Environmental Enrichment Prevents Gut Dysbiosis Progression and Enhances Glucose Metabolism in High-Fat Diet-Induced Obese Mice
Abstract
:1. Introduction
2. Results
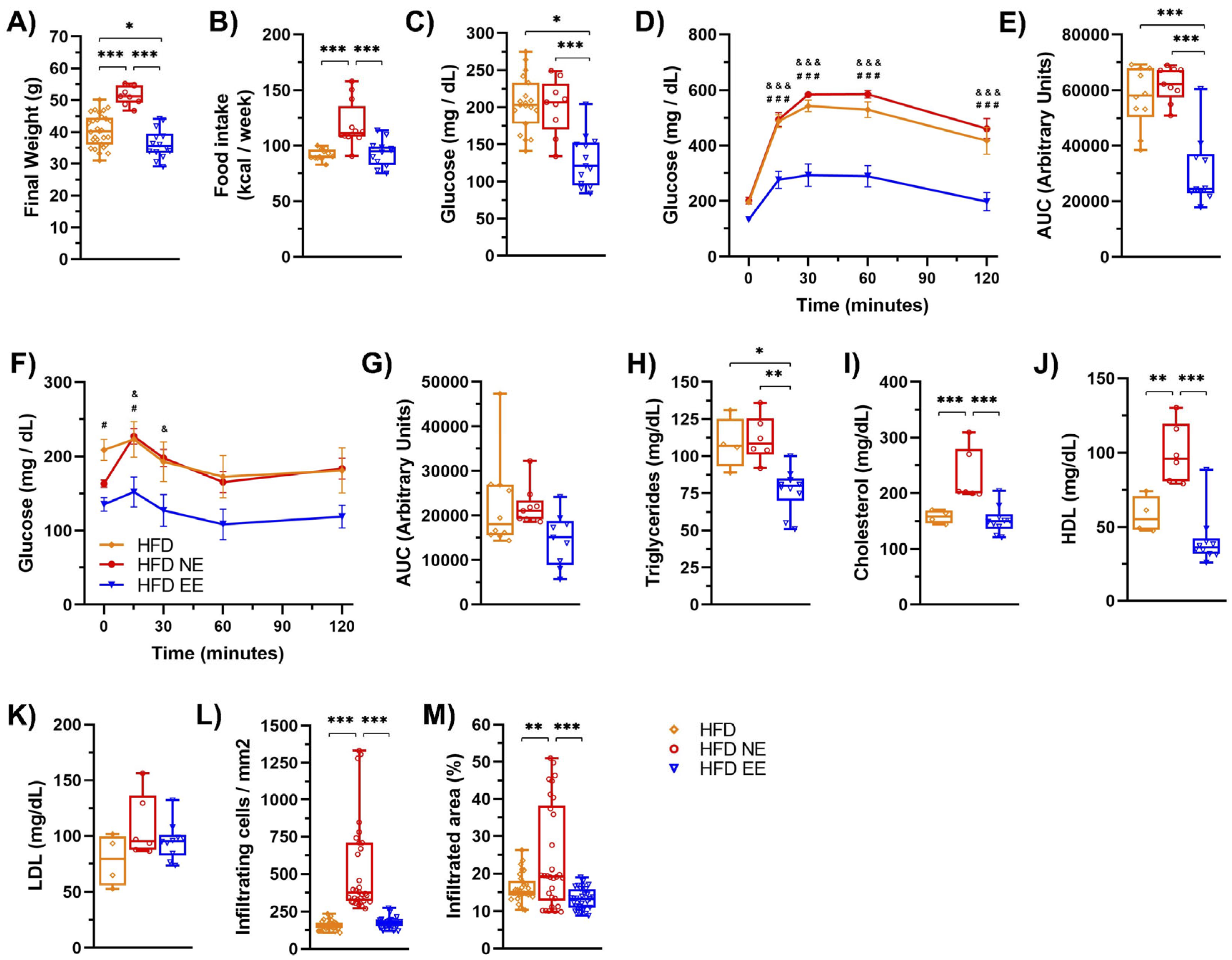
2.1. Environmental Enrichment Reverses the Metabolic Syndrome Induced by a High-Fat Diet Consumption
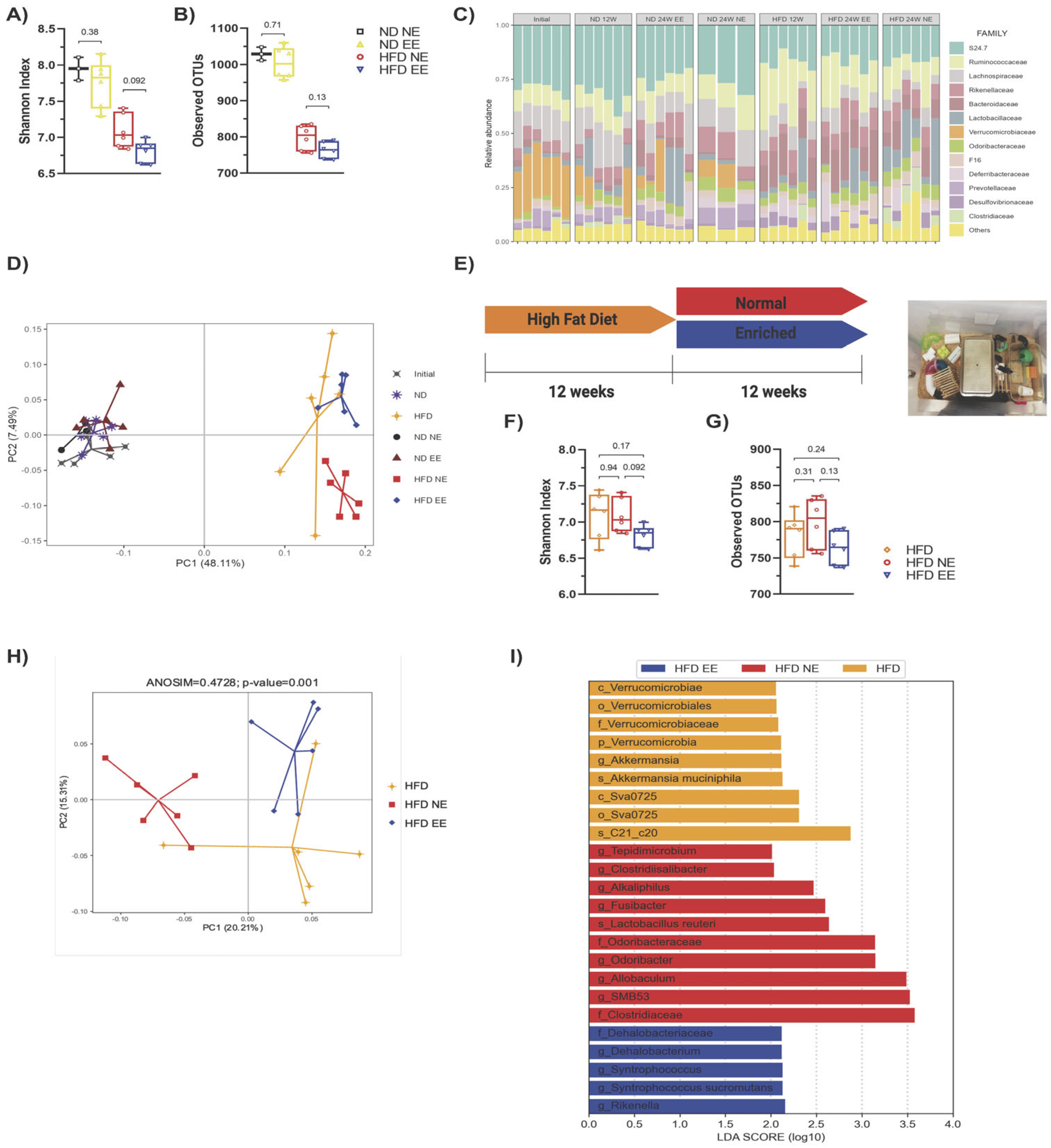
2.2. Protective Effects of Environmental Enrichment on Metabolism Correlates with Mitigating High-Fat Diet-Induced Dysbiosis
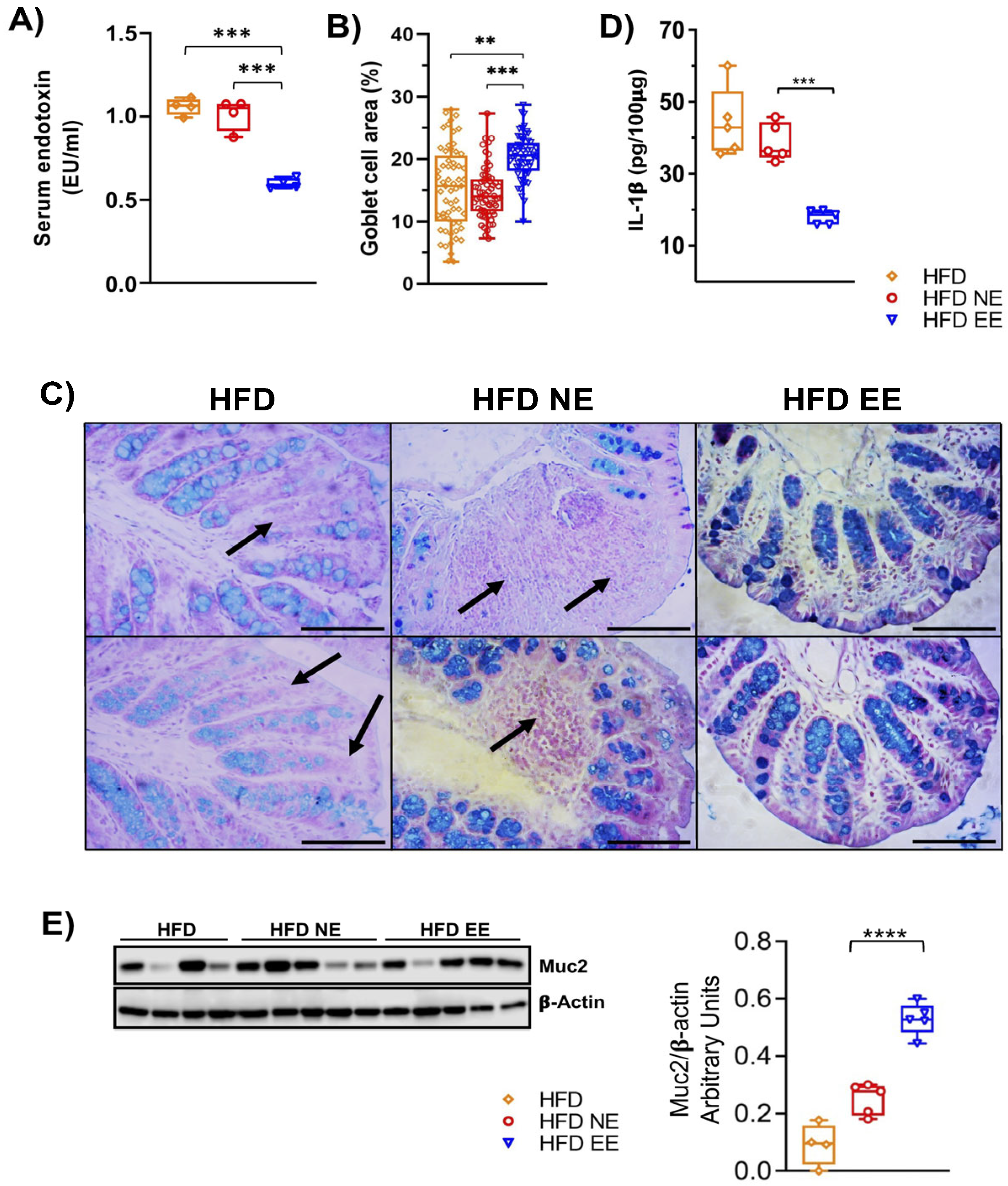
2.3. Environmental Enrichment Increases the Levels of Muc2 in the Colon of Mice Fed with a High-Fat Diet
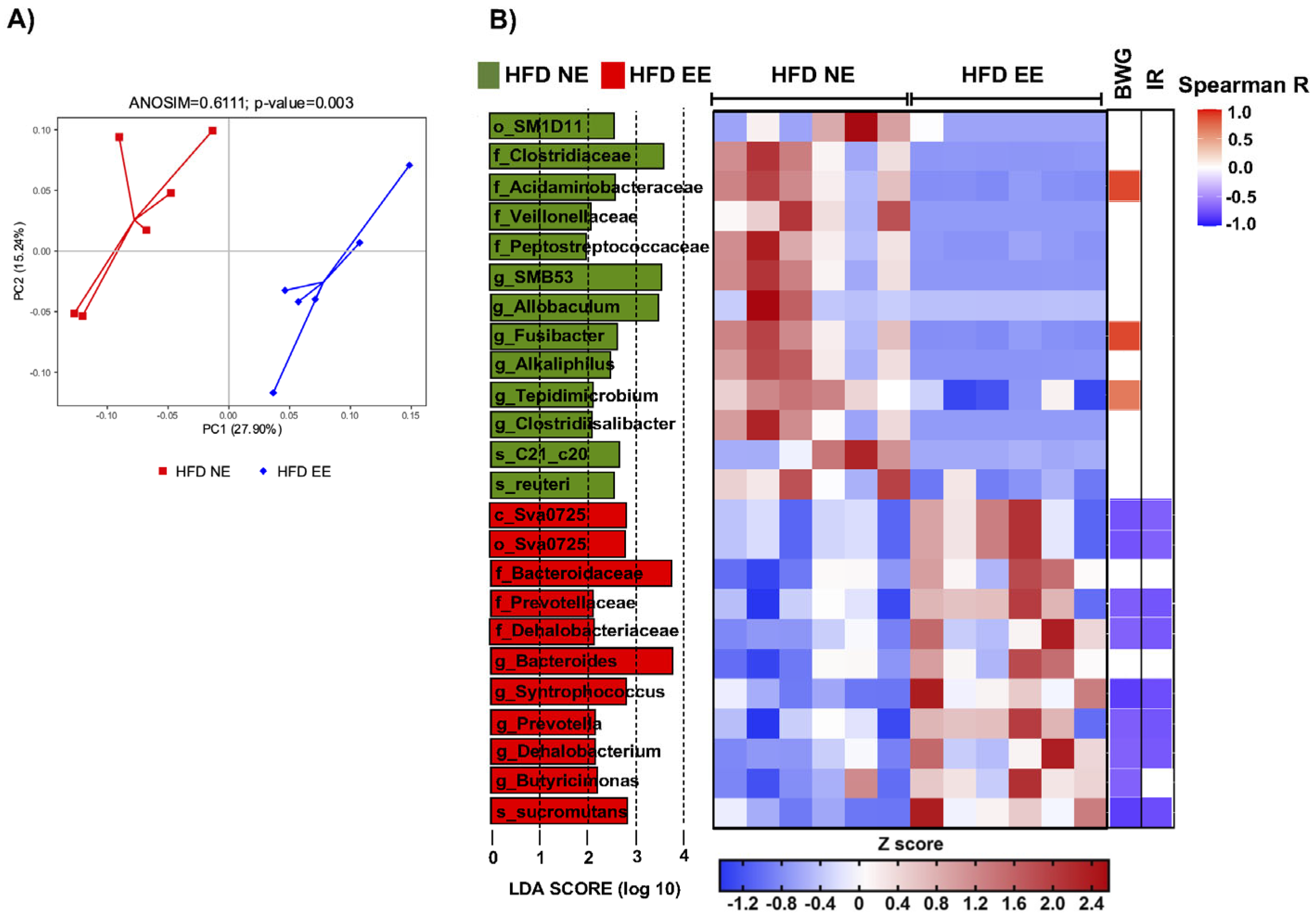
2.4. Environmental Enrichment Modulates Gut Microbiota Dysbiosis and Increases Specific Intestinal Taxa
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Basal Glucose Levels, Glucose Tolerance, and Insulin Resistance Test
4.3. Serum Harvesting
4.4. Lipids Determination
4.5. Mice Perfusion
4.6. Tissue Analysis
4.7. Total Protein Extraction and Western Blot Assay
4.8. ELISA
4.9. LPS Quantification
4.10. Stool Collection
4.11. Stool Preparation for 16S rRNA Sequencing
4.12. Bioinformatic Analysis of the 16S rRNA Profiling Data
4.13. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rohm, T.V.; Meier, D.T.; Olefsky, J.M.; Donath, M.Y. Inflammation in obesity, diabetes, and related disorders. Immunity 2022, 55, 31–55. [Google Scholar] [CrossRef] [PubMed]
- Cruz, N.G.; Sousa, L.P.; Sousa, M.O.; Pietrani, N.T.; Fernandes, A.P.; Gomes, K.B. The linkage between inflammation and Type 2 diabetes mellitus. Diabetes Res. Clin. Pract. 2013, 99, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.A.; Gu, W.; Lee, I.A.; Joh, E.H.; Kim, D.H. High Fat Diet-Induced Gut Microbiota Exacerbates Inflammation and Obesity in Mice via the TLR4 Signaling Pathway. PLoS ONE 2012, 7, e47713. [Google Scholar] [CrossRef] [PubMed]
- Moraes, J.C.; Coope, A.; Morari, J.; Cintra, D.E.; Roman, E.A.; Pauli, J.R.; Romanatto, T.; Carvalheira, J.B.; Oliveira, A.L.R.; Saad, M.J.; et al. High-fat diet induces apoptosis of hypothalamic neurons. PLoS ONE 2009, 4, e5045. [Google Scholar] [CrossRef]
- Monteiro, R.; Azevedo, I. Chronic inflammation in obesity and the metabolic syndrome. Mediat. Inflamm. 2010, 2010, 289645. [Google Scholar] [CrossRef]
- Posey, K.A.; Clegg, D.J.; Printz, R.L.; Byun, J.; Morton, G.J.; Vivekanandan-Giri, A.; Pennathur, S.; Baskin, D.G.; Heinecke, J.W.; Woods, S.C.; et al. Hypothalamic proinflammatory lipid accumulation, inflammation, and insulin resistance in rats fed a high-fat diet. Am. J. Physiol. Endocrinol. Metab. 2009, 296, 1003–1012. [Google Scholar] [CrossRef] [PubMed]
- Koyama, Y.; Brenner, D.A. Liver inflammation and fibrosis. J. Clin. Investig. 2017, 127, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Gregor, M.F.; Hotamisligil, G.S. Inflammatory mechanisms in obesity. Annu. Rev. Immunol. 2011, 29, 415–445. [Google Scholar] [CrossRef] [PubMed]
- Reilly, S.M.; Saltiel, A.R. Adapting to obesity with adipose tissue inflammation. Nat. Rev. Endocrinol. 2017, 13, 633–643. [Google Scholar] [CrossRef] [PubMed]
- De Luca, C.; Olefsky, J.M. Inflammation and insulin resistance. FEBS Lett. 2008, 582, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Moreira, A.P.B.; Texeira, T.F.S.; Ferreira, A.B.; Do Carmo Gouveia Peluzio, M.; De Cássia Gonçalves Alfenas, R. Influence of a high-fat diet on gut microbiota, intestinal permeability and metabolic endotoxaemia. Br. J. Nutr. 2012, 108, 801–809. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Cheng, D.; Peng, C.; Li, Y.; Zhu, Y.; Lu, N. High-fat diet induces dysbiosis of gastric microbiota prior to gut microbiota in association with metabolic disorders in mice. Front. Microbiol. 2018, 9, 639. [Google Scholar] [CrossRef] [PubMed]
- Murphy, E.A.; Velazquez, K.T.; Herbert, K.M. Influence of High-Fat-Diet on Gut Microbiota: A Driving Force for Chronic Disease Risk. Curr. Opin. Clin. Nutr. Metab. Care 2015, 18, 515–520. [Google Scholar] [CrossRef] [PubMed]
- Peterson, C.T.; Sharma, V.; Elmén, L.; Peterson, S.N. Immune homeostasis, dysbiosis and therapeutic modulation of the gut microbiota. Clin. Exp. Immunol. 2015, 179, 363–377. [Google Scholar] [CrossRef] [PubMed]
- Rabot, S.; Membrez, M.; Blancher, F.; Berger, B.; Moine, D.; Krause, L.; Bibiloni, R.; Bruneau, A.; Gérard, P.; Siddharth, J.; et al. High fat diet drives obesity regardless the composition of gut microbiota in mice. Sci. Rep. 2016, 6, 32484. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Ridaura, V.K.; Faith, J.J.; Rey, F.E.; Knight, R.; Gordon, J.I. The Effect of Diet on the Human Gut Microbiome: A Metagenomic Analysis in Humanized Gnotobiotic Mice Peter. Sci. Transl. Med. 2009, 1, 6ra14. [Google Scholar] [CrossRef] [PubMed]
- Bäckhed, F.; Ding, H.; Wang, T.; Hooper, L.V.; Gou, Y.K.; Nagy, A.; Semenkovich, C.F.; Gordon, J.I. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. USA 2004, 101, 15718–15723. [Google Scholar] [CrossRef] [PubMed]
- Cryan, J.F.; O’riordan, K.J.; Cowan, C.S.M.; Sandhu, K.V.; Bastiaanssen, T.F.S.; Boehme, M.; Codagnone, M.G.; Cussotto, S.; Fulling, C.; Golubeva, A.V.; et al. The microbiota-gut-brain axis. Physiol. Rev. 2019, 99, 1877–2013. [Google Scholar] [CrossRef] [PubMed]
- Gulas, E.; Wysiadecki, G.; Strzelecki, D.; Gawlik-Kotelnicka, O.; Polguj, M. Can microbiology affect psychiatry? A link between gut microbiota and psychiatric disorders. Psychiatr. Pol. 2018, 52, 1023–1039. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Liu, X.; Xu, S.; Hu, S.; Wang, S.; Shi, D.; Wang, K.; Wang, Z.; Lin, Q.; Li, S.; et al. Prevotella histicola Mitigated Estrogen Deficiency-Induced Depression via Gut Microbiota-Dependent Modulation of Inflammation in Ovariectomized Mice. Front. Nutr. 2022, 8, 805465. [Google Scholar] [CrossRef] [PubMed]
- Murros, K.E.; Huynh, V.A.; Takala, T.M.; Saris, P.E.J. Desulfovibrio Bacteria Are Associated with Parkinson’s Disease. Front. Cell. Infect. Microbiol. 2021, 11, 652617. [Google Scholar] [CrossRef] [PubMed]
- Sgritta, M.; Dooling, S.W.; Buffington, S.A.; Momin, E.N.; Francis, M.B.; Britton, R.A.; Costa-Mattioli, M. Mechanisms Underlying Microbial-Mediated Changes in Social Behavior in Mouse Models of Autism Spectrum Disorder. Neuron 2019, 101, 246–259.e6. [Google Scholar] [CrossRef] [PubMed]
- Nithianantharajah, J.; Hannan, A.J. Enriched environments, experience-dependent plasticity and disorders of the nervous system. Nat. Rev. Neurosci. 2006, 7, 697–709. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Choi, E.Y.; Liu, X.; Martin, A.; Wang, C.; Xu, X.; During, M.J. White to brown fat phenotypic switch induced by genetic and environmental activation of a hypothalamic-adipocyte axis. Cell Metab. 2011, 14, 324–338. [Google Scholar] [CrossRef] [PubMed]
- Díaz de León-Guerrero, S.; Salazar-León, J.; Meza-Sosa, K.F.; Valle-Garcia, D.; Aguilar-León, D.; Pedraza-Alva, G.; Pérez-Martínez, L. An enriched environment re-establishes metabolic homeostasis by reducing obesity-induced inflammation. Dis. Models Mech. 2022, 15, dmm048936. [Google Scholar] [CrossRef] [PubMed]
- Singh, Y.; El-Hadidi, M.; Admard, J.; Wassouf, Z.; Schulze-Hentrich, J.M.; Kohlhofer, U.; Quintanilla-Martinez, L.; Huson, D.; Riess, O.; Casadei, N. Enriched environmental conditions modify the gut microbiome composition and fecal markers of inflammation in parkinson’s disease. Front. Neurosci. 2019, 13, 1032. [Google Scholar] [CrossRef] [PubMed]
- Cuozzo, S.; de Moreno de LeBlanc, A.; LeBlanc, J.G.; Hoffmann, N.; Tortella, G.R. Streptomyces genus as a source of probiotics and its potential for its use in health. Microbiol. Res. 2023, 266, 127248. [Google Scholar] [CrossRef]
- Lim, T.; Lee, K.; Kim, R.H.; Cha, K.H.; Koo, S.Y.; Moon, E.C.; Hwang, K.T. Black raspberry extract can lower serum LDL cholesterol via modulation of gut microbial composition and serum bile acid profile in rats fed trimethylamine-N-oxide with a high-fat diet. Food Sci. Biotechnol. 2022, 31, 1041–1051. [Google Scholar] [CrossRef] [PubMed]
- Parker, B.J.; Wearsch, P.A.; Veloo, A.C.M.; Rodriguez-Palacios, A. The Genus Alistipes: Gut Bacteria with Emerging Implications to Inflammation, Cancer, and Mental Health. Front. Immunol. 2020, 11, 906. [Google Scholar] [CrossRef] [PubMed]
- Ball, N.J.; Mercado, E.; Orduña, I. Enriched environments as a potential treatment for developmental disorders: A critical assessment. Front. Psychol. 2019, 10, 466. [Google Scholar] [CrossRef] [PubMed]
- Clemenson, G.D.; Stark, S.M.; Rutledge, S.M.; Stark, C.E. Enriching hippocampal memory function in older adults through video games. Behav. Brain Res. 2020, 390, 112667. [Google Scholar] [CrossRef] [PubMed]
- Pradhan, S. The use of commercially available games for a combined physical and cognitive challenge during exercise for individuals with Parkinson’s disease—A case series report. Physiother. Theory Pract. 2019, 35, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Kimura, L.F.; Novaes, L.S.; Picolo, G.; Munhoz, C.D.; Cheung, C.W.; Camarini, R. How environmental enrichment balances out neuroinflammation in chronic pain and comorbid depression and anxiety disorders. Br. J. Pharmacol. 2022, 179, 1640–1660. [Google Scholar] [CrossRef] [PubMed]
- Binda, C.; Lopetuso, L.R.; Rizzatti, G.; Gibiino, G.; Cennamo, V.; Gasbarrini, A. Actinobacteria: A relevant minority for the maintenance of gut homeostasis. Dig. Liver Dis. 2018, 50, 421–428. [Google Scholar] [CrossRef] [PubMed]
- Khan, T.J.; Ahmed, Y.M.; Zamzami, M.A.; Mohamed, S.A.; Khan, I.; Baothman, O.A.S.; Mehanna, M.G.; Yasir, M. Effect of atorvastatin on the gut microbiota of high fat diet-induced hypercholesterolemic rats. Sci. Rep. 2018, 8, 662. [Google Scholar] [CrossRef] [PubMed]
- Million, M.; Maraninchi, M.; Henry, M.; Armougom, F.; Richet, H.; Carrieri, P.; Valero, R.; Raccah, D.; Vialettes, B.; Raoult, D. Obesity-associated gut microbiota is enriched in Lactobacillus reuteri and depleted in Bifidobacterium animalis and Methanobrevibacter smithii. Int. J. Obes. 2012, 36, 817–825. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Y.; Sun, J.; Xia, S.; Li, L.; Li, Y.; Wang, P.; Shi, Y.; Le, G. Effects of different Lactobacillus reuteri on inflammatory and fat storage in high-fat diet-induced obesity mice model. J. Funct. Foods 2015, 14, 424–434. [Google Scholar] [CrossRef]
- Wang, B.; Kong, Q.; Li, X.; Zhao, J.; Zhang, H.; Chen, W.; Wang, G. A high-fat diet increases gut microbiota biodiversity and energy expenditure due to nutrient difference. Nutrients 2020, 12, 3197. [Google Scholar] [CrossRef] [PubMed]
- Carloni, S.; Bertocchi, A.; Mancinelli, S.; Bellini, M.; Erreni, M.; Borreca, A.; Braga, D.; Giugliano, S.; Mozzarelli, A.M.; Manganaro, D.; et al. Identification of a choroid plexus vascular barrier closing during intestinal inflammation. Science 2021, 374, 439–448. [Google Scholar] [CrossRef] [PubMed]
- Obrenovich, M.E.M. Leaky gut, leaky brain? Microorganisms 2018, 6, 107. [Google Scholar] [CrossRef] [PubMed]
- Ahmadi, S.; Razazan, A.; Nagpal, R.; Jain, S.; Wang, B.; Mishra, S.P.; Wang, S.; Justice, J.; Ding, J.; McClain, D.A.; et al. Metformin reduces aging-related leaky gut and improves cognitive function by beneficially modulating gut microbiome/goblet cell/mucin axis. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2020, 75, E9–E21. [Google Scholar] [CrossRef] [PubMed]
- De Siqueira Valadares, L.T.; de Souza, L.S.B.; Salgado Júnior, V.A.; de Freitas Bonomo, L.; de Macedo, L.R.; Silva, M. Prevalence of metabolic syndrome in Brazilian adults in the last 10 years: A systematic review and meta-analysis. BMC Public Health 2022, 22, 327. [Google Scholar] [CrossRef] [PubMed]
- Strissel, K.J.; Stancheva, Z.; Miyoshi, H.; Perfield, J.W.; DeFuria, J.; Jick, Z.; Greenberg, A.S.; Obin, M.S. Adipocyte death, adipose tissue remodeling, and obesity complications. Diabetes 2007, 56, 2910–2918. [Google Scholar] [CrossRef] [PubMed]
- Murano, I.; Barbatelli, G.; Parisani, V.; Latini, C.; Muzzonigro, G.; Castellucci, M.; Cinti, S. Dead adipocytes, detected as crown-like structures, are prevalent in visceral fat depots of genetically obese mice. J. Lipid Res. 2008, 49, 1562–1568. [Google Scholar] [CrossRef] [PubMed]
- Inoue, M.; Ohtake, T.; Motomura, W.; Takahashi, N.; Hosoki, Y.; Miyoshi, S.; Suzuki, Y.; Saito, H.; Kohgo, Y.; Okumura, T. Increased expression of PPARγ in high fat diet-induced liver steatosis in mice. Biochem. Biophys. Res. Commun. 2005, 336, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Meli, R.; Mattace Raso, G.; Irace, C.; Simeoli, R.; Di Pascale, A.; Paciello, O.; Pagano, T.B.; Calignano, A.; Colonna, A.; Santamaria, R. High Fat Diet Induces Liver Steatosis and Early Dysregulation of Iron Metabolism in Rats. PLoS ONE 2013, 8, e66570. [Google Scholar] [CrossRef] [PubMed]
- Gauthier, M.-S.; Favier, R.; Lavoie, J.-M. Time course of the development of non-alcoholic hepatic steatosis in response to high-fat diet-induced obesity in rats. Br. J. Nutr. 2006, 95, 273–281. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Gao, J.; Ke, W.; Wang, J.; Li, D.; Liu, R.; Jia, Y.; Wang, X.; Chen, X.; Chen, F.; et al. Resveratrol reduces obesity in high-fat diet-fed mice via modulating the composition and metabolic function of the gut microbiota. Free Radic. Biol. Med. 2020, 156, 83–98. [Google Scholar] [CrossRef] [PubMed]
- De La Serre, C.B.; Ellis, C.L.; Lee, J.; Hartman, A.L.; Rutledge, J.C.; Raybould, H.E. Propensity to high-fat diet-induced obesity in rats is associated with changes in the gut microbiota and gut inflammation. Am. J. Physiol. Gastrointest. Liver Physiol. 2010, 299, 440–448. [Google Scholar] [CrossRef] [PubMed]
- Selmin, O.I.; Papoutsis, A.J.; Hazan, S.; Smith, C.; Greenfield, N.; Donovan, M.G.; Wren, S.N.; Doetschman, T.C.; Snider, J.M.; Snider, A.J.; et al. N-6 high fat diet induces gut microbiome dysbiosis and colonic inflammation. Int. J. Mol. Sci. 2021, 22, 6919. [Google Scholar] [CrossRef] [PubMed]
- Gulhane, M.; Murray, L.; Lourie, R.; Tong, H.; Sheng, Y.H.; Wang, R.; Kang, A.; Schreiber, V.; Wong, K.Y.; Magor, G.; et al. High Fat Diets Induce Colonic Epithelial Cell Stress and Inflammation that is Reversed by IL-22. Sci. Rep. 2016, 6, 28990. [Google Scholar] [CrossRef] [PubMed]
- Cani, P.D.; Bibiloni, R.; Knauf, C.; Neyrinck, A.M.; Delzenne, N.M. Changes in gut microbiota control metabolic diet–induced obesity and diabetes in mice. Diabetes 2008, 57, 1470–1481. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Hara, H. Dietary fat and bile juice, but not obesity, are responsible for the increase in small intestinal permeability induced through the suppression of tight junction protein expression in LETO and OLETF rats. Nutr. Metab. 2010, 7, 19. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, R.; Rah, B.; Bastola, D.; Dhawan, P.; Singh, A.B. Obesity-induces Organ and Tissue Specific Tight Junction Restructuring and Barrier Deregulation by Claudin Switching. Sci. Rep. 2017, 7, 5125. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.; Chi, M.M.; Scull, B.P.; Rigby, R.; Schwerbrock, N.M.J.; Magness, S.; Jobin, C.; Lund, P.K. High-fat diet: Bacteria interactions promote intestinal inflammation which precedes and correlates with obesity and insulin resistance in mouse. PLoS ONE 2010, 5, e12191. [Google Scholar] [CrossRef] [PubMed]
- Pendyala, S.; Walker, J.M.; Holt, P.R. A high-fat diet is associated with endotoxemia that originates from the gut. Gastroenterology 2012, 142, 1100–1101.e2. [Google Scholar] [CrossRef] [PubMed]
- Gersemann, M.; Becker, S.; Kübler, I.; Koslowski, M.; Wang, G.; Herrlinger, K.R.; Griger, J.; Fritz, P.; Fellermann, K.; Schwab, M.; et al. Differences in goblet cell differentiation between Crohn’s disease and ulcerative colitis. Differentiation 2009, 77, 84–94. [Google Scholar] [CrossRef] [PubMed]
- Sheng, Y.H.; Hasnain, S.Z.; Florin, T.H.J.; McGuckin, M.A. Mucins in inflammatory bowel diseases and colorectal cancer. J. Gastroenterol. Hepatol. 2012, 27, 28–38. [Google Scholar] [CrossRef] [PubMed]
- Zafar, H.; Saier, M.H. Gut Bacteroides species in health and disease. Gut Microbes 2021, 13, 1848158. [Google Scholar] [CrossRef] [PubMed]
- Fuller, A.K.; Bice, B.D.; Venancio, A.R.; Crowley, O.M.; Staab, A.M.; Georges, S.J.; Hidalgo, J.R.; Warncke, A.V.; Angus-Hill, M.L. A Method to Define the Effects of Environmental Enrichment on Colon Microbiome Biodiversity in a Mouse Colon Tumor Model. J. Vis. Exp. 2018, 2018, e57182. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Liang, L.; Yu, G.; Li, Q. Pumpkin polysaccharide modifies the gut microbiota during alleviation of type 2 diabetes in rats. Int. J. Biol. Macromol. 2018, 115, 711–717. [Google Scholar] [CrossRef] [PubMed]
- García-Legorreta, A.; Soriano-Pérez, L.A.; Flores-Buendía, A.M.; Medina-Campos, O.N.; Noriega, L.G.; Granados-Portillo, O.; Nambo-Venegas, R.; Tovar, A.R.; Mendoza-Vargas, A.; Barrera-Oviedo, D.; et al. Effect of dietary magnesium content on intestinal microbiota of rats. Nutrients 2020, 12, 2889. [Google Scholar] [CrossRef] [PubMed]
- Gallardo-Becerra, L.; Cornejo-Granados, F.; García-López, R.; Valdez-Lara, A.; Bikel, S.; Canizales-Quinteros, S.; López-Contreras, B.E.; Mendoza-Vargas, A.; Nielsen, H.; Ochoa-Leyva, A. Metatranscriptomic analysis to define the Secrebiome, and 16S rRNA profiling of the gut microbiome in obesity and metabolic syndrome of Mexican children. Microb. Cell Fact. 2020, 19, 61. [Google Scholar] [CrossRef] [PubMed]
- Devkota, S.; Wang, Y.; Musch, M.W.; Leone, V.; Fehlner-Peach, H.; Nadimpalli, A.; Antonopoulos, D.A.; Jabri, B.; Chang, E.B. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature 2012, 487, 104–108. [Google Scholar] [CrossRef] [PubMed]
- Loubinoux, J.; Bronowicki, J.P.; Pereira, I.A.C.; Mougenel, J.L.; Le Faou, A.E. Sulfate-reducing bacteria in human feces and their association with inflammatory bowel diseases. FEMS Microbiol. Ecol. 2002, 40, 107–112. [Google Scholar] [CrossRef]
- Horvath, A.; Durdevic, M.; Leber, B.; Di Vora, K.; Rainer, F.; Krones, E.; Douschan, P.; Spindelboeck, W.; Durchschein, F.; Zollner, G.; et al. Changes in the intestinal microbiome during a multispecies probiotic intervention in compensated cirrhosis. Nutrients 2020, 12, 1874. [Google Scholar] [CrossRef] [PubMed]
- Doré, J.; Bryant, M.P. Lipid growth requirement and influence of lipid supplement on fatty acid and aldehyde composition of Syntrophococcus sucromutans. Appl. Environ. Microbiol. 1989, 55, 927–933. [Google Scholar] [CrossRef] [PubMed]
- Bernard-Vailhé, M.; Besle, J.; Doré, J.; Jouany, J. Transformations of 14 C lignin cell walls of wheat by a fungus and by bacteria from the rumen. Ann. Zootech. 1994, 43, 268. [Google Scholar] [CrossRef]
- Dore, J.; Bryant, M.P. Metabolism of one-carbon compounds by the ruminal acetogen Syntrophococcus sucromutans. Appl. Environ. Microbiol. 1990, 56, 984–989. [Google Scholar] [CrossRef] [PubMed]
- Trinchese, G.; Cavaliere, G.; Canani, R.B.; Matamoros, S.; Bergamo, P.; De Filippo, C.; Aceto, S.; Gaita, M.; Cerino, P.; Negri, R.; et al. Human, donkey and cow milk differently affects energy efficiency and inflammatory state by modulating mitochondrial function and gut microbiota. J. Nutr. Biochem. 2015, 26, 1136–1146. [Google Scholar] [CrossRef] [PubMed]
- Van Hul, M.; Karnik, K.; Canene-Adams, K.; De Souza, M.; Van Den Abbeele, P.; Marzorati, M.; Delzenne, N.M.; Everard, A.; Cani, P.D. Comparison of the effects of soluble corn fiber and fructooligosaccharides on metabolism, inflammation, and gut microbiome of high-fat diet-fed mice. Am. J. Physiol. Endocrinol. Metab. 2020, 319, E779–E791. [Google Scholar] [CrossRef] [PubMed]
- Chassaing, B.; De Bodt, J.; Marzorati, M.; Van de Wiele, T.; Gewirtz, A.T. Dietary emulsifiers directly alter human microbiota composition and gene expression ex vivo potentiating intestinal inflammation. Gut 2017, 66, 1414–1427. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; An, J.; Kim, J.; Choi, D.; Song, Y.; Lee, C.K.; Kong, H.; Kim, S.B.; Kim, K. A Novel Bacterium, Butyricimonas virosa, Preventing HFD-Induced Diabetes and Metabolic Disorders in Mice via GLP-1 Receptor. Front. Microbiol. 2022, 13, 858192. [Google Scholar] [CrossRef] [PubMed]
- Pu, S.; Khazanehei, H.; Jones, P.J.; Khafipour, E. Interactions between obesity status and dietary intake of monounsaturated and polyunsaturated oils on human gut microbiome profiles in the canola oil multicenter intervention trial (COMIT). Front. Microbiol. 2016, 7, 1612. [Google Scholar] [CrossRef] [PubMed]
- Coppola, S.; Avagliano, C.; Calignano, A.; Berni Canani, R. The protective role of butyrate against obesity and obesity-related diseases. Molecules 2021, 26, 682. [Google Scholar] [CrossRef] [PubMed]
- Genser, L.; Aguanno, D.; Soula, H.A.; Dong, L.; Trystram, L.; Assmann, K.; Salem, J.E.; Vaillant, J.C.; Oppert, J.M.; Laugerette, F.; et al. Increased jejunal permeability in human obesity is revealed by a lipid challenge and is linked to inflammation and type 2 diabetes. J. Pathol. 2018, 246, 217–230. [Google Scholar] [CrossRef] [PubMed]
- Brenes, J.C.; Lackinger, M.; Höglinger, G.U.; Schratt, G.; Schwarting, R.K.W.; Wöhr, M. Differential effects of social and physical environmental enrichment on brain plasticity, cognition, and ultrasonic communication in rats. J. Comp. Neurol. 2016, 524, 1586–1607. [Google Scholar] [CrossRef] [PubMed]
- Prado Lima, M.G.; Schimidt, H.L.; Garcia, A.; Daré, L.R.; Carpes, F.P.; Izquierdo, I.; Mello-Carpes, P.B. Environmental enrichment and exercise are better than social enrichment to reduce memory deficits in amyloid beta neurotoxicity. Proc. Natl. Acad. Sci. USA 2018, 115, E2403–E2409. [Google Scholar] [CrossRef] [PubMed]
- Grégoire, C.A.; Bonenfant, D.; Le Nguyen, A.; Aumont, A.; Fernandes, K.J.L. Untangling the influences of voluntary running, environmental complexity, social housing and stress on adult hippocampal neurogenesis. PLoS ONE 2014, 9, e86237. [Google Scholar] [CrossRef]
- Langdon, K.D.; Corbett, D. Improved working memory following novel combinations of physical and cognitive activity. Neurorehabil. Neural Repair 2012, 26, 523–532. [Google Scholar] [CrossRef] [PubMed]
- Tai, M.M. A mathematical model for the determination of total area under glucose tolerance and other metabolic curves. Diabetes Care 1994, 17, 152–154. [Google Scholar] [CrossRef] [PubMed]
- Anandaraja, S.; Narang, R.; Godeswar, R.; Laksmy, R.; Talwar, K.K. Low-density lipoprotein cholesterol estimation by a new formula in Indian population. Int. J. Cardiol. 2005, 102, 117–120. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manzo, R.; Gallardo-Becerra, L.; Díaz de León-Guerrero, S.; Villaseñor, T.; Cornejo-Granados, F.; Salazar-León, J.; Ochoa-Leyva, A.; Pedraza-Alva, G.; Pérez-Martínez, L. Environmental Enrichment Prevents Gut Dysbiosis Progression and Enhances Glucose Metabolism in High-Fat Diet-Induced Obese Mice. Int. J. Mol. Sci. 2024, 25, 6904. https://doi.org/10.3390/ijms25136904
Manzo R, Gallardo-Becerra L, Díaz de León-Guerrero S, Villaseñor T, Cornejo-Granados F, Salazar-León J, Ochoa-Leyva A, Pedraza-Alva G, Pérez-Martínez L. Environmental Enrichment Prevents Gut Dysbiosis Progression and Enhances Glucose Metabolism in High-Fat Diet-Induced Obese Mice. International Journal of Molecular Sciences. 2024; 25(13):6904. https://doi.org/10.3390/ijms25136904
Chicago/Turabian StyleManzo, Rubiceli, Luigui Gallardo-Becerra, Sol Díaz de León-Guerrero, Tomas Villaseñor, Fernanda Cornejo-Granados, Jonathan Salazar-León, Adrian Ochoa-Leyva, Gustavo Pedraza-Alva, and Leonor Pérez-Martínez. 2024. "Environmental Enrichment Prevents Gut Dysbiosis Progression and Enhances Glucose Metabolism in High-Fat Diet-Induced Obese Mice" International Journal of Molecular Sciences 25, no. 13: 6904. https://doi.org/10.3390/ijms25136904








