Comparative Transcriptome Analysis Reveals Key Functions of MiMYB Gene Family in Macadamia Nut Pericarp Formation
Abstract
:1. Introduction
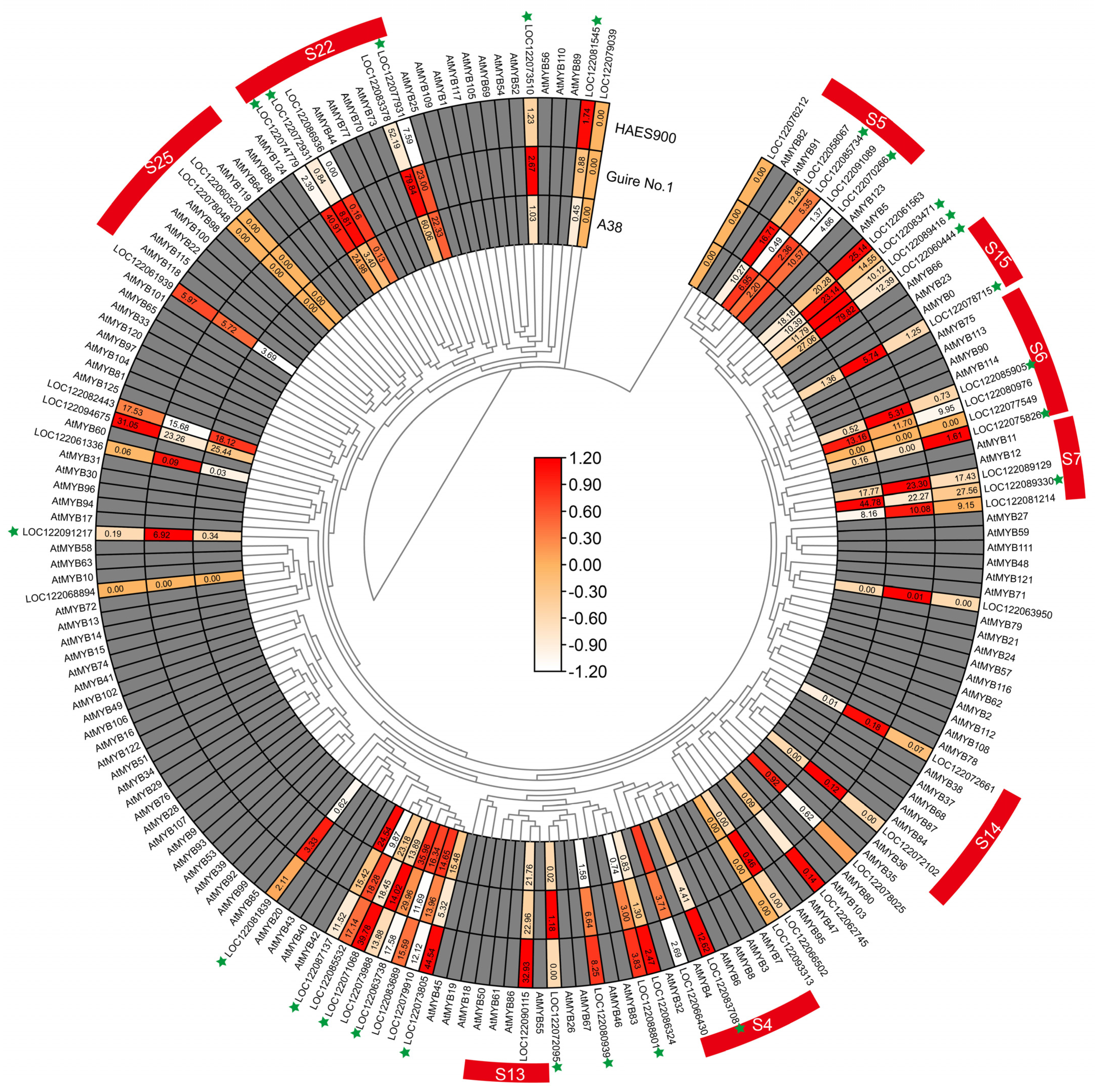
2. Results
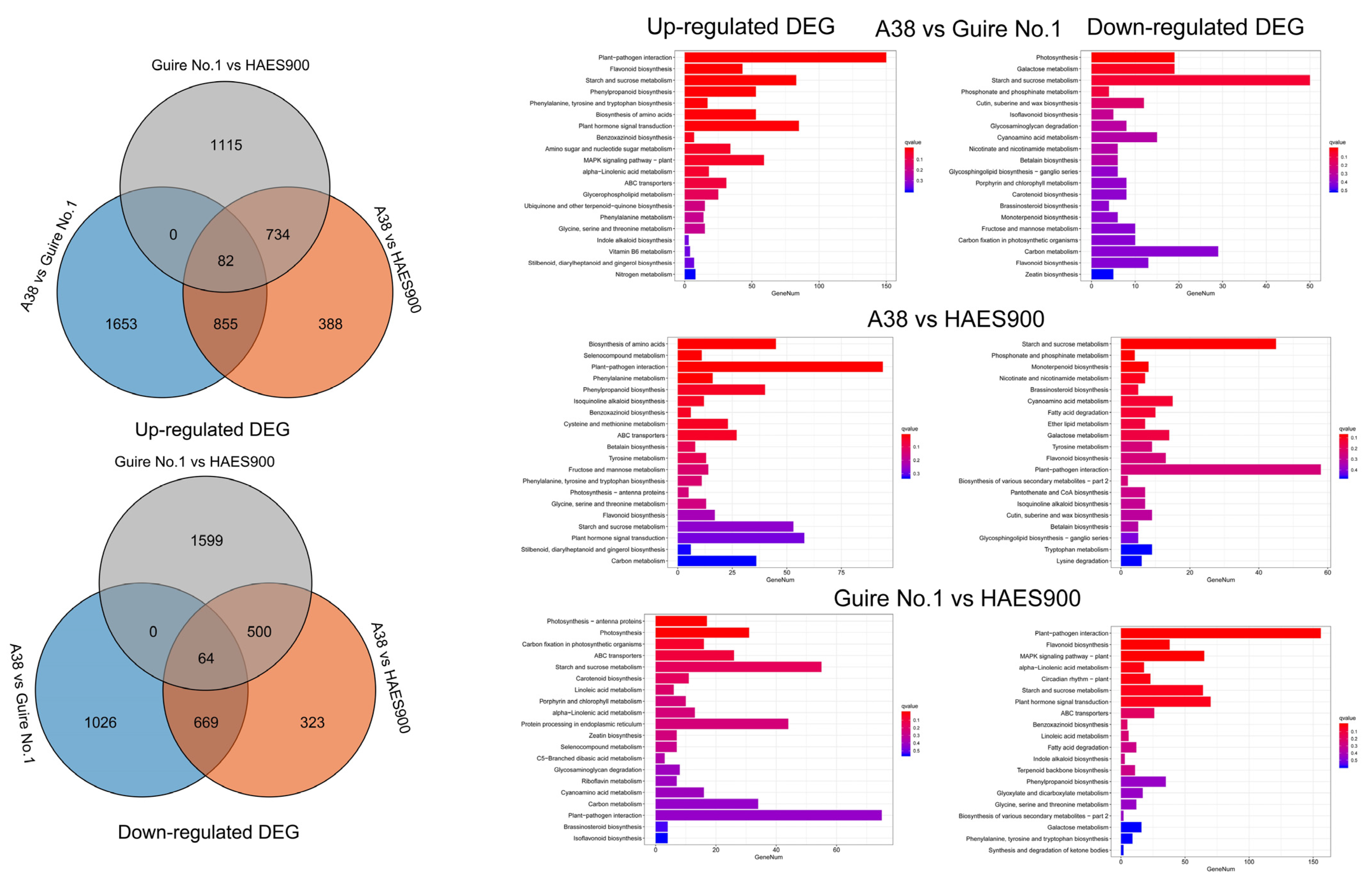
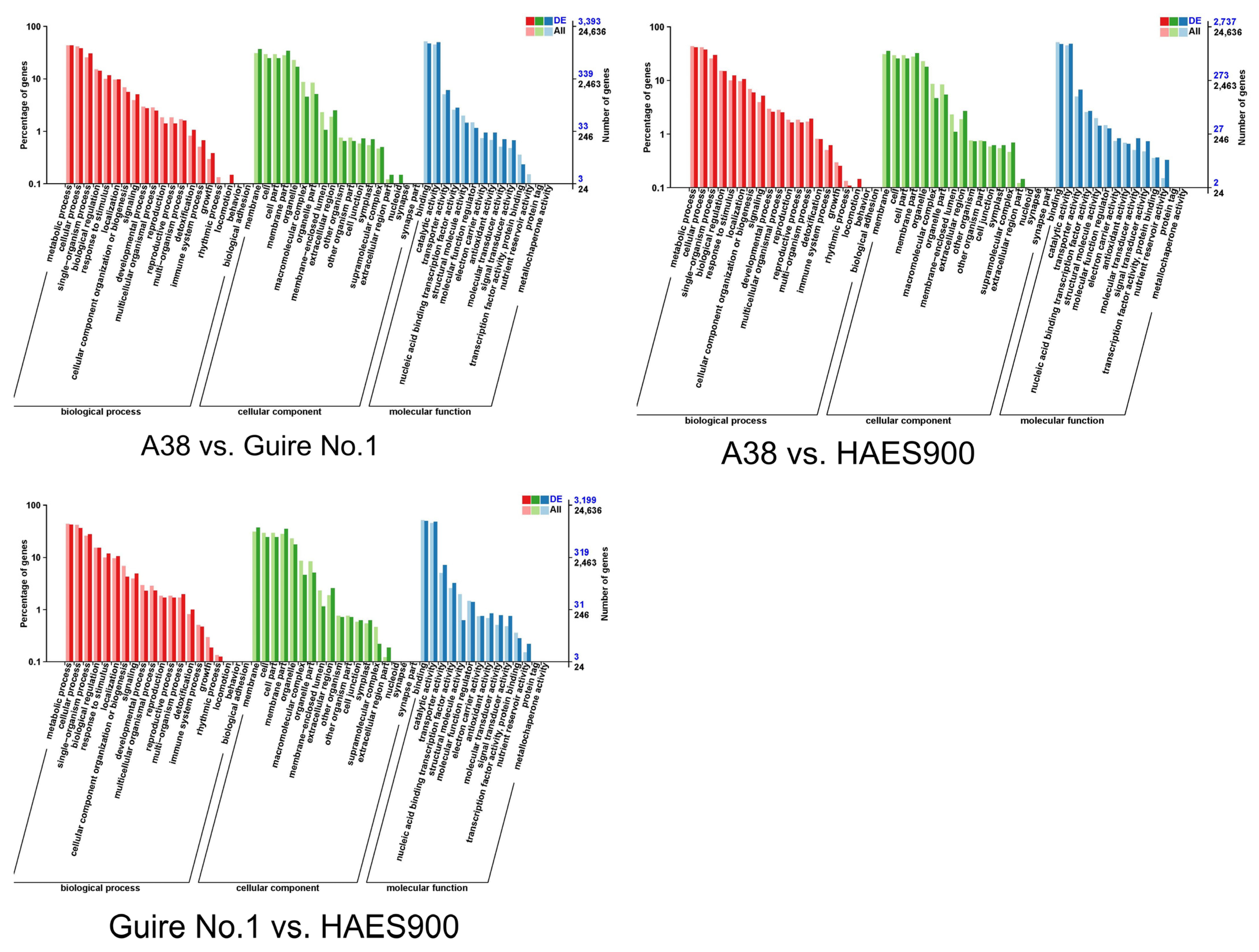
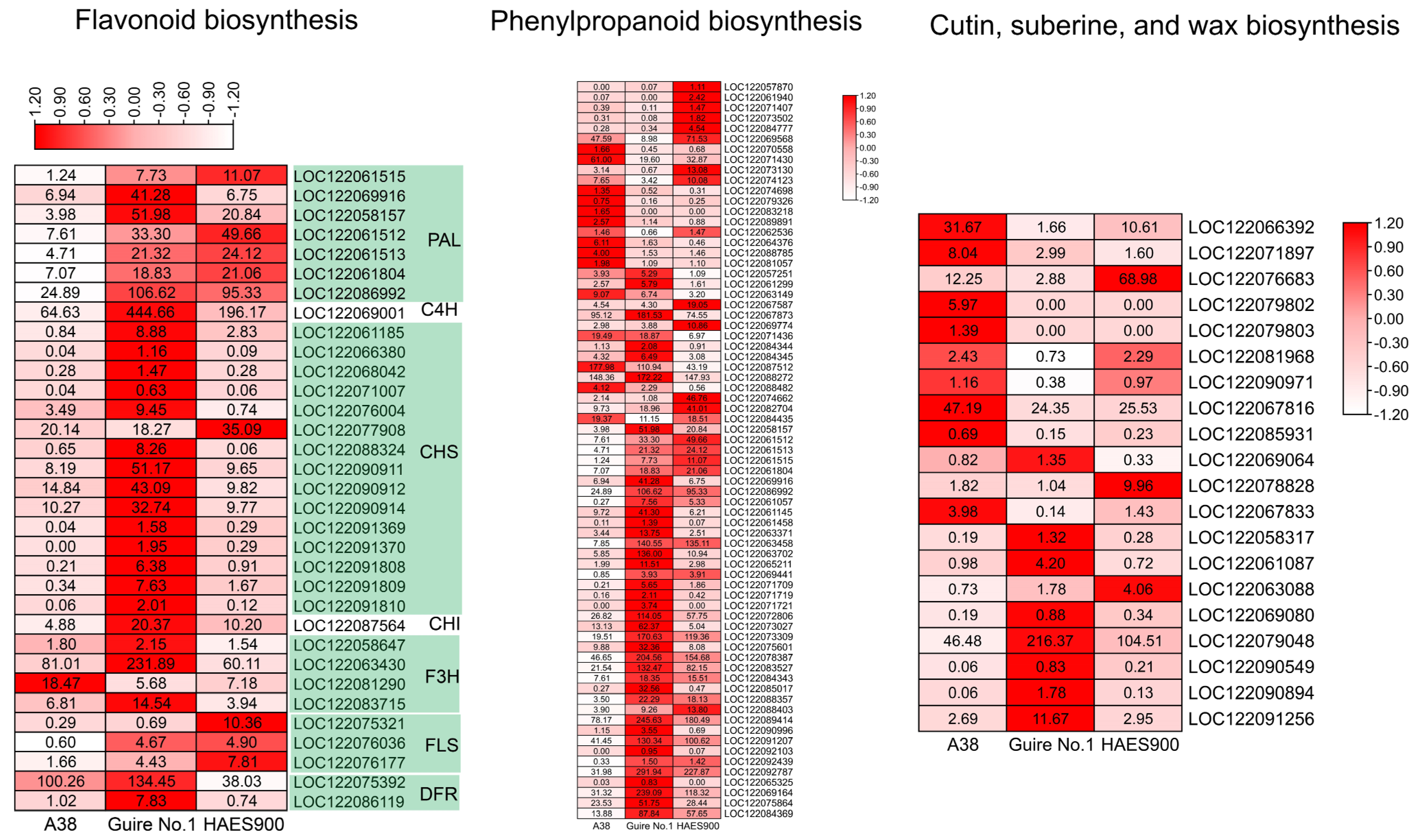
DEGs in the Three Macadamia Varieties with Different Pericarp Thicknesses
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Methods
4.2.1. Determination of Total Flavonoids
4.2.2. RNA Extraction and RNA-Seq Analysis
4.2.3. cDNA Synthesis and RT-qPCR Verification
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nock, C.J.; Baten, A.; Mauleon, R.; Langdon, K.S.; Topp, B.; Hardner, C.; Furtado, A.; Henry, R.J.; King, G.J. Chromosome-Scale Assembly and Annotation of the Macadamia Genome (Macadamia integrifolia HAES 741). G3 Genes Genomes Genet. 2020, 10, 3497–3504. [Google Scholar] [CrossRef] [PubMed]
- Abubaker, M.; Hawary, S.S.E.; Mahrous, E.A.; El-Kader, E.M.A. Study of Nutritional Contents of Macadamia integrifolia Maiden and Betche Leaves, Kernel and Pericarp Cultivated in Egypt. Int. J. Pharmacogn. Phytochem. Res. 2018, 9, 1442–1445. [Google Scholar] [CrossRef]
- Somwongin, S.; Sirilun, S.; Chantawannakul, P.; Anuchapreeda, S.; Yawootti, A.; Chaiyana, W. Ultrasound-assisted green extraction methods: An approach for cosmeceutical compounds isolation from Macadamia integrifolia pericarp. Ultrason. Sonochem. 2023, 92, 106266. [Google Scholar] [CrossRef] [PubMed]
- Sen, D.; Fernández, A.; Crozier, D.; Henrich, B.; Sokolov, A.V.; Scully, M.O.; Rooney, W.L.; Verhoef, A.J. Non-Destructive Direct Pericarp Thickness Measurement of Sorghum Kernels Using Extended-Focus Optical Coherence Microscopy. Sensors 2023, 23, 707. [Google Scholar] [CrossRef] [PubMed]
- Xiong, C.; Pei, H.; Zhang, Y.; Ren, W.; Ma, Z.; Tang, Y.; Huang, J. Integrative analysis of transcriptome and miRNAome reveals molecular mechanisms regulating pericarp thickness in sweet corn during kernel development. Front. Plant Sci. 2022, 13, 945379. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Wang, B.; Xie, F.; Zhang, L.; Gong, J.; Zhu, W.; Li, X.; Feng, F.; Huang, J. QTL mapping and transcriptome analysis identify candidate genes regulating pericarp thickness in sweet corn. BMC Plant Biol. 2020, 20, 117. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Su, Y.; Li, J.; Jia, B.; Cao, Z.; Qin, G. Physiological adjustment of pomegranate pericarp responding to sunburn and its underlying molecular mechanisms. BMC Plant Biol. 2022, 22, 169. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liao, B.; Wang, Y.; Luo, H.; Wang, S.; Li, C.; Song, W.; Zhang, K.; Yang, B.; Lu, S.; et al. Transcriptome and metabolome analyses provide insights into the relevance of pericarp thickness variations in Camellia drupifera and Camellia oleifera. Front. Plant Sci. 2022, 13, 1016475. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.-H.; Yang, J.; Ha, S.-H.; Kim, J.-K.; Lee, J.-Y.; Lim, S.-H. An OsKala3, R2R3 MYB TF, Is a Common Key Player for Black Rice Pericarp as Main Partner of an OsKala4, bHLH TF. Front. Plant Sci. 2021, 12, 765049. [Google Scholar] [CrossRef]
- Liu, R.; Song, J.; Liu, S.; Chen, C.; Zhang, S.; Wang, J.; Xiao, Y.; Cao, B.; Lei, J.; Zhu, Z. Genome-wide identification of the Capsicum bHLH transcription factor family: Discovery of a candidate regulator involved in the regulation of species-specific bioactive metabolites. BMC Plant Biol. 2021, 21, 262. [Google Scholar] [CrossRef]
- Wang, J.; Liu, Y.; Tang, B.; Dai, X.; Xie, L.; Liu, F.; Zou, X. Genome-Wide Identification and Capsaicinoid Biosynthesis-Related Expression Analysis of the R2R3-MYB Gene Family in Capsicum annuum L. Front. Genet. 2020, 11, 598183. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Man, Y.; Wang, Y. Light- and Temperature-Induced Expression of an R2R3-MYB Gene Regulates Anthocyanin Biosynthesis in Red-Fleshed Kiwifruit. Int. J. Mol. Sci. 2019, 20, 5228. [Google Scholar] [CrossRef]
- Wang, L.; Tang, W.; Hu, Y.; Zhang, Y.; Sun, J.; Guo, X.; Lu, H.; Yang, Y.; Fang, C.; Niu, X.; et al. A MYB/bHLH complex regulates tissue-specific anthocyanin biosynthesis in the inner pericarp of red-centered kiwifruit Actinidia chinensis cv. Hongyang. Plant J. 2019, 99, 359–378. [Google Scholar] [CrossRef]
- Gan, L.; Song, M.; Wang, X.; Yang, N.; Li, H.; Liu, X.; Li, Y. Cytokinins are involved in regulation of tomato pericarp thickness and fruit size. Hortic. Res. 2022, 9, uhab041. [Google Scholar] [CrossRef] [PubMed]
- Damasceno Junior, C.V.; Godoy, S.; Gonela, A.; Scapim, C.A.; Grandis, A.; Dos Santos, W.D.; Mangolin, C.A.; Buckeridge, M.S.; Machado, M.d.F.P.S. Biochemical composition of the pericarp cell wall of popcorn inbred lines with different popping expansion. Curr. Res. Food Sci. 2021, 5, 102–106. [Google Scholar] [CrossRef]
- Zhang, L.; Duan, Z.; Ma, S.; Sun, S.; Sun, M.; Xiao, Y.; Ni, N.; Irfan, M.; Chen, L.; Sun, Y. SlMYB7, an AtMYB4-Like R2R3-MYB Transcription Factor, Inhibits Anthocyanin Accumulation in Solanum lycopersicum Fruits. J. Agric. Food Chem. 2023, 71, 18758–18768. [Google Scholar] [CrossRef]
- Velten, J.; Cakir, C.; Youn, E.; Chen, J.; Cazzonelli, C.I. Transgene silencing and transgene-derived siRNA production in tobacco plants homozygous for an introduced AtMYB90 construct. PLoS ONE 2012, 7, e30141. [Google Scholar] [CrossRef] [PubMed]
- Velten, J.; Cakir, C.; Cazzonelli, C.I. A spontaneous dominant-negative mutation within a 35S::AtMYB90 transgene inhibits flower pigment production in tobacco. PLoS ONE 2010, 5, e9917. [Google Scholar] [CrossRef]
- Pandey, A.; Misra, P.; Trivedi, P.K. Constitutive expression of Arabidopsis MYB transcription factor, AtMYB11, in tobacco modulates flavonoid biosynthesis in favor of flavonol accumulation. Plant Cell Rep. 2015, 34, 1515–1528. [Google Scholar] [CrossRef]
- Petroni, K.; Falasca, G.; Calvenzani, V.; Allegra, D.; Stolfi, C.; Fabrizi, L.; Altamura, M.M.; Tonelli, C. The AtMYB11 gene from Arabidopsis is expressed in meristematic cells and modulates growth in planta and organogenesis in vitro. J. Exp. Bot. 2008, 59, 1201–1213. [Google Scholar] [CrossRef]
- Woźniak, M.; Waśkiewicz, A.; Ratajczak, I. The Content of Phenolic Compounds and Mineral Elements in Edible Nuts. Molecules 2022, 27, 4326. [Google Scholar] [CrossRef] [PubMed]
- Winkel-Shirley, B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001, 126, 485–493. [Google Scholar] [CrossRef] [PubMed]
- Xia, C.; Jiang, S.; Tan, Q.; Wang, W.; Zhao, L.; Zhang, C.; Bao, Y.; Liu, Q.; Xiao, J.; Deng, K.; et al. Chromosomal-level genome of macadamia (Macadamia integrifolia). Trop. Plants 2022, 1, 3. [Google Scholar] [CrossRef]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Ogata, K.; Kanei-Ishii, C.; Sasaki, M.; Hatanaka, H.; Nagadoi, A.; Enari, M.; Nakamura, H.; Nishimura, Y.; Ishii, S.; Sarai, A. The cavity in the hydrophobic core of Myb DNA-binding domain is reserved for DNA recognition and trans-activation. Nat. Struct. Biol. 1996, 3, 178–187. [Google Scholar] [CrossRef] [PubMed]
- Rowan, D.D.; Cao, M.; Lin-Wang, K.; Cooney, J.M.; Jensen, D.J.; Austin, P.T.; Hunt, M.B.; Norling, C.; Hellens, R.P.; Schaffer, R.J.; et al. Environmental regulation of leaf colour in red 35S:PAP1 Arabidopsis thaliana. New Phytol. 2009, 182, 102–115. [Google Scholar] [CrossRef] [PubMed]
- Li, S.F.; Parish, R.W. Isolation of two novel myb-like genes from Arabidopsis and studies on the DNA-binding properties of their products. Plant J. 1995, 8, 963–972. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, H.; Chong, S.L.; Priyanka, B.; Han, X.; Lin, E.; Tong, Z.; Huang, H. Full-length transcriptomic identification of R2R3-MYB family genes related to secondary cell wall development in Cunninghamia lanceolata (Chinese fir). BMC Plant Biol. 2021, 21, 581. [Google Scholar] [CrossRef] [PubMed]
- Huo, D.; Liu, X.; Zhang, Y.; Duan, J.; Zhang, Y.; Luo, J. A Novel R2R3-MYB Transcription Factor PqMYB4 Inhibited Anthocyanin Biosynthesis in Paeonia qiui. Int. J. Mol. Sci. 2020, 21, 5878. [Google Scholar] [CrossRef]
- Matsui, K.; Oshima, Y.; Mitsuda, N.; Sakamoto, S.; Nishiba, Y.; Walker, A.R.; Ohme-Takagi, M.; Robinson, S.P.; Yasui, Y.; Mori, M.; et al. Buckwheat R2R3 MYB transcription factor FeMYBF1 regulates flavonol biosynthesis. Plant Sci. 2018, 274, 466–475. [Google Scholar] [CrossRef]
- Lai, B.; Du, L.-N.; Hu, B.; Wang, D.; Huang, X.-M.; Zhao, J.-T.; Wang, H.-C.; Hu, G.-B. Characterization of a novel litchi R2R3-MYB transcription factor that involves in anthocyanin biosynthesis and tissue acidification. BMC Plant Biol. 2019, 19, 62. [Google Scholar] [CrossRef] [PubMed]
- Fahima, A.; Levinkron, S.; Maytal, Y.; Hugger, A.; Lax, I.; Huang, X.; Eyal, Y.; Lichter, A.; Goren, M.; Stern, R.A.; et al. Cytokinin treatment modifies litchi fruit pericarp anatomy leading to reduced susceptibility to post-harvest pericarp browning. Plant Sci. 2019, 283, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Landoni, M.; Puglisi, D.; Cassani, E.; Borlini, G.; Brunoldi, G.; Comaschi, C.; Pilu, R. Phlobaphenes modify pericarp thickness in maize and accumulation of the fumonisin mycotoxins. Sci. Rep. 2020, 10, 1417. [Google Scholar] [CrossRef] [PubMed]
- Pielot, R.; Kohl, S.; Manz, B.; Rutten, T.; Weier, D.; Tarkowská, D.; Rolčík, J.; Strnad, M.; Volke, F.; Weber, H.; et al. Hormone-mediated growth dynamics of the barley pericarp as revealed by magnetic resonance imaging and transcript profiling. J. Exp. Bot. 2015, 66, 6927–6943. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Chen, L.; Shi, Q.; Ren, Z. SlMYB102, an R2R3-type MYB gene, confers salt tolerance in transgenic tomato. Plant Sci. 2020, 291, 110356. [Google Scholar] [CrossRef]
- Piao, W.; Sakuraba, Y.; Paek, N.-C. Transgenic expression of rice MYB102 (OsMYB102) delays leaf senescence and decreases abiotic stress tolerance in Arabidopsis thaliana. BMB Rep. 2019, 52, 653–658. [Google Scholar] [CrossRef]
- Benamar, H.; Rached, W.; Derdour, A.; Marouf, A. Screening of Algerian Medicinal Plants for Acetylcholinesterase Inhibitory Activity. J. Biol. Sci. 2010, 10, 1–9. [Google Scholar] [CrossRef]
- Guo, B.; Liu, M.; Yang, H.; Dai, L.; Wang, L. Brassinosteroids Regulate the Water Deficit and Latex Yield of Rubber Trees. Int. J. Mol. Sci. 2023, 24, 12857. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Jin, J.; Tian, F.; Yang, D.-C.; Meng, Y.-Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.-T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tan, Q.; Huan, X.; Pan, Z.; Yang, X.; Wei, Y.; Zhou, C.; Wang, W.; Wang, L. Comparative Transcriptome Analysis Reveals Key Functions of MiMYB Gene Family in Macadamia Nut Pericarp Formation. Int. J. Mol. Sci. 2024, 25, 6840. https://doi.org/10.3390/ijms25136840
Tan Q, Huan X, Pan Z, Yang X, Wei Y, Zhou C, Wang W, Wang L. Comparative Transcriptome Analysis Reveals Key Functions of MiMYB Gene Family in Macadamia Nut Pericarp Formation. International Journal of Molecular Sciences. 2024; 25(13):6840. https://doi.org/10.3390/ijms25136840
Chicago/Turabian StyleTan, Qiujin, Xiuju Huan, Zhenzhen Pan, Xiaozhou Yang, Yuanrong Wei, Chunheng Zhou, Wenlin Wang, and Lifeng Wang. 2024. "Comparative Transcriptome Analysis Reveals Key Functions of MiMYB Gene Family in Macadamia Nut Pericarp Formation" International Journal of Molecular Sciences 25, no. 13: 6840. https://doi.org/10.3390/ijms25136840






