Assessing the Potential of an Enzymatically Liberated Salmon Oil to Support Immune Health Recovery from Acute SARS-CoV-2 Infection via Change in the Expression of Cytokine, Chemokine and Interferon-Related Genes
Abstract
:1. Introduction
2. Results
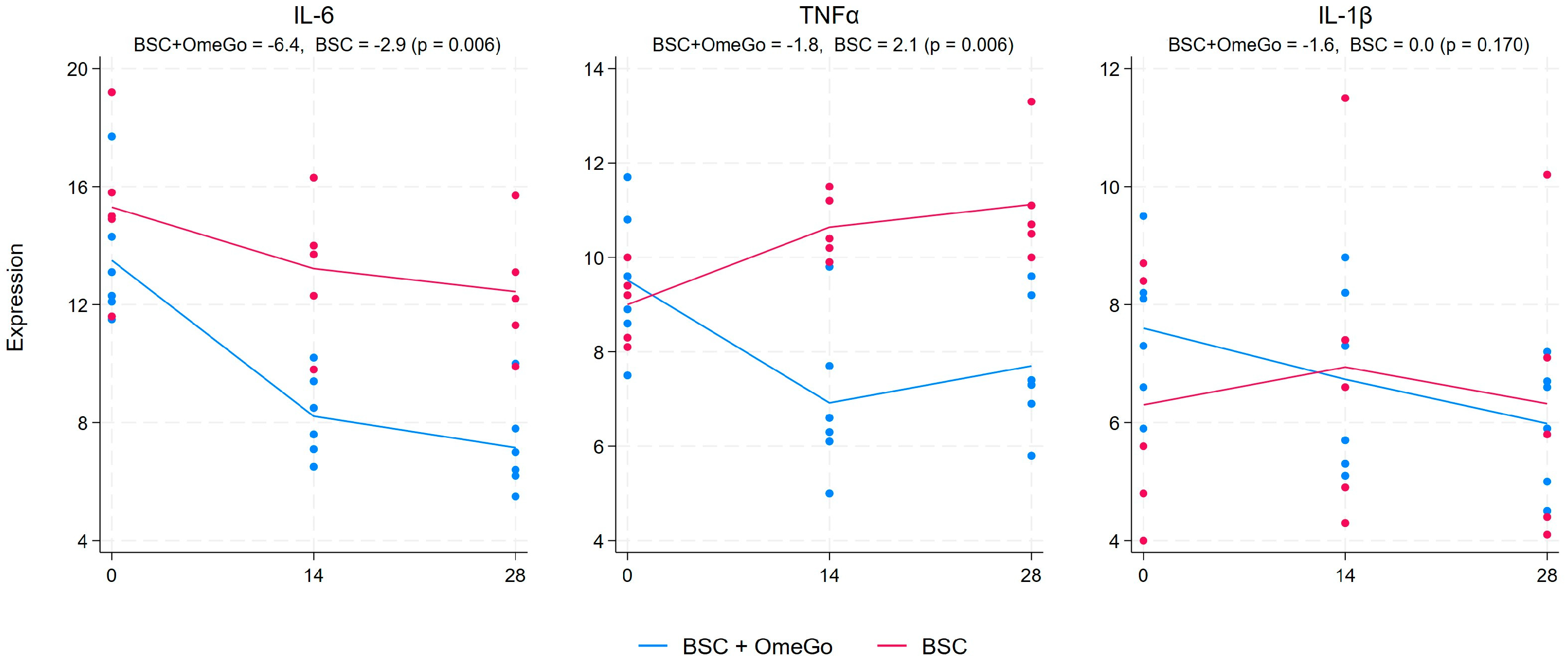
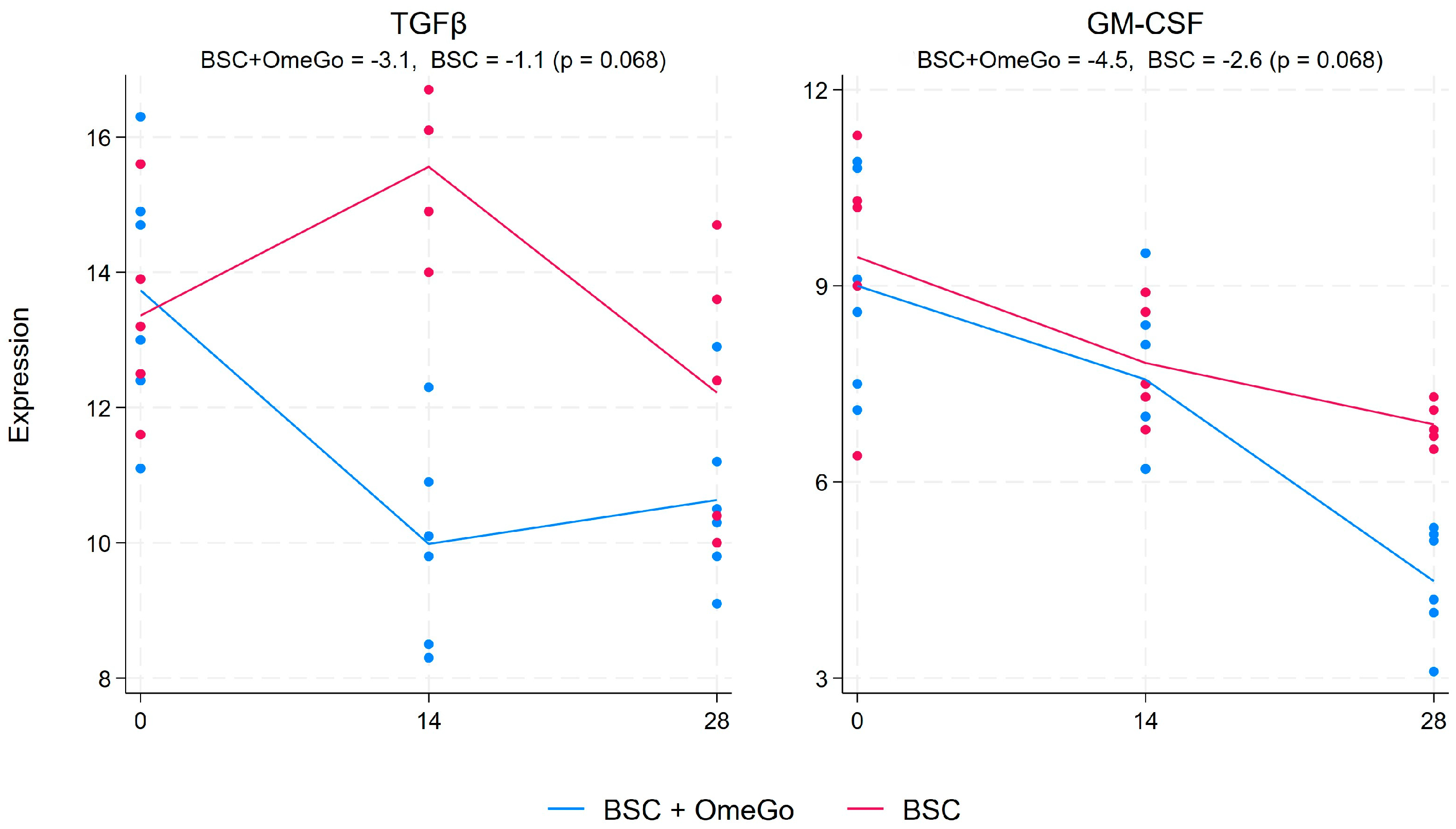
2.1. Change in Cytokine Gene Expression Levels
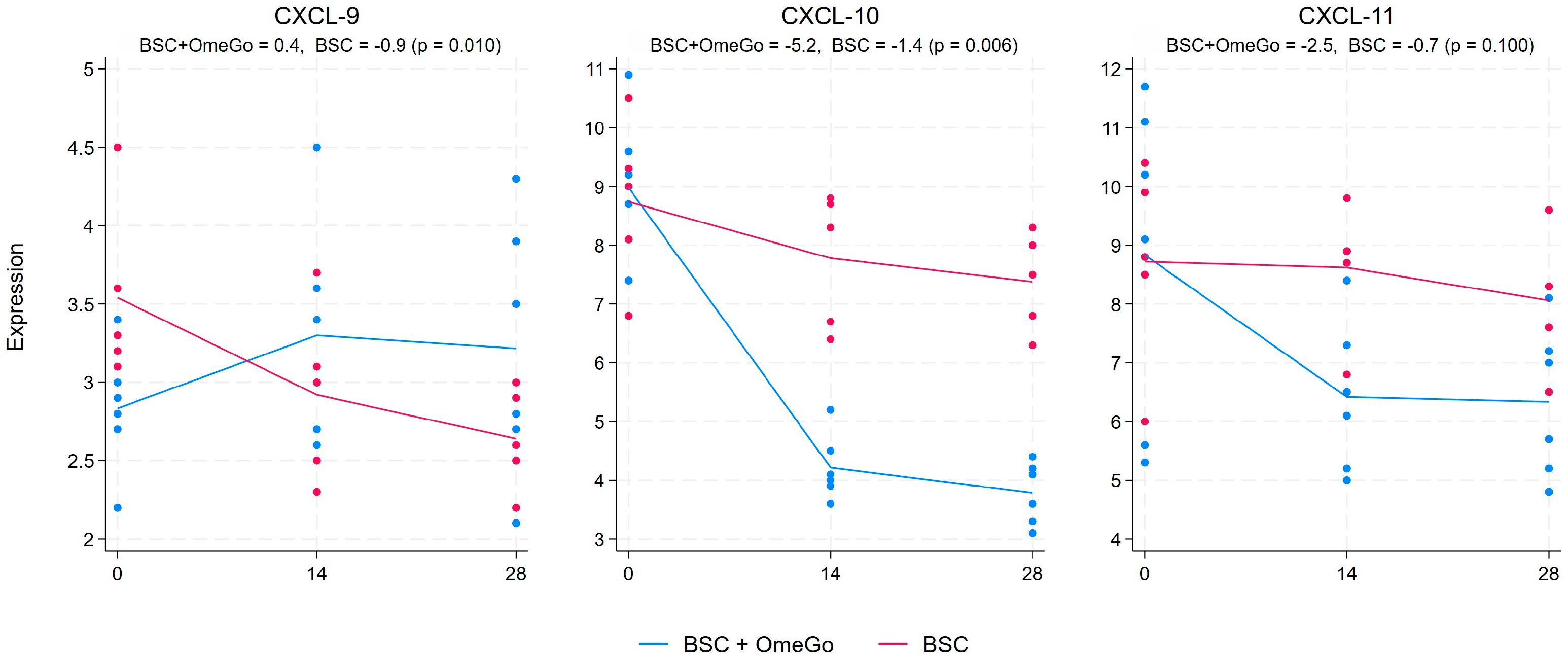
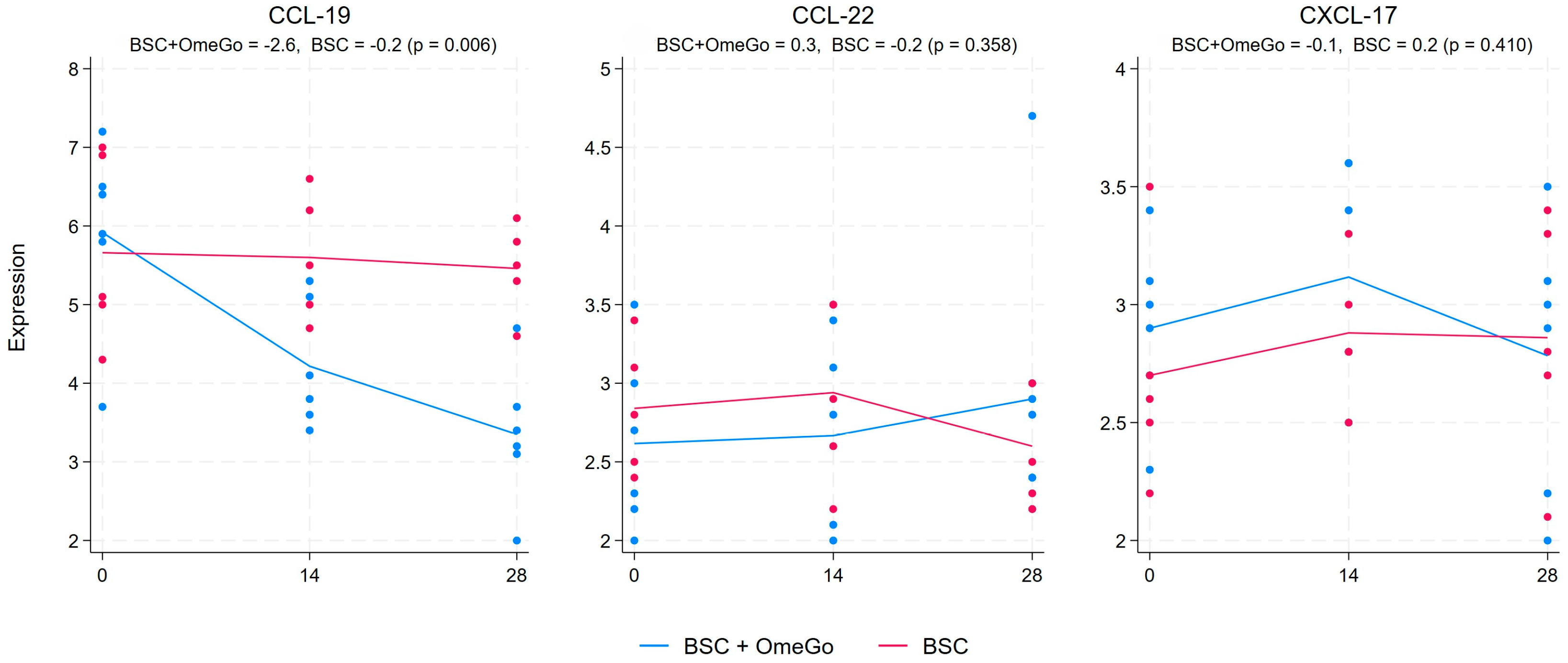
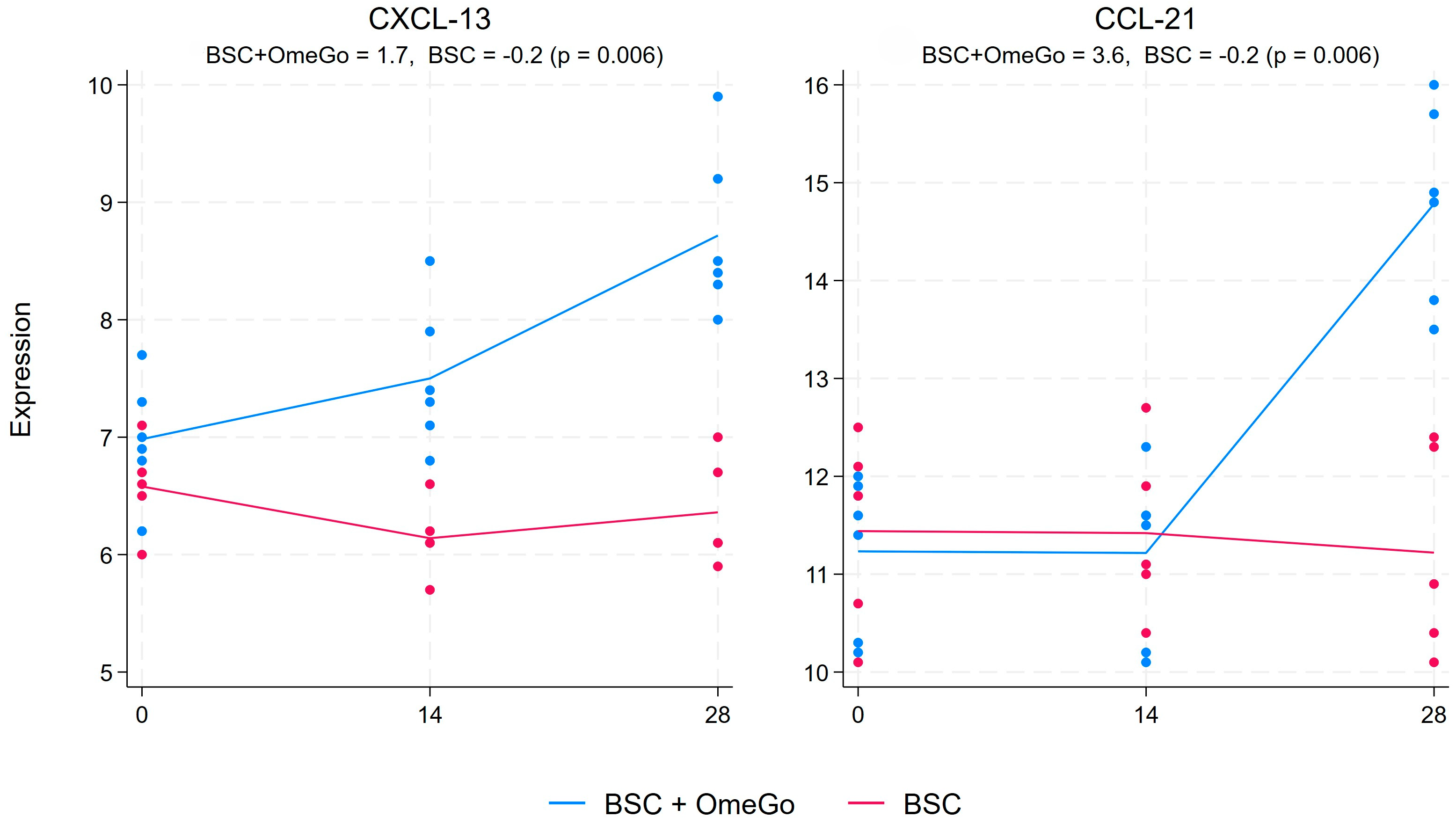
2.2. Change in Chemokine Gene Expression Levels
2.3. Change in Interferon and Interferon-Related Factors Gene Expression Levels
3. Discussion
4. Materials and Methods
4.1. Study Design and Study Subjects
4.2. Gene Expression Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Queiroz, M.A.F.; Moura da Neves, P.F.; Lima, S.S.; Lopes, J.D.C.; Torres, M.K.D.S.; Vallinoto, I.M.V.C.; Bichara, C.D.A.; Santos, E.F.D.; de Brito, M.T.F.M.; da Silva, A.L.S.; et al. Cytokine Profiles Associated With Acute COVID-19 and Long COVID-19 Syndrome. Front. Cell Infect. Microbiol. 2022, 12, 922422. [Google Scholar] [CrossRef] [PubMed]
- Scultheis, C.; Wilscher, E.; Paschold, L.; Gottschick, C.; Klee, B.; Henkes, S.S.; Bosurgi, L.; Dutzmann, J.; Sedding, D.; Frese, T.; et al. The IL-1, IL-6 and TNF cytokine triad is associated with post-acute sequelae of COVID-19. Cell Rep. Med. 2022, 6, 100663. [Google Scholar] [CrossRef]
- Gudowska-Sawczuk, M.; Mroczko, B. What is Currently Known about the Role of CXCL10 in SARS-CoV-2 Infection? Int. J. Mol. Sci. 2022, 7, 3673. [Google Scholar] [CrossRef] [PubMed]
- Unterman, A.; Sumida, T.S.; Nouri, N.; Yan, X.; Zhao, A.Y.; Gasque, V.; Schupp, J.C.; Asashima, H.; Liu, Y.; Cosme, C., Jr.; et al. Single cell multi-omics reveals dyssynchrony of the innate and adaptive immune system in progressive COVID-19. Nat. Commun. 2022, 13, 440. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, Y.; Liu, Q.; Yao, Q.; Wang, X.; Zhang, H.; Chen, R.; Ren, L.; Min, J.; Deng, F.; et al. SARS-CoV-2 cell tropism and multiorgan infection. Cell Discov. 2021, 1, 17. [Google Scholar] [CrossRef] [PubMed]
- Eberhardt, N.; Noval, M.G.; Kaur, R.; Amadori, L.; Gildea, M.; Sajja, S.; Das, D.; Cilhoroz, B.; Stewart, O.J.; Fernandez, D.M.; et al. SARS-CoV-2 infection triggers pro-atherogenic inflammatory responses in human coronary vessels. Nat. Cardiovasc. Res. 2023, 2, 889–916. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Meol, D.; Nilsson-Payant, E.; Liu, W.C.; Uhl, S.; Hoagland, D.; Møller, R.; Jordan, T.X.; Oishi, K.; Panis, M.; Sachs, D.; et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell 2020, 5, 1036–1045. [Google Scholar] [CrossRef]
- Janeway, C.A.; Medzihitov, R. Innate Immune Recognition. Ann. Rev. Immunol. 2002, 20, 197–216. [Google Scholar] [CrossRef]
- Lazaer, H.M.; Schoggins, J.W.; Diamond, M.S. Shared and Distinct Functions of Type I and III Interferons. Immunity 2019, 4, 907–923. [Google Scholar] [CrossRef]
- Vivier, E.; Tomasello, E.; Baratin, M.; Walzer, T.; Ugolini, S. Functions of natural killer cells. Nat. Immunol. 2008, 9, 503–510. [Google Scholar] [CrossRef]
- Witkowski, M.; Tizian, C.; Ferreira-Gomes, M.; Niemeyer, D.; Jones, T.C.; Heinrich, F.; Frischbutter, S.; Angermair, S.; Hohnstein, T.; Mattiola, I.; et al. Untimely TGFβ responses in COVID-19 limit antiviral functions of NK cells. Nature 2021, 600, 295–300. [Google Scholar] [CrossRef]
- Terjerina, F.; Catalan, P.; Rodriguez-Grande, C.; Adan, J.; Rodriguez-Gonzalez, C.; Muñoz, P.; Aldamiz, T.; Diez, C.; Perez, L.; Fanciulli, C.; et al. Post-COVID-19 syndrome. SARS-CoV-2 RNA detection in plasma, stool, and urine in patients with persistent symptoms after COVID-19. BMC Infect. Dis. 2022, 22, 211. [Google Scholar]
- Davis, H.E.; McCorkell, L.; Vogel, J.M.; Topol, E.J. Long COVID: Major findings, mechanisms and recommendations. Nat. Rev. Microbiol. 2023, 21, 133–146. [Google Scholar] [CrossRef] [PubMed]
- Merad, M.; Blish, C.A.; Sallusto, F.; Iwasaki, A. The immunology and immunopathology of COVID-19. Science 2022, 375, 1122–1127. [Google Scholar] [CrossRef]
- Harris, W.S.; Tintle, N.L.; Sathyanarayanan, S.P.; Westra, J. Association between blood N-3 fatty acid levels and the risk of coronavirus disease 2019 in the UK Biobank. Am. J. Clin. Nutr. 2023, 117, 357–363. [Google Scholar] [CrossRef] [PubMed]
- Chiang, N.; Serhan, C.N. Specialized pro-resolving mediator network: An update on production and actions. Essays Biochem. 2020, 3, 443–462. [Google Scholar]
- Chang, C.S.; Sun, H.L.; Lii, C.K.; Chen, H.W.; Chen, P.Y.; Liu, K.L. Gamma-linolenic acid inhibits inflammatory responses by regulating NF-kappaB and AP-1 activation in lipopolysaccharide-induced RAW 264.7 macrophages. Inflammation 2010, 1, 46–57. [Google Scholar] [CrossRef] [PubMed]
- De Souza, C.O.; Valenzuela, C.A.; Baker, E.J.; Miles, E.A.; Rosa Neto, J.C.; Calder, P.C. Palmitoleic acid has stronger anti-inflammatory potential in human endothelial cells compared to oleic and palmitic acids. Mol. Nutr. Food Res. 2018, 20, e1800322. [Google Scholar] [CrossRef]
- Venn-Watson, S.; Lumpkin, R.; Dennis, E.A. Efficacy of dietary odd-chain saturated fatty acid pentadecanoic acid parallels broad associated health benefits in humans: Could it be essential? Sci. Rep. 2020, 10, 8161. [Google Scholar] [CrossRef]
- Framroze, B.; Heggdal, H. An in vitro study to explore the modulation of eosinophil effector function in human allergic peripheral blood eosinophils using enzymatically extracted salmonid oil. Funct. Foods Health Dis. 2020, 10, 357. [Google Scholar] [CrossRef]
- Frediani, J.K.; Parsons, R.; McLendon, K.B.; Westbrook, A.L.; Lam, W.; Martin, G.; Pollock, N.R. The New Normal: Delayed Peak SARS-CoV-2 Viral Loads Relative to Symptom Onset and Implications for COVID-19 Testing Programs. Clin. Infect Dis. 2024, 78, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Faes, C.; Abrams, S.; van Beckhoven, D.; Meyfroidt, G.; Vlieghe, E.; Hens, N.; Belgian Collaborative Group on COVID-19 Hospital Surveillance. Time between Symptom Onset, Hospitalisation and Recovery or Death: Statistical Analysis of Belgian COVID-19 Patients. Int. J. Environ. Res. Public Health 2020, 17, 7560. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Guo, L.; Yuan, J.; Xu, Z.; Gu, Y.; Zhang, J.; Guan, Y.; Liang, J.; Lu, H.; Liu, Y. Viral and antibody dynamics of acute infection with SARS-CoV-2 omicron variant (B.1.1.529): A prospective cohort study from Shezhen, China. Lancet 2023, 4, e632–e641. [Google Scholar] [CrossRef] [PubMed]
- Woodruff, M.C.; Bonham, K.S.; Anam, F.A.; Walker, T.A.; Faliti, C.E.; Ishii, Y.; Kaminski, C.Y.; Ruunstrom, M.C.; Cooper, K.R.; Truong, A.D.; et al. Chronic inflammation, neutrophil activity, and autoreactivity splits long COVID. Nat. Commun. 2023, 14, 4201. [Google Scholar] [CrossRef] [PubMed]
- Donlan, A.N.; Sutherland, T.E.; Marie, C.; Preissner, S.; Bradley, B.T.; Carpenter, R.M.; Sturek, J.M.; Ma, J.Z.; Moreau, G.B.; Donowitz, J.R.; et al. IL-13 is a driver of COVID-19 severity. JCI Insight 2021, 15, e150107. [Google Scholar] [CrossRef] [PubMed]
- Bonaventura, A.; Vecchie, A.; Wang, T.S.; Lee, E.; Cremer, P.C.; Carey, B.; Rajendram, P.; Hudock, K.M.; Korbee, L.; Van Tassell, B.W.; et al. Targeting GM-CSF in COVID-19 Pneumonia: Rationale and Strategies. Front. Immunol. 2020, 11, 01625. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.M.; Kim, Y.; Seo, J.W.; Lee, J.; Park, U.; Ha, N.Y.; Koh, J.; Park, H.; Lee, J.W.; Ro, H.J.; et al. Enhanced eosinophil-mediated inflammation associated with antibody and complement-dependent pneumonic insults in critical COVID-19. Cell. Rep. 2021, 1, 109798. [Google Scholar] [CrossRef]
- Peart, M.J.; Smyth, G.K.; van Laar, R.K.; Bowtell, D.D.; Richon, V.M.; Marks, P.A.; Holloway, A.J.; Johnstone, R.W. Identification and functional significance of genes regulated by structurally different histone deacetylase inhibitors. Proc. Natl Acad. Sci. USA 2005, 102, 3697–3702. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, D.J.; Smyth, G.K. Testing significance relative to a fold-change threshold is a TREAT. Bioinformatics 2009, 6, 765–771. [Google Scholar] [CrossRef]
- Cperchini, F.; Chiovato, L.; Rotondo, M. Interleukin-6, CXCL10 and Infiltrating Macrophages in COVID-19-Related Cytokine Storm: Not One for All But All for One! Front. Immunol. 2021, 12, 668507. [Google Scholar] [CrossRef]
- Noto, A.; Joo, V.; Mancarella, A.; Suffiotti, M.; Pellaton, C.; Fenwick, C.; Perreau, M.; Pantaleo, G. CXCL-12 and CXCL-13 Cytokine Serum Levels Are Associated with the Magnitude and the Quality of SARS-CoV-2 Humoral Responses. Viruses 2022, 14, 2665. [Google Scholar] [CrossRef] [PubMed]
- Kozai, M.; Kubo, Y.; Katakai, T.; Kondo, H.; Kiyonari, H.; Schaeuble, K.; Luther, S.A.; Ishimaru, N.; Ohigashi, I.; Takahama, Y. Essential role of CCL21 in establishment of central self-tolerance in T cells. J. Exp. Med. 2017, 7, 1925–1935. [Google Scholar] [CrossRef] [PubMed]
- Perreau, M.; Suffiotti, M.; Marques-Vidal, P.; Wiedemann, A.; Levy, Y.; Laouénan, C.; Ghosn, J.; Fenwick, C.; Comte, D.; Roger, T.; et al. The cytokines HGF and CXCL13 predict the severity and the mortality in COVID-19 patients. Nat. Commun. 2021, 12, 4888. [Google Scholar] [CrossRef] [PubMed]
- Celik, N.; Celik, O.; Laloglu, E.; Özkaya, A. The CXCL9/10/11-CXCR3 axis as a predictor of COVID-19 progression: A prospective case-control study. Rev. Soc. Bras. Med. Trop. 2023, 56, e0128–e2023. [Google Scholar] [CrossRef]
- Xia, H.; Cao, Z.; Xie, X.; Zhang, X.; Chen, J.Y.C.; Wang, H.; Menachery, V.D.; Rajsbaum, R.; Shi, P.Y. Evasion of Type I Interferon by SARS-CoV-2. Cell. Rep. 2020, 33, 08234. [Google Scholar] [CrossRef] [PubMed]
- Lei, X.; Dong, X.; Ma, R.; Wang, W.; Xiao, X.; Tian, Z.; Wang, C.; Wang, Y.; Li, L.; Ren, L.; et al. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 2020, 1, 3810. [Google Scholar] [CrossRef] [PubMed]
- Le Messurier, K.S.; Häcker, H.; Chi, L.; Tuomanen, E.; Redecke, V. Type I interferon protects against pneumococcal invasive disease by inhibiting bacterial transmigration across the lung. PLoS Pathog. 2013, 9, e1003727. [Google Scholar]
- Odendall, C.; Voak, A.A.; Kagan, J.C. Type III IFNs are commonly induced by bacteria-sensing TLRs and reinforce epithelial barriers during infection. J. Immunol. 2017, 199, 3270–3279. [Google Scholar] [CrossRef] [PubMed]
- Reis, G.; Moreira Silva, E.A.S.; Meideros Silva, D.C.; Thabane, L.; Campos, V.H.; Ferreira, T.S.; Santos, C.V.; Nogueira, A.M.; Almeida, A.P.; Savassi, L.C.; et al. Early treatment with pegylated interferon lambda for COVID-19. N. Engl. J. Med. 2023, 388, 518–528. [Google Scholar] [CrossRef]
- Myasnikov, A.L.; Berns, S.A.; Talyzin, P.A.; Ershov, F.I. Interferon gamma in the treatment of patients with moderate COVID-19. Vopr. Virusol. 2021, 66, 47–54. [Google Scholar] [CrossRef]
- Van Laarhoven, A.; Kurver, L.; Overheul, G.J.; Kooistra, E.J.; Abdo, W.F.; van Crevel, R.; Duivenvoorden, R.; Kox, M.; Oever, J.T.; Schouten, J.; et al. Interferon gamma immunotherapy in five critically ill COVID-19 patients with impaired cellular immunity: A case series. Med 2021, 2, 1163–1170. [Google Scholar] [CrossRef] [PubMed]
- Hilligan, K.L.; Namasivayam, S.; Clancy, C.S.; Baker, P.J.; Old, S.I.; Peluf, V.; Amaral, E.P.; Oland, S.D.; O’Mard, D.; Laux, J.; et al. Bacterial-induced or passively administered interferon gamma conditions the lung for early control of SARS-CoV-2. Nat. Commun. 2023, 14, 8829. [Google Scholar] [CrossRef] [PubMed]
- Schoggins, J.W. ISGs: What do they do? Ann. Rev. Virol. 2019, 1, 567–584. [Google Scholar] [CrossRef] [PubMed]
- Vladimer, G.I.; Gorner, M.W.; Superti-Furga, G. IFITS: Emerging roles as key antiviral proteins. Front. Immunol. 2014, 5, 94. [Google Scholar] [CrossRef] [PubMed]
- Honda, K.; Takaoka, A.; Taniguchi, T. Type I interferon gene induction by the interferon regulatory factor family of transcription factors. Immunity 2006, 25, 349–360. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Huang, G.; Wang, Z.; Wang, L.; Gao, Q. IRF7: Role and regulation in immunity and autoimmunity. Front. Immunol. 2023, 14, 1236923. [Google Scholar] [CrossRef] [PubMed]
- Honda, K.; Yanai, H.; Negishi, H.; Asagiri, M.; Sato, M.; Mizutani, T.; Shimada, N.; Ohba, Y.; Takaoka, A.; Yoshida, N.; et al. Irf-7 is the master regulator of type-I interferon-dependent immune responses. Nature 2005, 7034, 772–777. [Google Scholar] [CrossRef]
- Bohnacker, S.; Hartung, F.; Henkel, F.; Quaranta, A.; Kolmert, J.; Priller, A.; Ud-Dean, M.; Giglberger, J.; Kugler, L.M.; Pechtold, L.; et al. Mild COVID-19 imprints a long-term inflammatory eicosanoid-and chemokine memory in monocyte-derived macrophages. Mucosal Immunol. 2022, 15, 515–524. [Google Scholar] [CrossRef]
- Palmas, F.; Clarke, J.; Colas, R.A.; Gomez, E.A.; Keogh, A.; Boylan, M.; McEvoy, N.; McElvaney, O.J.; McElvaney, O.; Alalqam, R.; et al. Dysregulated plasma lipid mediator profiles in critically ill COVID-19 patients. PLoS ONE 2021, 8, e0256226. [Google Scholar] [CrossRef]
- Koenis, D.S.; Beegun, I.; Jouvene, C.C.; Aguirre, G.A.; Souza, P.R.; Gonzalez-Nunez, M.; Ly, L.; Pistorius, K.; Kocher, H.M.; Ricketts, W.; et al. Disrupted Resolution Mechanisms Favor Altered Phagocyte Response in COVID-19. Circ. Res. 2021, 129, e54–e71. [Google Scholar] [CrossRef]
- Xie, Y.; Xu, E.; Bowe, B.; Al-Aly, Z. Long-term cardiovascular outcomes of COVID-19. Nat. Med. 2022, 28, 583–590. [Google Scholar] [CrossRef] [PubMed]
- Monje, M.; Iwasaki, A. The neurobiology of long COVID. Neuron 2022, 110, 3484–3496. [Google Scholar] [CrossRef]
- Lai, Y.J.; Liu, S.H.; Manachevakul, S.; Lee, T.A.; Kuo, C.T.; Bello, D. Biomarkers in long COVID-19: A systematic review. Front. Med. 2023, 10, 1085988. [Google Scholar] [CrossRef]
- Calder, P.C. Marine omega-3 fatty acids and inflammatory processes: Effects, mechanisms and clinical relevance. Biochem. Biophys. Acta. 2015, 1851, 469–484. [Google Scholar] [CrossRef]
- Regidor, P.-A.; Mueller, A.; Sailer, M.; Gonzalez Santos, F.; Rizo, J.M.; Egea, F.M. Chronic inflammation in PCOS: The potential benefits of specialized pro-resolving lipid mediators (SPMs) in the improvement of the resolutive response. Int. J. Mol. Sci. 2020, 22, 384. [Google Scholar] [CrossRef]
- Chiang, N.; Serhan, C.N. Structural elucidation and physiologic functions of specialized pro-resolving mediators and their receptors. Mol. Aspects Med. 2017, 58, 114–129. [Google Scholar] [CrossRef]
- Morita, M.; Kuba, K.; Ichikawa, A.; Nakayama, M.; Katahira, J.; Iwamoto, R.; Watanebe, T.; Sakabe, S.; Daidoji, T.; Nakamura, S.; et al. The lipid mediator protectin D1 inhibits influenza virus replication and improves severe influenza. Cell 2013, 1, 112–125. [Google Scholar] [CrossRef] [PubMed]
- Brunvoll, S.H.; Nygaard, B.; Ellingjord-Dale, M.; Holland, P.; Istre, M.S.; Kalleberg, K.T.; Søraas, C.L.; Holven, K.B.; Ulven, S.M.; Hjartåker, A.; et al. Prevention of COVID-19 and other acute respiratory infections with cod liver oil supplementation, a low dose vitamin D supplement: Quadruple blinded, randomised placebo-controlled trial. BMJ 2022, 378, e071245. [Google Scholar] [CrossRef]
- Velotti, F.; Costantini, L.; Merendino, N. Omega-3 Polyunsaturated Fatty Acids (n-3 PUFAs) for Immunomodulation in COVID-19 Related Acute Respiratory Distress Syndrome (ARDS). J. Clin. Med. 2022, 1, 304. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.V.; Dalli, J.; Serhan, C.N. The novel lipid mediator PD1n-3 DPA: An overview of the structural elucidation, synthesis, biosynthesis and bioactions. Prostaglandins 2017, 133, 103–110. [Google Scholar] [CrossRef]
- Pistorius, K.; Souza, P.R.; De Matteis, R.; Austin-Williams, S.; Primdahl, K.G.; Vik, A.; Mazzacuva, F.; Colas, R.A.; Marques, R.M.; Hansen, T.V.; et al. PDn-3 DPA Pathway Regulates Human Monocyte Differentiation and Macrophage Function. Cell Chem. Biol. 2018, 6, 749–760 e9. [Google Scholar] [CrossRef] [PubMed]
- Droin, G.; Rioux, V.; Legrand, P. The n-3 docosapentaenoic acid (DPA): A new player in the n-3 long chain polyunsaturated fatty acid family. Biochemie 2019, 159, 36–48. [Google Scholar] [CrossRef]
- Santa-María, C.; López-Enríquez, S.; la Paz, S.M.-D.; Geniz, I.; Reyes-Quiroz, M.E.; Moreno, M.; Palomares, F.; Sobrino, F.; Alba, G. Update on Anti-Inflammatory Molecular Mechanism Induced by Oleic Acid. Nutrients 2023, 1, 224. [Google Scholar] [CrossRef]
- Venn-Watson, S.K.; Butterworth, C.N. Broader and safer clinically-relevant activities of pentadecanoic acid compared to omega-3: Evaluation of emerging essential fatty acid across twelve primary human cell-based disease systems. PLoS ONE. 2022, 5, e0268778. [Google Scholar] [CrossRef] [PubMed]
- Deo, P.; Dhillon, V.S.; Thomas, P.; Fenech, M. Oleic Acid Status Positively Correlates with the Soluble Receptor for Advanced Glycation End-Products (sRAGE) in Healthy Adults Who Are Homozygous for G Allele of RAGE G82S Polymorphism. Cells 2023, 12, 1662. [Google Scholar] [CrossRef]
- Holms, R.D. Long COVID (PASC) Is Maintained by a Self-Sustaining Pro-Inflammatory TLR4/RAGE-Loop of S100A8/A9 >TLR4/RAGE Signalling, Inducing Chronic Expression of IL-1b, IL-6 and TNFa: Anti-Inflammatory Ezrin Peptides as Potential Therapy. Immuno 2022, 2, 512–533. [Google Scholar] [CrossRef]
- Currie, C.; Framroze, B.; Singh, D.; Sharma, D.; Bjerknes, C.; Hermansen, E. Pharmacological evaluation of the effects of enzymatically liberated fish oil on eosinophilic inflammation in animal models. Biotechnol. Appl. Biochem. 2023, 1, 157–163. [Google Scholar] [CrossRef]
- Currie, C.; Framroze, B.; Singh, D.; Lea, S.; Bjerknes, C.; Hermansen, E. Assessing the Anti-Inflammatory Effects of an Orally Dosed Enzymatically Liberated Fish Oil in a House Dust Model of Allergic Asthma. Biomedicines 2022, 10, 2574. [Google Scholar] [CrossRef]
- Guo, Y.; Xiao, P.; Lei, S.; Deng, F.; Xiao, G.G.; Liu, Y.; Chen, X.; Li, L.; Wu, S.; Chen, Y. How is mRNA expression predictive for protein expression? A correlation study on human circulating monocytes. Acta Biochim. Biophys. Sin. 2008, 40, 426–436. [Google Scholar] [CrossRef] [PubMed]
- Koussounadis, A.; Langdon, S.P.; Um, I.H.; Harrison, D.J.; Smith, V.A. Relationship between differentially expressed mRNA and mRNA-protein correlations in a xenograft mouse model system. Sci. Rep. 2015, 5, 10775. [Google Scholar] [CrossRef]
- Zlei, M.; Sidorov, I.A.; Joosten, S.A.; Heemskerk, M.H.M.; Myeni, S.K.; Pothast, C.R.; de Brouwer, C.S.; der Zanden, A.L.B.-V.; van Meijgaarden, K.E.; Morales, S.T.; et al. Immune Determinants of Viral Clearance in Hospitalised COVID-19 Patients: Reduced Circulating Naïve CD4+ Cell Counts Correspond with Delayed Viral Clearance. Cells 2022, 11, 2743. [Google Scholar] [CrossRef] [PubMed]



| Age Range (Years) | Number of Subjects | Subjects on OmeGo and BSC | Subjects on BSC Only |
|---|---|---|---|
| 20–29 | 6 | 2 | 4 |
| 30–39 | 3 | 3 | 0 |
| 40–49 | 0 | 0 | 0 |
| 50–59 | 2 | 1 | 1 |
| Mean age (years) | 33.8 | 29.4 | |
| Ethnicity | |||
| Caucasian | 6 | 3 | |
| Southeast Asian | 0 | 2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Currie, C.; Myklebust, T.Å.; Bjerknes, C.; Framroze, B. Assessing the Potential of an Enzymatically Liberated Salmon Oil to Support Immune Health Recovery from Acute SARS-CoV-2 Infection via Change in the Expression of Cytokine, Chemokine and Interferon-Related Genes. Int. J. Mol. Sci. 2024, 25, 6917. https://doi.org/10.3390/ijms25136917
Currie C, Myklebust TÅ, Bjerknes C, Framroze B. Assessing the Potential of an Enzymatically Liberated Salmon Oil to Support Immune Health Recovery from Acute SARS-CoV-2 Infection via Change in the Expression of Cytokine, Chemokine and Interferon-Related Genes. International Journal of Molecular Sciences. 2024; 25(13):6917. https://doi.org/10.3390/ijms25136917
Chicago/Turabian StyleCurrie, Crawford, Tor Åge Myklebust, Christian Bjerknes, and Bomi Framroze. 2024. "Assessing the Potential of an Enzymatically Liberated Salmon Oil to Support Immune Health Recovery from Acute SARS-CoV-2 Infection via Change in the Expression of Cytokine, Chemokine and Interferon-Related Genes" International Journal of Molecular Sciences 25, no. 13: 6917. https://doi.org/10.3390/ijms25136917
APA StyleCurrie, C., Myklebust, T. Å., Bjerknes, C., & Framroze, B. (2024). Assessing the Potential of an Enzymatically Liberated Salmon Oil to Support Immune Health Recovery from Acute SARS-CoV-2 Infection via Change in the Expression of Cytokine, Chemokine and Interferon-Related Genes. International Journal of Molecular Sciences, 25(13), 6917. https://doi.org/10.3390/ijms25136917






