A Quantitative Human Red Blood Cell Agglutination Assay for Characterisation of Galectin Inhibitors
Abstract
1. Introduction
2. Results
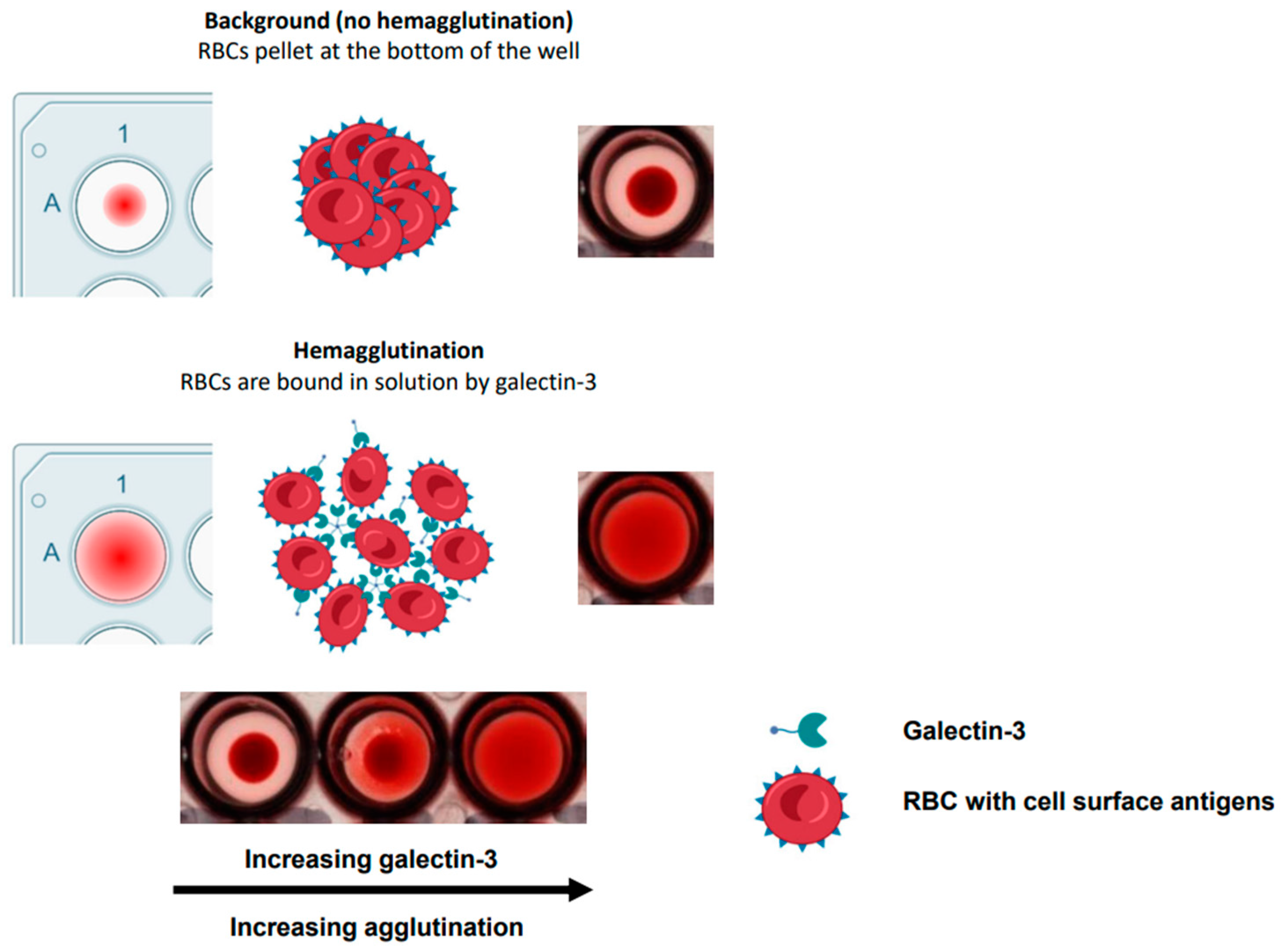
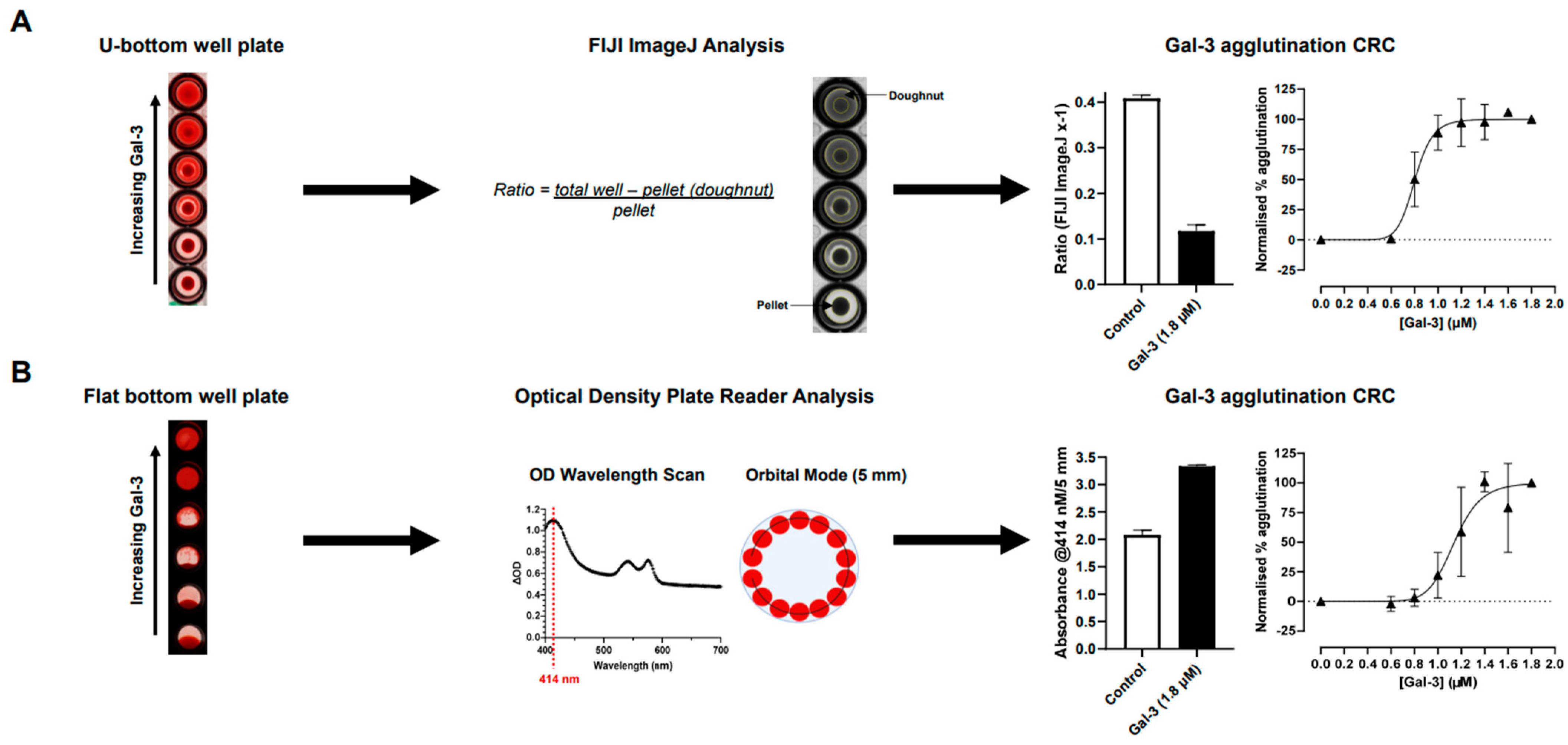
2.1. Optimisation of Hemagglutination Assay
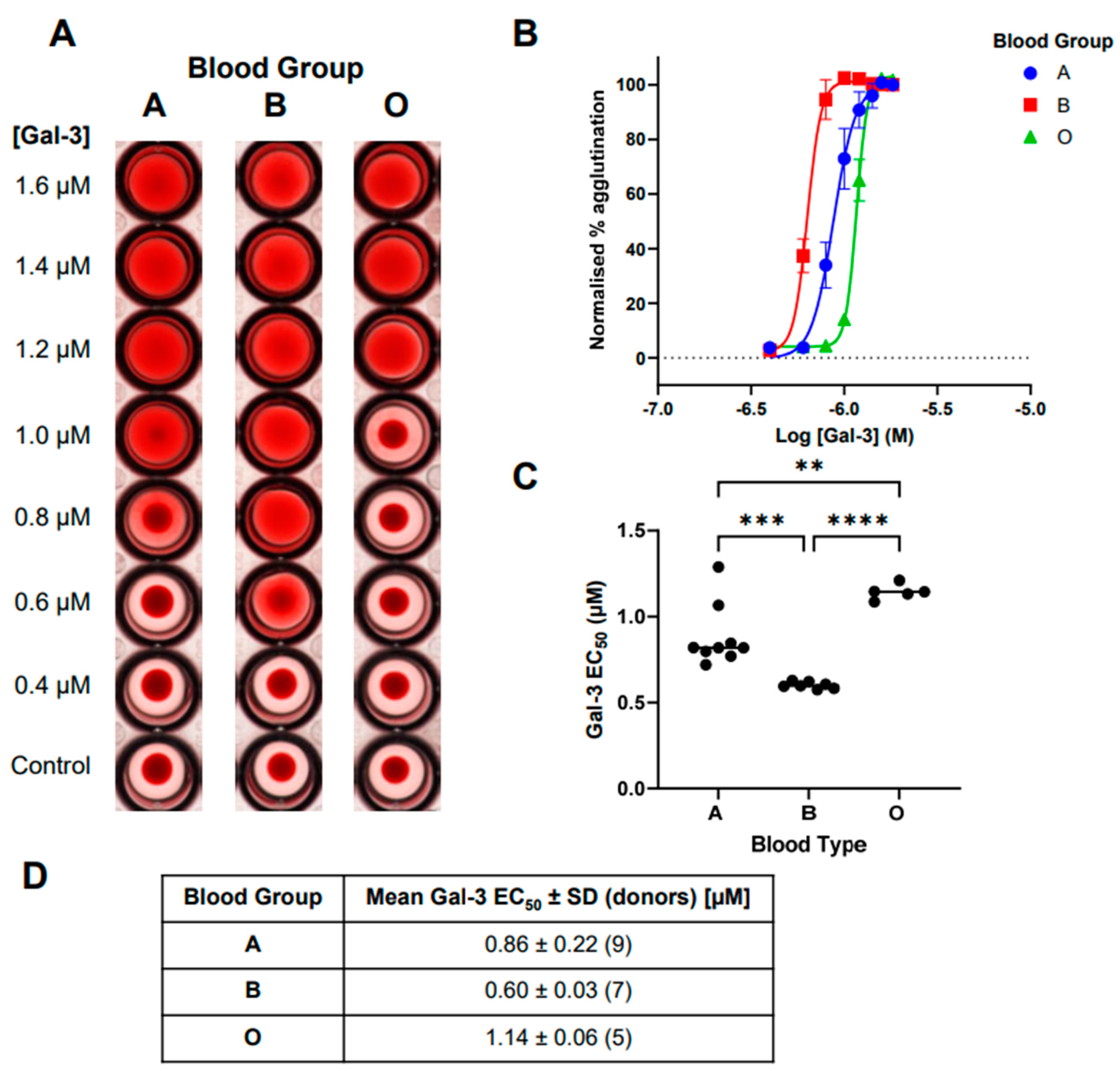
2.2. Comparison of Galectin-3-Induced Hemagglutination across Blood Groups A, B, and O
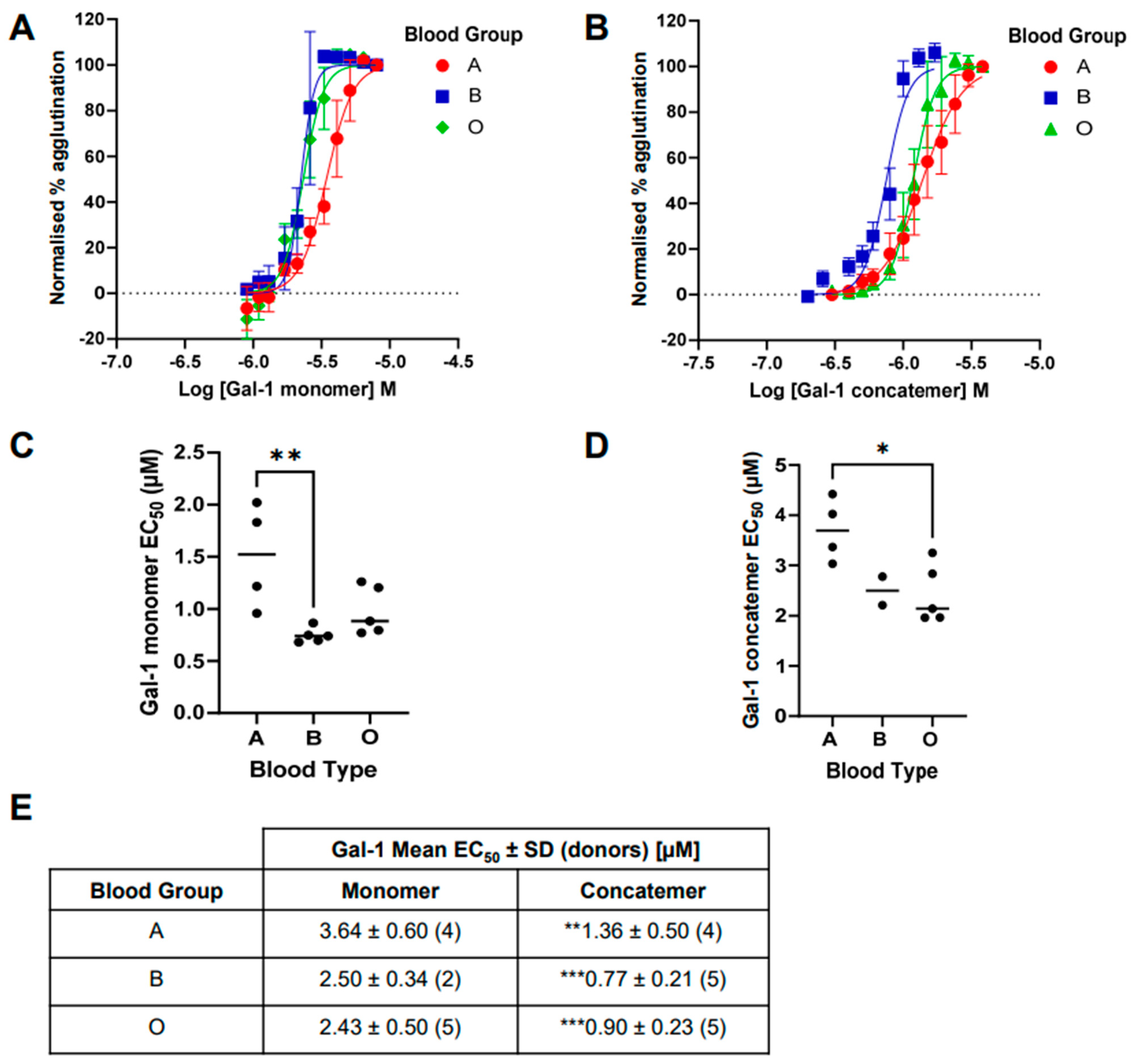
2.3. Comparison of Galectin-1-Induced Hemagglutination across Blood Groups A, B, and O
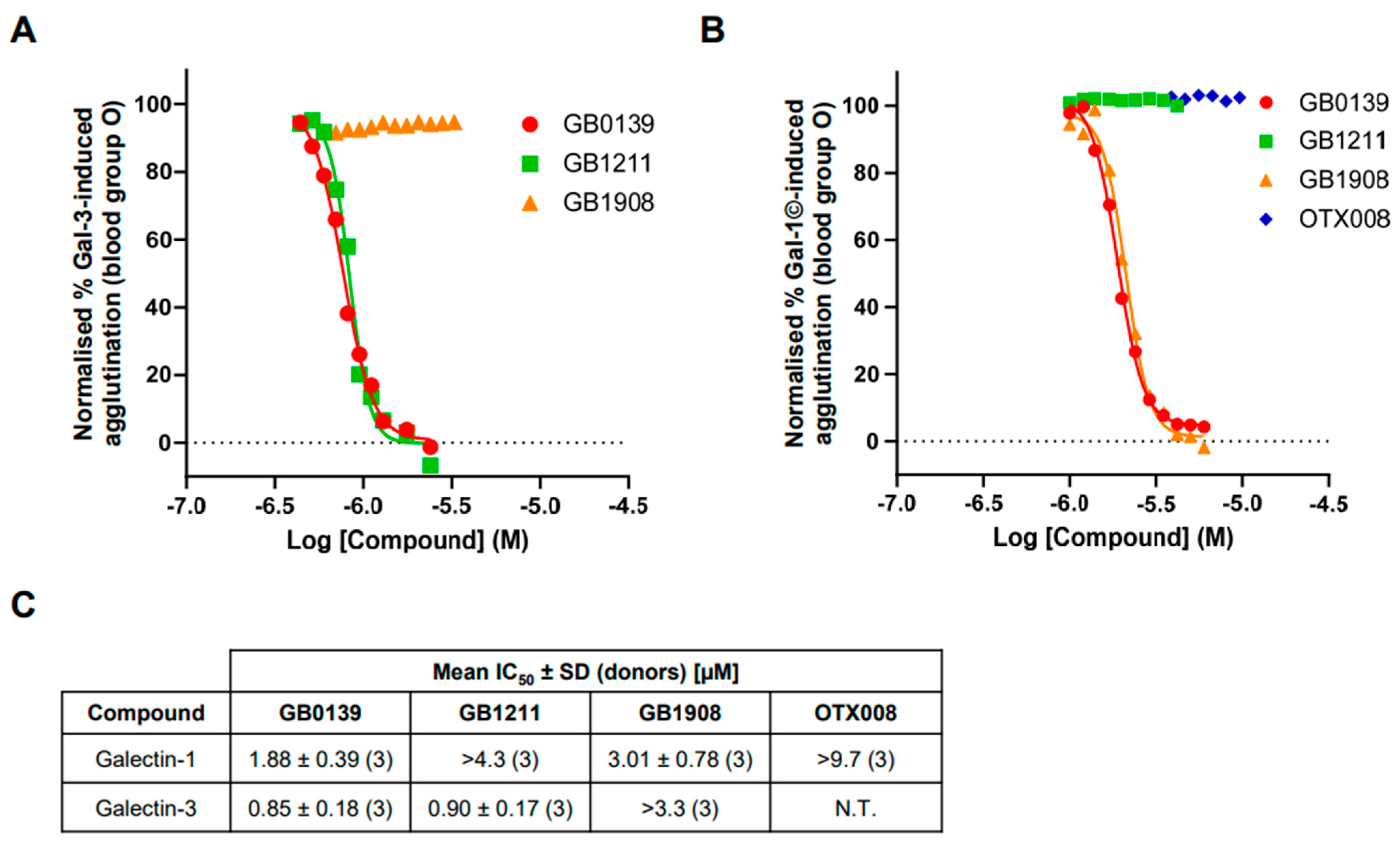
2.4. Inhibition of Galectin-Induced Hemagglutination with Small-Molecule Glycomimetic and Allosteric Inhibitors
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Red Blood Cell Preparation
4.3. Hemagglutination Assay
4.3.1. Optimisation of Hemagglutination Quantification
4.3.2. Determining Galectin Inhibitor Hemagglutination IC50 Values
4.4. Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yang, R.-Y.; Rabinovich, G.A.; Liu, F.-T. Galectins: Structure, function and therapeutic potential. Expert Rev. Mol. Med. 2008, 10, e17. [Google Scholar] [CrossRef]
- Slack, R.J.; Mills, R.; Mackinnon, A.C. The therapeutic potential of galectin-3 inhibition in fibrotic disease. Int. J. Biochem. Cell Biol. 2020, 130, 105881. [Google Scholar] [CrossRef]
- Ruvolo, P.P. Galectin 3 as a guardian of the tumor microenvironment. Biochim. Biophys. Acta (BBA)—Mol. Cell Res. 2016, 1863, 427–437. [Google Scholar] [CrossRef]
- Nabi, I.R.; Shankar, J.; Dennis, J.W. The galectin lattice at a glance. J. Cell Sci. 2015, 128, 2213–2219. [Google Scholar] [CrossRef]
- Troncoso, M.F.; Elola, M.T.; Blidner, A.G.; Sarrias, L.; Espelt, M.V.; Rabinovich, G.A. The universe of galectin-binding partners and their functions in health and disease. J. Biol. Chem. 2023, 299, 105400. [Google Scholar] [CrossRef]
- Mariño, K.V.; Cagnoni, A.J.; Croci, D.O.; Rabinovich, G.A. Targeting galectin-driven regulatory circuits in cancer and fibrosis. Nat. Rev. Drug Discov. 2023, 22, 295–316. [Google Scholar] [CrossRef]
- Huang, Y.; Wang, H.-C.; Zhao, J.; Wu, M.-H.; Shih, T.-C. Immunosuppressive Roles of Galectin-1 in the Tumor Microenvironment. Biomolecules 2021, 11, 1398. [Google Scholar] [CrossRef]
- Stowell, S.R.; Arthur, C.M.; Mehta, P.; Slanina, K.A.; Blixt, O.; Leffler, H.; Smith, D.F.; Cummings, R.D. Galectin-1, -2, and -3 Exhibit Differential Recognition of Sialylated Glycans and Blood Group Antigens. J. Biol. Chem. 2008, 283, 10109–10123. [Google Scholar] [CrossRef]
- Stegmayr, J.; Lepur, A.; Kahl-Knutson, B.; Aguilar-Moncayo, M.; Klyosov, A.A.; Field, R.A.; Oredsson, S.; Nilsson, U.J.; Leffler, H. Low or No Inhibitory Potency of the Canonical Galectin Carbohydrate-binding Site by Pectins and Galactomannans. J. Biol. Chem. 2016, 291, 13318–13334. [Google Scholar] [CrossRef]
- Shu, Z.; Li, J.; Mu, N.; Gao, Y.; Huang, T.; Zhang, Y.; Wang, Z.; Li, M.; Hao, Q.; Li, W.; et al. Expression, purification and characterization of galectin-1 in Escherichia coli. Protein Expr. Purif. 2014, 99, 58–63. [Google Scholar] [CrossRef]
- Sörme, P.; Kahl-Knutsson, B.; Huflejt, M.; Nilsson, U.J.; Leffler, H. Fluorescence polarization as an analytical tool to evaluate galectin–ligand interactions. Anal. Biochem. 2004, 334, 36–47. [Google Scholar] [CrossRef]
- Delaine, T.; Collins, P.; MacKinnon, A.; Sharma, G.; Stegmayr, J.; Rajput, V.K.; Mandal, S.; Cumpstey, I.; Larumbe, A.; Salameh, B.A.; et al. Galectin-3-Binding Glycomimetics that Strongly Reduce Bleomycin-Induced Lung Fibrosis and Modulate Intracellular Glycan Recognition. ChemBioChem 2016, 17, 1759–1770. [Google Scholar] [CrossRef]
- Zetterberg, F.R.; MacKinnon, A.; Brimert, T.; Gravelle, L.; Johnsson, R.E.; Kahl-Knutson, B.; Leffler, H.; Nilsson, U.J.; Pedersen, A.; Peterson, K.; et al. Discovery and Optimization of the First Highly Effective and Orally Available Galectin-3 Inhibitors for Treatment of Fibrotic Disease. J. Med. Chem. 2022, 65, 12626–12638. [Google Scholar] [CrossRef]
- Zetterberg, F.R.; Peterson, K.; Nilsson, U.J.; Dahlgren, K.A.; Diehl, C.; Holyer, I.; Håkansson, M.; Khabut, A.; Kahl-Knutson, B.; Leffler, H.; et al. Discovery of the selective and orally available galectin-1 inhibitor GB1908 as a potential treatment for lung cancer. J. Med. Chem. 2024, 67, 9374–9388. [Google Scholar] [CrossRef]
- Dings, R.P.; Miller, M.C.; Nesmelova, I.; Astorgues-Xerri, L.; Kumar, N.; Serova, M.; Chen, X.; Raymond, E.; Hoye, T.R.; Mayo, K.H. Antitumor Agent Calixarene 0118 Targets Human Galectin-1 as an Allosteric Inhibitor of Carbohydrate Binding. J. Med. Chem. 2012, 55, 5121–5129. [Google Scholar] [CrossRef]
- Pozder, C.; Screever, E.M.; van der Velde, A.R.; Silljé, H.H.; Suwijn, J.; de Rond, S.; Kleber, M.E.; Delgado, G.; Schuringa, J.J.; van Gilst, W.H.; et al. Galectin-3 and Blood Group: Binding Properties, Effects on Plasma Levels, and Consequences for Prognostic Performance. Int. J. Mol. Sci. 2023, 24, 4415. [Google Scholar] [CrossRef]
- MacKinnon, A.C.; Gibbons, M.A.; Farnworth, S.L.; Leffler, H.; Nilsson, U.J.; Delaine, T.; Simpson, A.J.; Forbes, S.J.; Hirani, N.; Gauldie, J.; et al. Regulation of transforming growth factor-β1-driven lung fibrosis by galectin-3. Am. J. Respir. Crit. Care Med. 2012, 185, 537–546. [Google Scholar] [CrossRef]
- Hirani, N.; MacKinnon, A.C.; Nicol, L.; Ford, P.; Schambye, H.; Pedersen, A.; Nilsson, U.J.; Leffler, H.; Sethi, T.; Tantawi, S.; et al. Target inhibition of galectin-3 by inhaled TD139 in patients with idiopathic pulmonary fibrosis. Eur. Respir. J. 2021, 57, 2002559. [Google Scholar] [CrossRef]
- Aslanis, V.; Slack, R.J.; MacKinnon, A.C.; McClinton, C.; Tantawi, S.; Gravelle, L.; Nilsson, U.J.; Leffler, H.; Brooks, A.; Khindri, S.K.; et al. Safety and pharmacokinetics of GB1211, an oral galectin-3 inhibitor: A single- and multiple-dose first-in-human study in healthy participants. Cancer Chemother. Pharmacol. 2023, 91, 267–280. [Google Scholar] [CrossRef]
- Salomonsson, E.; Larumbe, A.; Tejler, J.; Tullberg, E.; Rydberg, H.; Sundin, A.; Khabut, A.; Frejd, T.; Lobsanov, Y.D.; Rini, J.M.; et al. Monovalent Interactions of Galectin-1. Biochemistry 2010, 49, 9518–9532. [Google Scholar] [CrossRef] [PubMed]
- Salomonsson, E.; Carlsson, M.C.; Osla, V.; Hendus-Altenburger, R.; Kahl-Knutson, B.; Öberg, C.T.; Sundin, A.; Nilsson, R.; Nordberg-Karlsson, E.; Nilsson, U.J.; et al. Mutational Tuning of Galectin-3 Specificity and Biological Function. J. Biol. Chem. 2010, 285, 35079–35091. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gasson, R.; Roper, J.A.; Slack, R.J. A Quantitative Human Red Blood Cell Agglutination Assay for Characterisation of Galectin Inhibitors. Int. J. Mol. Sci. 2024, 25, 6756. https://doi.org/10.3390/ijms25126756
Gasson R, Roper JA, Slack RJ. A Quantitative Human Red Blood Cell Agglutination Assay for Characterisation of Galectin Inhibitors. International Journal of Molecular Sciences. 2024; 25(12):6756. https://doi.org/10.3390/ijms25126756
Chicago/Turabian StyleGasson, Rhianna, James A. Roper, and Robert J. Slack. 2024. "A Quantitative Human Red Blood Cell Agglutination Assay for Characterisation of Galectin Inhibitors" International Journal of Molecular Sciences 25, no. 12: 6756. https://doi.org/10.3390/ijms25126756
APA StyleGasson, R., Roper, J. A., & Slack, R. J. (2024). A Quantitative Human Red Blood Cell Agglutination Assay for Characterisation of Galectin Inhibitors. International Journal of Molecular Sciences, 25(12), 6756. https://doi.org/10.3390/ijms25126756








