexed imaging mass spectrometry of the extracellular matrix using serial enzyme digests from formalin-fixed paraffin-embedded tissue sections. Anal. Bioanal. Chem. 2021, 413, 2709–2719. [Google Scholar] [CrossRef]
- Clift, C.L.; McLaughlin, S.; Munoz, M.; Suuronen, E.J.; Rotstein, B.H.; Mehta, A.S.; Drake, R.R.; Alarcon, E.I.; Angel, P.M. Evaluation of Therapeutic Collagen-Based Biomaterials in the Infarcted Mouse Heart by Extracellular Matrix Targeted MALDI Imaging Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2021, 32, 2746–2754. [Google Scholar] [CrossRef]
- Rujchanarong, D.; Lefler, J.; Saunders, J.E.; Pippin, S.; Spruill, L.; Bethard, J.R.; Ball, L.E.; Mehta, A.S.; Drake, R.R.; Ostrowski, M.C.; et al. Defining the Tumor Microenvironment by Integration of Immunohistochemistry and Extracellular Matrix Targeted Imaging Mass Spectrometry. Cancers 2021, 13, 4419. [Google Scholar] [CrossRef]
- Sprague, B.L.; Vacek, P.M.; Mulrow, S.E.; Evans, M.F.; Trentham-Dietz, A.; Herschorn, S.D.; James, T.A.; Surachaicharn, N.; Keikhosravi, A.; Eliceiri, K.W.; et al. Collagen Organization in Relation to Ductal Carcinoma In Situ Pathology and Outcomes. Cancer Epidemiol. Biomark. Prev. 2021, 30, 80–88. [Google Scholar] [CrossRef]
- Angel, P.M.; Zambrzycki, S.C. Predictive value of collagen in cancer. Adv. Cancer Res. 2022, 154, 15–45. [Google Scholar] [CrossRef]
- Toss, M.S.; Miligy, I.M.; Gorringe, K.L.; AlKawaz, A.; Mittal, K.; Aneja, R.; Ellis, I.O.; Green, A.R.; Roxanis, I.; Rakha, E.A. Geometric characteristics of collagen have independent prognostic significance in breast ductal carcinoma in situ: An image analysis study. Mod. Pathol. 2019, 32, 1473–1485. [Google Scholar] [CrossRef]
- Sweeney, S.M.; Orgel, J.P.; Fertala, A.; McAuliffe, J.D.; Turner, K.R.; Di Lullo, G.A.; Chen, S.; Antipova, O.; Perumal, S.; Ala-Kokko, L.; et al. Candidate cell and matrix interaction domains on the collagen fibril, the predominant protein of vertebrates. J. Biol. Chem. 2008, 283, 21187–21197. [Google Scholar] [CrossRef]
- Di Lullo, G.A.; Sweeney, S.M.; Korkko, J.; Ala-Kokko, L.; San Antonio, J.D. Mapping the ligand-binding sites and disease-associated mutations on the most abundant protein in the human, type I collagen. J. Biol. Chem. 2002, 277, 4223–4231. [Google Scholar] [CrossRef]
- San Antonio, J.D.; Jacenko, O.; Fertala, A.; Orgel, J. Collagen Structure-Function Mapping Informs Applications for Regenerative Medicine. Bioengineering 2020, 8, 3. [Google Scholar] [CrossRef]
- Shoulders, M.D.; Raines, R.T. Collagen structure and stability. Annu. Rev. Biochem. 2009, 78, 929–958. [Google Scholar] [CrossRef]
- Kamil, M.; Shinsato, Y.; Higa, N.; Hirano, T.; Idogawa, M.; Takajo, T.; Minami, K.; Shimokawa, M.; Yamamoto, M.; Kawahara, K.; et al. High filamin-C expression predicts enhanced invasiveness and poor outcome in glioblastoma multiforme. Br. J. Cancer 2019, 120, 819–826. [Google Scholar] [CrossRef]
- Qi, Y.; Xu, F.; Chen, L.; Li, Y.; Xu, Z.; Zhang, Y.; Wei, W.; Su, N.; Zhang, T.; Fan, F.; et al. Quantitative proteomics reveals FLNC as a potential progression marker for the development of hepatocellular carcinoma. Oncotarget 2016, 7, 68242–68252. [Google Scholar] [CrossRef]
- UniProt, C. UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023, 51, D523–D531. [Google Scholar] [CrossRef]
- Quinn, C.M.; Ostrowski, J.L. Cytological and architectural heterogeneity in ductal carcinoma in situ of the breast. J. Clin. Pathol. 1997, 50, 596–599. [Google Scholar] [CrossRef]
- Van Seijen, M.; Jóźwiak, K.; Pinder, S.E.; Hall, A.; Krishnamurthy, S.; Thomas, J.S.; Collins, L.C.; Bijron, J.; Bart, J.; Cohen, D.; et al. Variability in grading of ductal carcinoma in situ among an international group of pathologists. J. Pathol. Clin. Res. 2021, 7, 233–242. [Google Scholar] [CrossRef]
- Le Cao, K.A.; Boitard, S.; Besse, P. Sparse PLS discriminant analysis: Biologically relevant feature selection and graphical displays for multiclass problems. BMC Bioinform. 2011, 12, 253. [Google Scholar] [CrossRef]
- Davies, E.L.; Gee JM, W.; Cochrane, R.A.; Jiang, W.G.; Sharma, A.K.; Nicholson, R.I.; Mansel, R.E. The immunohistochemical expression of desmoplakin and its role in vivo in the progression and metastasis of breast cancer. Eur. J. Cancer 1999, 35, 902–907. [Google Scholar] [CrossRef]
- Piacente, F.; Caffa, I.; Nencioni, A. Nicotinic acid: A case for a vitamin that moonlights for cancer? Cell Cycle 2017, 16, 1635–1636. [Google Scholar] [CrossRef]
- Rivenbark, A.G.; Coleman, W.B. Field cancerization in mammary carcinogenesis—Implications for prevention and treatment of breast cancer. Exp. Mol. Pathol. 2012, 93, 391–398. [Google Scholar] [CrossRef]
- Forsberg, L.A.; Rasi, C.; Pekar, G.; Davies, H.; Piotrowski, A.; Absher, D.; Razzaghian, H.R.; Ambicka, A.; Halaszka, K.; Przewoznik, M.; et al. Signatures of post-zygotic structural genetic aberrations in the cells of histologically normal breast tissue that can predispose to sporadic breast cancer. Genome Res. 2015, 25, 1521–1535. [Google Scholar] [CrossRef]
- Asioli, S.; Morandi, L.; Cavatorta, C.; Cucchi, M.C.; Foschini, M.P. The impact of field cancerization on the extent of duct carcinoma in situ (DCIS) in breast tissue after conservative excision. Eur. J. Surg. Oncol. 2016, 42, 1806–1813. [Google Scholar] [CrossRef]
- Muse, M.E.; Titus, A.J.; Salas, L.A.; Wilkins, O.M.; Mullen, C.; Gregory, K.J.; Schneider, S.S.; Crisi, G.M.; Jawale, R.M.; Otis, C.N.; et al. Enrichment of CpG island shore region hypermethylation in epigenetic breast field cancerization. Epigenetics 2020, 15, 1093–1106. [Google Scholar] [CrossRef]
- Srour, M.K.; Gao, B.; Dadmanesh, F.; Carlson, K.; Qu, Y.; Deng, N.; Cui, X.; Giuliano, A.E. Gene expression comparison between primary triple-negative breast cancer and paired axillary and sentinel lymph node metastasis. Breast J. 2020, 26, 904–910. [Google Scholar] [CrossRef]
- Paul, A.M.; George, B.; Saini, S.; Pillai, M.R.; Toi, M.; Costa, L.; Kumar, R. Delineation of Pathogenomic Insights of Breast Cancer in Young Women. Cells 2022, 11, 1927. [Google Scholar] [CrossRef]
- Yang, F.; Lin, L.; Li, X.; Wen, R.; Zhang, X. Silencing of COL3A1 represses proliferation, migration, invasion, and immune escape of triple negative breast cancer cells via down-regulating PD-L1 expression. Cell Biol. Int. 2022, 46, 1959–1969. [Google Scholar] [CrossRef]
- Salesse, S.; Odoul, L.; Chazee, L.; Garbar, C.; Duca, L.; Martiny, L.; Mahmoudi, R.; Debelle, L. Elastin molecular aging promotes MDA-MB-231 breast cancer cell invasiveness. FEBS Open Bio 2018, 8, 1395–1404. [Google Scholar] [CrossRef]
- Casasent, A.K.; Schalck, A.; Gao, R.; Sei, E.; Long, A.; Pangburn, W.; Casasent, T.; Meric-Bernstam, F.; Edgerton, M.E.; Navin, N.E. Multiclonal Invasion in Breast Tumors Identified by Topographic Single Cell Sequencing. Cell 2018, 172, 205–217.e12. [Google Scholar] [CrossRef]
- Sinha, V.C.; Piwnica-Worms, H. Intratumoral Heterogeneity in Ductal Carcinoma In Situ: Chaos and Consequence. J. Mammary Gland Biol. Neoplasia 2018, 23, 191–205. [Google Scholar] [CrossRef]
- Conklin, M.W.; Gangnon, R.E.; Sprague, B.L.; Van Gemert, L.; Hampton, J.M.; Eliceiri, K.W.; Bredfeldt, J.S.; Liu, Y.; Surachaicharn, N.; Newcomb, P.A. Collagen alignment as a predictor of recurrence after ductal carcinoma in situ. Cancer Epidemiol. Biomark. Prev. 2018, 27, 138–145. [Google Scholar] [CrossRef]
- Li, H.; Bera, K.; Toro, P.; Fu, P.; Zhang, Z.; Lu, C.; Feldman, M.; Ganesan, S.; Goldstein, L.J.; Davidson, N.E. Collagen fiber orientation disorder from H&E images is prognostic for early stage breast cancer: Clinical trial validation. NPJ Breast Cancer 2021, 7, 104. [Google Scholar]
- Conklin, M.W.; Eickhoff, J.C.; Riching, K.M.; Pehlke, C.A.; Eliceiri, K.W.; Provenzano, P.P.; Friedl, A.; Keely, P.J. Aligned Collagen Is a Prognostic Signature for Survival in Human Breast Carcinoma. Am. J. Pathol. 2011, 178, 1221–1232. [Google Scholar] [CrossRef]
- Di Martino, J.S.; Nobre, A.R.; Mondal, C.; Taha, I.; Farias, E.F.; Fertig, E.J.; Naba, A.; Aguirre-Ghiso, J.A.; Bravo-Cordero, J.J. A tumor-derived type III collagen-rich ECM niche regulates tumor cell dormancy. Nat. Cancer 2022, 3, 90–107. [Google Scholar] [CrossRef]
- Sun, X.; Wu, B.; Chiang, H.-C.; Deng, H.; Zhang, X.; Xiong, W.; Liu, J.; Rozeboom, A.M.; Harris, B.T.; Blommaert, E.; et al. Tumour DDR1 promotes collagen fibre alignment to instigate immune exclusion. Nature 2021, 599, 673–678. [Google Scholar] [CrossRef]
- Shi, R.; Gao, S.; Zhang, J.; Xu, J.; Graham, L.M.; Yang, X.; Li, C. Collagen prolyl 4-hydroxylases modify tumor progression. Acta Biochim. Biophys. Sin. 2021, 53, 805–814. [Google Scholar] [CrossRef]
- Gilkes, D.M.; Chaturvedi, P.; Bajpai, S.; Wong, C.C.; Wei, H.; Pitcairn, S.; Hubbi, M.E.; Wirtz, D.; Semenza, G.L. Collagen Prolyl Hydroxylases Are Essential for Breast Cancer Metastasis. Cancer Res. 2013, 73, 3285–3296. [Google Scholar] [CrossRef]
- Toss, M.S.; Miligy, I.M.; Gorringe, K.L.; AlKawaz, A.; Khout, H.; Ellis, I.O.; Green, A.R.; Rakha, E.A. Prolyl-4-hydroxylase A subunit 2 (P4HA2) expression is a predictor of poor outcome in breast ductal carcinoma in situ (DCIS). Br. J. Cancer 2018, 119, 1518–1526. [Google Scholar] [CrossRef]
- Curtius, K.; Wright, N.A.; Graham, T.A. An evolutionary perspective on field cancerization. Nat. Rev. Cancer 2018, 18, 19–32. [Google Scholar] [CrossRef]
- Shlyakhtina, Y.; Moran, K.L.; Portal, M.M. Genetic and Non-Genetic Mechanisms Underlying Cancer Evolution. Cancers 2021, 13, 1380. [Google Scholar] [CrossRef]
- Lips, E.H.; Kumar, T.; Megalios, A.; Visser, L.L.; Sheinman, M.; Fortunato, A.; Shah, V.; Hoogstraat, M.; Sei, E.; Mallo, D.; et al. Genomic analysis defines clonal relationships of ductal carcinoma in situ and recurrent invasive breast cancer. Nat. Genet. 2022, 54, 850–860. [Google Scholar] [CrossRef]
- Harrison, B.T.; Hwang, E.S.; Partridge, A.H.; Thompson, A.M.; Schnitt, S.J. Variability in diagnostic threshold for comedo necrosis among breast pathologists: Implications for patient eligibility for active surveillance trials of ductal carcinoma in situ. Mod. Pathol. 2019, 32, 1257–1262. [Google Scholar] [CrossRef]
- Kerlikowske, K.; Molinaro, A.; Cha, I.; Ljung, B.M.; Ernster, V.L.; Stewart, K.; Chew, K.; Moore, D.H., 2nd; Waldman, F. Characteristics associated with recurrence among women with ductal carcinoma in situ treated by lumpectomy. J. Natl. Cancer Inst. 2003, 95, 1692–1702. [Google Scholar] [CrossRef]
- Liu, Y.; Colditz, G.A.; Gehlert, S.; Goodman, M. Racial disparities in risk of second breast tumors after ductal carcinoma in situ. Breast Cancer Res. Treat. 2014, 148, 163–173. [Google Scholar] [CrossRef]
- Lester, S.C.; Bose, S.; Chen, Y.-Y.; Connolly, J.L.; De Baca, M.E.; Fitzgibbons, P.L.; Hayes, D.F.; Kleer, C.; O’Malley, F.P.; Page, D.L.; et al. Protocol for the Examination of Specimens From Patients With Ductal Carcinoma In Situ of the Breast. Arch. Pathol. Lab. Med. 2009, 133, 15–25. [Google Scholar] [CrossRef]
- Powers, T.W.; Jones, E.E.; Betesh, L.R.; Romano, P.R.; Gao, P.; Copland, J.A.; Mehta, A.S.; Drake, R.R. Matrix assisted laser desorption ionization imaging mass spectrometry workflow for spatial profiling analysis of N-linked glycan expression in tissues. Anal. Chem. 2013, 85, 9799–9806. [Google Scholar] [CrossRef]
- Powers, T.W.; Neely, B.A.; Shao, Y.; Tang, H.; Troyer, D.A.; Mehta, A.S.; Haab, B.B.; Drake, R.R. MALDI imaging mass spectrometry profiling of N-glycans in formalin-fixed paraffin embedded clinical tissue blocks and tissue microarrays. PLoS ONE 2014, 9, e106255. [Google Scholar] [CrossRef]
- Powers, T.W.; Holst, S.; Wuhrer, M.; Mehta, A.S.; Drake, R.R. Two-Dimensional N-Glycan Distribution Mapping of Hepatocellular Carcinoma Tissues by MALDI-Imaging Mass Spectrometry. Biomolecules 2015, 5, 2554–2572. [Google Scholar] [CrossRef]
- Nesvizhskii, A.I.; Keller, A.; Kolker, E.; Aebersold, R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003, 75, 4646–4658. [Google Scholar] [CrossRef]
- Keller, A.; Nesvizhskii, A.I.; Kolker, E.; Aebersold, R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 2002, 74, 5383–5392. [Google Scholar] [CrossRef]
- Lu, Y.; Pang, Z.; Xia, J. Comprehensive investigation of pathway enrichment methods for functional interpretation of LC-MS global metabolomics data. Brief Bioinform. 2023, 24, bbac553. [Google Scholar] [CrossRef]
- Goedhart, J.; Luijsterburg, M.S. VolcaNoseR is a web app for creating, exploring, labeling and sharing volcano plots. Sci. Rep. 2020, 10, 20560. [Google Scholar] [CrossRef]



| Age of Dx, Mean (SD) | n = 13 Patients | |
|---|---|---|
| 58.4 (SD = 13.6) | ||
| Pathology, n (%) | n = 18 specimens | |
| DCIS only | 7 (38.9%) | |
| Mixed DCIS-IDC | 6 (33.3%) | |
| IDC | 4 (22.2%) | |
| Inflammatory foci | 1 (5.6%) | |
| Surgical Treatment, n (%) | n = 11 patients | |
| Lumpectomy | 5 (45.5%) | |
| Partial mastectomy | 3 (27.3%) | |
| Mastectomy | 3 (27.3%) | |
| Race, n (%) | n = 6 patients | |
| African American | 1 (16.7%) | |
| White | 5 (83.3%) | |
| DCIS | IDC | |
| Nuclear Grade, n (%) | ||
| 1 | 0 (0%) | 0 (0%) |
| 2 | 3 (23.1%) | 2 (20%) |
| 3 | 10 (76.9%) | 8 (80%) |
| Architecture, n (%) | Note some DCIS samples have mixed pathology | |
| Solid | 11 (84.6%) | NA |
| Cribriform | 3 (23.1%) | NA |
| Comedo necrosis | 6 (46.2%) | NA |
| Micropapillary | 1 (7.7%) | NA |
| Pathological tumor size (cm), Mean (SD) | n = 4 * | n = 13 |
| 5.4 (SD = 2.9) | 3.3 (SD = 2.6) | |
| Marker Status, n (%) | n = 8 | n = 13 |
| ER(+) | 3 (37.5%) | 7 (53.8%) |
| PR(+) | 2 (25.0%) | 7 (53.8%) |
| HER2(+) | 3 (37.5%) | 7 (53.8%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hulahan, T.S.; Spruill, L.; Wallace, E.N.; Park, Y.; West, R.B.; Marks, J.R.; Hwang, E.S.; Drake, R.R.; Angel, P.M. Extracellular Microenvironment Alterations in Ductal Carcinoma In Situ and Invasive Breast Cancer Pathologies by Multiplexed Spatial Proteomics. Int. J. Mol. Sci. 2024, 25, 6748. https://doi.org/10.3390/ijms25126748
Hulahan TS, Spruill L, Wallace EN, Park Y, West RB, Marks JR, Hwang ES, Drake RR, Angel PM. Extracellular Microenvironment Alterations in Ductal Carcinoma In Situ and Invasive Breast Cancer Pathologies by Multiplexed Spatial Proteomics. International Journal of Molecular Sciences. 2024; 25(12):6748. https://doi.org/10.3390/ijms25126748
Chicago/Turabian StyleHulahan, Taylor S., Laura Spruill, Elizabeth N. Wallace, Yeonhee Park, Robert B. West, Jeffrey R. Marks, E. Shelley Hwang, Richard R. Drake, and Peggi M. Angel. 2024. "Extracellular Microenvironment Alterations in Ductal Carcinoma In Situ and Invasive Breast Cancer Pathologies by Multiplexed Spatial Proteomics" International Journal of Molecular Sciences 25, no. 12: 6748. https://doi.org/10.3390/ijms25126748
APA StyleHulahan, T. S., Spruill, L., Wallace, E. N., Park, Y., West, R. B., Marks, J. R., Hwang, E. S., Drake, R. R., & Angel, P. M. (2024). Extracellular Microenvironment Alterations in Ductal Carcinoma In Situ and Invasive Breast Cancer Pathologies by Multiplexed Spatial Proteomics. International Journal of Molecular Sciences, 25(12), 6748. https://doi.org/10.3390/ijms25126748






