Effect of HVEM/CD160 Variations on the Clear Cell Renal Carcinoma Risk and Overall Survival
Abstract
:1. Introduction
2. Results
2.1. Correlation of HVEM and CD160 Gene SNPs with ccRCC Susceptibility
2.2. Multifactorial Regression Analysis
2.3. Haplotype Analysis
2.4. Sex-Dependent Association of HVEM and CD160 Polymorphisms and ccRCC Risk
2.5. Association of HVEM and CD160 Polymorphisms with Clinical Features of ccRCC
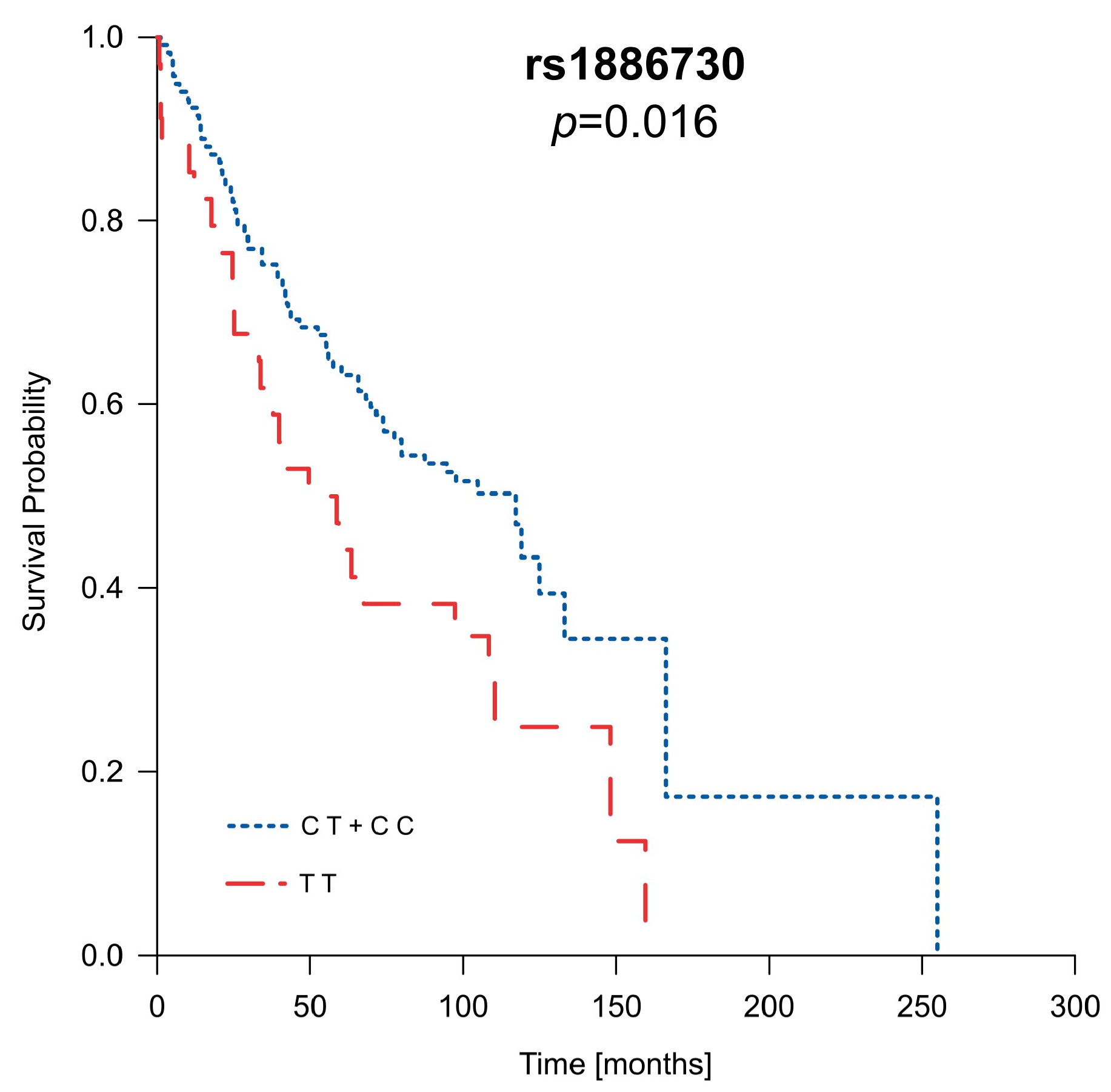
2.6. Analysis of Patients’ Survival in Context of Clinical Parameters as Well as HVEM and CD160 Gene Polymorphisms
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. Controls
4.3. Selection of SNPs
4.4. DNA Isolation and SNP Genotyping
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bukavina, L.; Bensalah, K.; Bray, F.; Carlo, M.; Challacombe, B.; Karam, J.A.; Kassouf, W.; Mitchell, T.; Montironi, R.; O’Brien, T.; et al. Epidemiology of renal cell carcinoma: 2022 update. Eur. Urol. 2022, 82, 529–542. [Google Scholar] [CrossRef] [PubMed]
- Padala, S.A.; Barsouk, A.; Thandra, K.C.; Saginala, K.; Mohammed, A.; Vakiti, A.; Rawla, P.; Barsouk, A. Epidemiology of Renal Cell Carcinoma. World J. Oncol. 2020, 11, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Muglia, V.F.; Prando, A. Renal cell carcinoma: Histological classification and correlation with imaging findings. Radiol. Bras. 2015, 48, 166–174. [Google Scholar] [CrossRef] [PubMed]
- Cancer Today, Global Cancer Observatory. Available online: https://gco.iarc.fr/today/home (accessed on 2 February 2024).
- Dyck, L.; Mills, K.H.G. Immune checkpoints and their inhibition in cancer and infectious diseases. Eur. J. Immunol. 2017, 47, 765–779. [Google Scholar] [CrossRef] [PubMed]
- Topalian, S.L.; Taube, J.M.; Anders, R.A.; Pardoll, D.M. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat. Rev. Cancer 2016, 16, 275–287. [Google Scholar] [CrossRef] [PubMed]
- Del Rio, M.L.; Lucas, C.L.; Buhler, L.; Rayat, G.; Rodriguez-Barbosa, J.I. HVEM/LIGHT/BTLA/CD160 cosignaling pathways as targets for immune regulation. J. Leukoc. Biol. 2009, 87, 223–235. [Google Scholar] [CrossRef] [PubMed]
- Andrzejczak, A.; Karabon, L. BTLA biology in cancer: From bench discoveries to clinical potentials. Biomark. Res. 2024, 12, 8. [Google Scholar] [CrossRef] [PubMed]
- Murphy, K.M.; Nelson, C.A.; Šedý, J.R. Balancing co-stimulation and inhibition with BTLA and HVEM. Nat. Rev. Immunol. 2006, 6, 671–681. [Google Scholar] [CrossRef]
- Lan, X.; Li, S.; Gao, H.; Nanding, A.; Quan, L.; Yang, C.; Ding, S.; Xue, Y. Increased BTLA and HVEM in gastric cancer are associated with progression and poor prognosis. OncoTargets Ther. 2017, 10, 919–926. [Google Scholar] [CrossRef]
- Ren, S.; Tian, Q.; Amar, N.; Yu, H.; Rivard, C.J.; Caldwell, C.; Ng, T.L.; Tu, M.; Liu, Y.; Gao, D.; et al. The immune checkpoint, HVEM may contribute to immune escape in non-small cell lung cancer lacking PD-L1 expression. Lung Cancer 2018, 125, 115–120. [Google Scholar] [CrossRef]
- Malissen, N.; Macagno, N.; Granjeaud, S.; Granier, C.; Moutardier, V.; Gaudy-Marqueste, C.; Habel, N.; Mandavit, M.; Guillot, B.; Pasero, C.; et al. HVEM has a broader expression than PD-L1 and constitutes a negative prognostic marker and potential treatment target for melanoma. OncoImmunology 2019, 8, e1665976. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.-T.; Giustiniani, J.; Farren, T.; Jia, L.; Bensussan, A.; Gribben, J.G.; Agrawal, S.G. CD160 signaling mediates PI3K-dependent survival and growth signals in chronic lymphocytic leukemia. Blood 2010, 115, 3079–3088. [Google Scholar] [CrossRef] [PubMed]
- Gauci, M.-L.; Giustiniani, J.; Lepelletier, C.; Garbar, C.; Thonnart, N.; Dumaz, N.; Foussat, A.; Lebbé, C.; Bensussan, A.; Marie-Cardine, A. The soluble form of CD160 acts as a tumor mediator of immune escape in melanoma. Cancer Immunol. Immunother. 2022, 71, 2731–2742. [Google Scholar] [CrossRef] [PubMed]
- Farren, T.W.; Giustiniani, J.; Liu, F.-T.; Tsitsikas, D.A.; Macey, M.G.; Cavenagh, J.D.; Oakervee, H.E.; Taussig, D.; Newland, A.C.; Calaminici, M.; et al. Differential and tumor-specific expression of CD160 in B-cell malignancies. Blood 2011, 118, 2174–2183. [Google Scholar] [CrossRef] [PubMed]
- Shastry, B.S. SNPs: Impact on gene function and phenotype. In Single Nucleotide Polymorphisms; Komar, A.A., Ed.; Humana Press: Totowa, NJ, USA, 2009; Volume 578, pp. 3–22. ISBN 978-1-60327-410-4. [Google Scholar]
- Li, D.; Fu, Z.; Chen, S.; Yuan, W.; Liu, Y.; Li, L.; Pang, D.; Li, D. HVEM Gene Polymorphisms Are Associated with Sporadic Breast Cancer in Chinese Women. PLoS ONE 2013, 8, e71040. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Cao, R.; Liu, C.; Tang, W.; Kang, M. Investigation of IL-4, IL-10, and HVEM polymorphisms with esophageal squamous cell carcinoma: A case–control study involving 1929 participants. Biosci. Rep. 2020, 40, BSR20193895. [Google Scholar] [CrossRef] [PubMed]
- Svejgaard, A.; Ryder, L.P. HLA and disease associations: Detecting the strongest association. Tissue Antigens 1994, 43, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Stadler, Z.K.; Vijai, J.; Thom, P.; Kirchhoff, T.; Hansen, N.A.L.; Kauff, N.D.; Robson, M.; Offit, K. Genome-wide association studies of cancer predisposition. Hematol. Oncol. Clin. N. Am. 2010, 24, 973–996. [Google Scholar] [CrossRef] [PubMed]
- Peltomäki, P. Mutations and epimutations in the origin of cancer. Exp. Cell Res. 2012, 318, 299–310. [Google Scholar] [CrossRef]
- Foulkes, W.D. Inherited Susceptibility to Common Cancers. N. Engl. J. Med. 2008, 359, 2143–2153. [Google Scholar] [CrossRef]
- Testa, U.; Pelosi, E.; Castelli, G. Genetic alterations in renal cancers: Identification of the mechanisms underlying cancer initiation and progression and of therapeutic targets. Medicines 2020, 7, 44. [Google Scholar] [CrossRef] [PubMed]
- Braga, E.A.; Fridman, M.V.; Loginov, V.I.; Dmitriev, A.A.; Morozov, S.G. Molecular mechanisms in clear cell renal cell carcinoma: Role of miRNAs and hypermethylated miRNA genes in crucial oncogenic pathways and processes. Front. Genet. 2019, 10, 320. [Google Scholar] [CrossRef] [PubMed]
- Rathmell, W.K.; Chen, S. VHL inactivation in renal cell carcinoma: Implications for diagnosis, prognosis and treatment. Expert Rev. Anticancer Ther. 2008, 8, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Shulman, M.; Shi, R.; Zhang, Q. Von Hippel-Lindau tumor suppressor pathways & corresponding therapeutics in kidney cancer. J. Genet. Genom. 2021, 48, 552–559. [Google Scholar] [CrossRef] [PubMed]
- Szegedi, K.; Szabó, Z.; Kállai, J.; Király, J.; Szabó, E.; Bereczky, Z.; Juhász, É.; Dezső, B.; Szász, C.; Zsebik, B.; et al. Potential role of VHL, PTEN, and BAP1 mutations in renal tumors. J. Clin. Med. 2023, 12, 4538. [Google Scholar] [CrossRef] [PubMed]
- Knudson, A.G. Two genetic hits (more or less) to cancer. Nat. Rev. Cancer 2001, 1, 157–162. [Google Scholar] [CrossRef]
- Andrzejczak, A.; Partyka, A.; Wiśniewski, A.; Porębska, I.; Pawełczyk, K.; Ptaszkowski, K.; Kuśnierczyk, P.; Jasek, M.; Karabon, L. The association of BTLA gene polymorphisms with non-small lung cancer risk in smokers and never-smokers. Front. Immunol. 2023, 13, 1006639. [Google Scholar] [CrossRef] [PubMed]
- Andrzejczak, A.; Tupikowski, K.; Tomkiewicz, A.; Małkiewicz, B.; Ptaszkowski, K.; Domin, A.; Szydełko, T.; Karabon, L. The Variations’ in Genes Encoding TIM-3 and Its Ligand, Galectin-9, Influence on ccRCC Risk and Prognosis. Int. J. Mol. Sci. 2023, 24, 2042. [Google Scholar] [CrossRef] [PubMed]
- Partyka, A.; Tupikowski, K.; Kolodziej, A.; Zdrojowy, R.; Halon, A.; Malkiewicz, B.; Dembowski, J.; Frydecka, I.; Karabon, L. Association of 3′ nearby gene BTLA polymorphisms with the risk of renal cell carcinoma in the Polish population. Urol. Oncol. Semin. Orig. Investig. 2016, 34, 419.e13–419.e19. [Google Scholar] [CrossRef]
- Darvin, P.; Toor, S.M.; Sasidharan Nair, V.; Elkord, E. Immune checkpoint inhibitors: Recent progress and potential biomarkers. Exp. Mol. Med. 2018, 50, 1–11. [Google Scholar] [CrossRef]
- Wagner, M.; Jasek, M.; Karabon, L. Immune checkpoint molecules—Inherited variations as markers for cancer risk. Front. Immunol. 2021, 11, 606721. [Google Scholar] [CrossRef]
- Tupikowski, K.; Partyka, A.; Pawlak, E.A.; Ptaszkowski, K.; Zdrojowy, R.; Frydecka, I.; Hałoń, A.; Karabon, L. Variation in the gene encoding the co-inhibitory molecule BTLA is associated with survival in patients treated for clear cell renal carcinoma—Results of a prospective cohort study. Arch. Med. Sci. 2023, 19, 1454–1462. [Google Scholar] [CrossRef]
- Wagner, M.; Tupikowski, K.; Jasek, M.; Tomkiewicz, A.; Witkowicz, A.; Ptaszkowski, K.; Karpinski, P.; Zdrojowy, R.; Halon, A.; Karabon, L. SNP-SNP interaction in genes encoding PD-1/PD-L1 axis as a potential risk factor for clear cell renal cell carcinoma. Cancers 2020, 12, 3521. [Google Scholar] [CrossRef] [PubMed]
- Tupikowski, K.; Partyka, A.; Kolodziej, A.; Dembowski, J.; Debinski, P.; Halon, A.; Zdrojowy, R.; Frydecka, I.; Karabon, L. CTLA-4 and CD28 genes’ polymorphisms and renal cell carcinoma susceptibility in the Polish population -a prospective study. Tissue Antigens 2015, 86, 353–361. [Google Scholar] [CrossRef]
- Cheung, T.C.; Oborne, L.M.; Steinberg, M.W.; Macauley, M.G.; Fukuyama, S.; Sanjo, H.; D’Souza, C.; Norris, P.S.; Pfeffer, K.; Murphy, K.M.; et al. T cell intrinsic heterodimeric complexes between HVEM and BTLA determine receptivity to the surrounding microenvironment. J. Immunol. 2009, 183, 7286–7296. [Google Scholar] [CrossRef]
- Shrestha, R.; Garrett-Thomson, S.C.; Liu, W.; Almo, S.C.; Fiser, A. Redesigning HVEM interface for selective binding to LIGHT, BTLA, and CD160. Structure 2020, 28, 1197–1205.e2. [Google Scholar] [CrossRef]
- Cai, G.; Freeman, G.J. The CD160, BTLA, LIGHT/HVEM pathway: A bidirectional switch regulating T-cell activation. Immunol. Rev. 2009, 229, 244–258. [Google Scholar] [CrossRef] [PubMed]
- Tu, T.C.; Brown, N.K.; Kim, T.-J.; Wroblewska, J.; Yang, X.; Guo, X.; Lee, S.H.; Kumar, V.; Lee, K.-M.; Fu, Y.-X. CD160 is essential for NK-mediated IFN-γ production. J. Exp. Med. 2015, 212, 415–429. [Google Scholar] [CrossRef] [PubMed]
- Šedý, J.R.; Bjordahl, R.L.; Bekiaris, V.; Macauley, M.G.; Ware, B.C.; Norris, P.S.; Lurain, N.S.; Benedict, C.A.; Ware, C.F. CD160 activation by herpesvirus entry mediator augments inflammatory cytokine production and cytolytic function by NK cells. J. Immunol. 2013, 191, 828–836. [Google Scholar] [CrossRef]
- Liu, W.; Garrett, S.C.; Fedorov, E.V.; Ramagopal, U.A.; Garforth, S.J.; Bonanno, J.B.; Almo, S.C. Structural basis of CD160:HVEM recognition. Structure 2019, 27, 1286–1295.e4. [Google Scholar] [CrossRef]
- Lee, W.-C. Searching for disease-susceptibility loci by testing for Hardy-Weinberg disequilibrium in a gene bank of affected individuals. Am. J. Epidemiol. 2003, 158, 397–400. [Google Scholar] [CrossRef] [PubMed]
- Makino, T.; Kadomoto, S.; Izumi, K.; Mizokami, A. Epidemiology and prevention of renal cell carcinoma. Cancers 2022, 14, 4059. [Google Scholar] [CrossRef] [PubMed]
- Fang, M.; Huang, W.; Mo, D.; Zhao, W.; Huang, R. Association of five snps in cytotoxic T-lymphocyte antigen 4 and cancer susceptibility: Evidence from 67 studies. Cell. Physiol. Biochem. 2018, 47, 414–427. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Liu, Y.; Zheng, S.; Geng, P.; He, T.; Lu, L.; Feng, Y.; Jiang, Q. Associations of PD-1 and PD-L1 gene polymorphisms with cancer risk: A meta-analysis based on 50 studies. Aging 2024, 16, 6068–6097. [Google Scholar] [CrossRef] [PubMed]
- Jo, B.-S.; Choi, S.S. Introns: The functional benefits of introns in genomes. Genomics Inform. 2015, 13, 112. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, K.; Yang, H.; Han, Z.; Tao, J.; Chen, H.; Ge, Y.; Guo, M.; Suo, C.; Wei, J.-F.; et al. Associations between HVEM/LIGHT/BTLA/CD160 polymorphisms and the occurrence of antibody-mediate rejection in renal transplant recipients. Oncotarget 2017, 8, 100079–100094. [Google Scholar] [CrossRef]
- He, W.; Zhao, J.; Liu, X.; Li, S.; Mu, K.; Zhang, J.; Zhang, J. Associations between CD160 polymorphisms and autoimmune thyroid disease: A case-control study. BMC Endocr. Disord. 2021, 21, 148. [Google Scholar] [CrossRef]
| SNP | Genotype | Case | Control | OR (95% CI) | p Value * | ||
|---|---|---|---|---|---|---|---|
| N | % | N | % | ||||
| rs1886730T > C | TT | 49 | 20.59 | 140 | 26.87 | 1 | 0.17 |
| CT | 130 | 54.62 | 257 | 49.33 | 1.44 (0.98–2.12) | ||
| CC | 59 | 24.79 | 124 | 23.80 | 1.36 (0.87–2.12) | ||
| CT + CC | 189 | 79.41 | 381 | 73.13 | 1.41 (0.98–2.04) | 0.06 | |
| TT + CT | 179 | 75.21 | 397 | 76.20 | 0.94 (0.66–1.35) | 0.77 | |
| rs2234167G > A | GG | 167 | 70.17 | 404 | 77.54 | 1 | 0.06 |
| AG | 68 | 28.57 | 108 | 20.73 | 1.52 (1.07–2.17) | ||
| AA | 3 | 1.26 | 9 | 1.73 | 0.89 (0.26–3.07) | ||
| AG + AA | 71 | 29.83 | 117 | 22.46 | 1.47 (1.04–2.07) | 0.03 | |
| GG + AG | 235 | 98.74 | 512 | 98.27 | 1.25 (0.36–4.29) | 0.63 | |
| rs8725G > A | GG | 52 | 21.85 | 145 | 27.83 | 1 | 0.22 |
| AG | 125 | 52.52 | 252 | 48.37 | 1.38 (0.94–2.02) | ||
| AA | 61 | 25.63 | 124 | 23.80 | 1.37 (0.88–2.12) | ||
| AG + AA | 186 | 78.15 | 376 | 72.17 | 1.37 (0.96–1.97) | 0.08 | |
| GG + AG | 177 | 74.37 | 397 | 76.20 | 0.90 (0.64–1.29) | 0.59 | |
| rs744877C > A | CC | 77 | 32.35 | 173 | 33.21 | 1 | 0.95 |
| AC | 118 | 49.58 | 258 | 49.52 | 1.03 (0.73–1.45) | ||
| AA | 43 | 18.07 | 90 | 17.27 | 1.08 (0.69–1.69) | ||
| AC + AA | 161 | 67.65 | 348 | 66.79 | 1.04 (0.75–1.44) | 0.82 | |
| CC + AC | 195 | 81.93 | 431 | 82.73 | 0.94 (0.63–1.40) | 0.79 | |
| rs2231375C > T | CC | 78 | 33.05 | 206 | 39.62 | 1 | 0.03 |
| CT | 130 | 55.08 | 233 | 44.81 | 1.47 (1.05–2.06) | ||
| TT | 28 | 11.86 | 81 | 15.58 | 0.92 (0.56–1.52) | ||
| CT + TT | 158 | 66.95 | 314 | 60.38 | 1.33 (0.96–1.83) | 0.08 | |
| CC + CT | 208 | 88.14 | 439 | 84.42 | 1.36 (0.86–2.14) | 0.18 | |
| ccRCC (n = 238) | Control (n = 521) | ||||
|---|---|---|---|---|---|
| A+, B+ | 39 | 57 | |||
| A+, B− | 28 | 51 | |||
| A−, B+ | 91 | 177 | |||
| A−, B− | 78 | 236 | |||
| Test | OR | p value * | 95% CI | Comparison | Individual association |
| (a) A+ | 1.53 | 0.02 | 1.08–2.18 | ||
| (b) B+ | 1.58 | 0.04 | 1.16–2.15 | ||
| (c) A+ B+ vs. A−B+ | 1.33 | 0.24 | 0.82–2.15 | A in B positive | A association |
| (d) A+ B− vs. A −B − | 1.85 | 0.02 | 1.09–3.13 | A in B negative | |
| (e) A+ B+ vs. A+ B− | 1.25 | 0.48 | 0.67–2.31 | B in A positive | B association |
| (f) A− B + vs. A − B− | 1.56 | 0.02 | 1.09–2.23 | B in A negative | |
| (g) A + B− vs. A− B+ | 1.07 | 0.81 | 0.63–1.81 | Differences between A and B | |
| (h) A +B + vs. A− B− | 2.07 | 0.003 | 1.30–3.35 | Combined association | |
| Logistic Regression | Regression Coefficient | Standard Error | p-Value * | OR | CI 95% | |
|---|---|---|---|---|---|---|
| Unifactorial Model | ||||||
| rs2234167 | AA + AG | 0.38 | 0.18 | 0.029 | 1.47 | 1.04–2.07 |
| rs8725 | G G | −0.32 | 0.18 | 0.082 | 0.72 | 0.50–1.04 |
| rs1886730 | T T | −0.35 | 0.19 | 0.064 | 0.71 | 0.49–1.02 |
| rs2231375 | C T | 0.41 | 0.16 | 0.010 | 1.50 | 1.10–2.05 |
| Multifactorial Model | ||||||
| rs2234167 | AA +AG | 0.26 | 0.19 | 0.16 | 1.30 | 0.90–1.88 |
| rs8725 | G G | −0.09 | 0.33 | 0.80 | 0.92 | 0.48–1.75 |
| rs1886730 | T T | −0.18 | 0.34 | 0.60 | 0.84 | 0.43–1.62 |
| rs2231375 | C T | 0.39 | 0.16 | 0.015 | 1.47 | 1.08–2.01 |
| Haplotype * | Case (%) | Control (%) | Odds Ratio [95% CI] | p Value ** |
|---|---|---|---|---|
| CAAAC | 15.23 (0.032) | 41.05 (0.039) | 0.79 [0.434~1.438] | 0.44 |
| CAACT | 44.30 (0.094) | 42.11 (0.040) | 2.41 [1.55~3.73] | 5.78 × 10−5 |
| CGAAC | 79.55 (0.179) | 156.46 (0.150) | 1.11 [0.83~1.50] | 0.48 |
| CGACC | 29.73 (0.063) | 83.12 (0.080) | 0.75 [0.49~1.16] | 0.20 |
| CGACT | 50.36 (0.107) | 111.32 (0.107) | 0.97 [0.68~1.38] | 0.86 |
| TGGAC | 92.68 (0.196) | 189.24 (0.182) | 1.07 [0.81~1.41] | 0.65 |
| TGGCC | 39.02 (0.083) | 102.89 (0.099) | 0.80 [0.54~1.17] | 0.25 |
| TGGCT | 80.71 (0.171) | 201.04 (0.193) | 0.83 [0.62~1.11] | 0.21 |
| Global χ2 = 20.33, df = 7, p = 0.005 | ||||
| N (Case/Control) | OR (95% CI); p Value * | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs1886730 | TT | CT | CC | CT + CC vs. TT | CT + TT vs. CC | CT vs. TT | CT vs. TT + CC | ||||
| 15/50 | 46/77 | 25/44 | 1.92 (1.01–3.64); | 0.04 | 0.84 (0.47–1.49); | 0.57 | 1.95 (0.99–3.84); | 0.05 | 1.40 (0.83–2.36); | 0.20 | |
| rs2234167 | GG | AG | AA | AG + AA vs. GG | GG + AG vs. AA | AG vs. GG | AG vs. GG + AA | ||||
| 55/142 | 37/80 | 1/2 | 2.63 (1.46–4.73); | 0.001 | 0.84 (0.11–6.47); | 1.00 | 2.73 (1.50–4.96); | 0.001 | 2.76 (1.51–5.02); | 0.001 | |
| rs8725 | GG | AG | AA | AG + AA vs. GG | GG + AG vs. AA | AG vs. GG | AG vs. GG + AA | ||||
| 14/56 | 47/74 | 25/41 | 2.45 (1.28–4.67); | 0.01 | 0.77 (0.43–1.37); | 0.38 | 2.48 (1.26–4.91); | 0.01 | 1.58 (0.94–2.66); | 0.09 | |
| rs2231375 | CC | CT | TT | TT + CT vs. CC | CC + CT vs. TT | CT vs. CC | CT vs. CC + TT | ||||
| 28/77 | 47/65 | 10/28 | 1.67 (0.97–2.87); | 0.06 | 1.44 (0.67–3.08); | 0.32 | 1.97 (1.12–3.48); | 0.02 | 2.00 (1.18–3.39); | 0.01 | |
| Variable | All N = 238 | Male N = 151 | Female N = 86 |
|---|---|---|---|
| Age at diagnosis | |||
| Median | 62 | 61 | 63 |
| Mean | 62.61 | 62.01 | 63.67 |
| Q1–Q3 | 56–70 | 56–68 | 58–71 |
| Min, Max | 21, 85 | 21, 85 | 24, 85 |
| BMI | |||
| Median | 27.70 | 27.70 | 27.75 |
| Mean | 28.29 | 28.26 | 28.33 |
| Q1–Q3 | 24.6–31.5 | 25.1–30.7 | 23.85–31.2 |
| Min, Max | 19.1, 43.8 | 19.7, 43.8 | 19.1, 43.8 |
| Stage of disease | N (%) | N (%) | N (%) |
| I | 108 (45.57) | 63 (41.72) | 45 (52.33) |
| II | 26 (10.97) | 20 (13.25) | 6 (6.98) |
| III | 26 (10.97) | 16 (10.60) | 10 (11.63) |
| IV | 76 (32.07) | 51 (33.77) | 25 (29.07) |
| Unknown | 1 (0.42) | 1 (0.66) | 0 (0) |
| Metastasis | |||
| No | 165 (69.62) | 101 (66.89) | 64 (74.42) |
| Present | 53 (22.36) | 35 (23.18) | 18 (20.93) |
| Unknown | 19 (8.02) | 15 (9.93) | 4 (4.65) |
| Necrosis | |||
| No | 118 (59.00) | 71 (55.47) | 47 (65.28) |
| Present | 82 (41.00) | 57 (44.53) | 25 (34.72) |
| Unknown | 0 (0) | 0 (0) | 0 (0) |
| Tumor size | |||
| <70 mm | 143 (60.34) | 87 (57.61) | 56 (65.12) |
| >70 mm | 65 (27.42) | 48 (31.79) | 17 (19.77) |
| Unknown | 29 (12.24) | 16 (10.60) | 13 (15.11) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andrzejczak, A.; Małkiewicz, B.; Tupikowski, K.; Ptaszkowski, K.; Szydełko, T.; Karabon, L. Effect of HVEM/CD160 Variations on the Clear Cell Renal Carcinoma Risk and Overall Survival. Int. J. Mol. Sci. 2024, 25, 6860. https://doi.org/10.3390/ijms25136860
Andrzejczak A, Małkiewicz B, Tupikowski K, Ptaszkowski K, Szydełko T, Karabon L. Effect of HVEM/CD160 Variations on the Clear Cell Renal Carcinoma Risk and Overall Survival. International Journal of Molecular Sciences. 2024; 25(13):6860. https://doi.org/10.3390/ijms25136860
Chicago/Turabian StyleAndrzejczak, Anna, Bartosz Małkiewicz, Krzysztof Tupikowski, Kuba Ptaszkowski, Tomasz Szydełko, and Lidia Karabon. 2024. "Effect of HVEM/CD160 Variations on the Clear Cell Renal Carcinoma Risk and Overall Survival" International Journal of Molecular Sciences 25, no. 13: 6860. https://doi.org/10.3390/ijms25136860





