Cumulative Dose from Recurrent CT Scans: Exploring the DNA Damage Response in Human Non-Transformed Cells
Abstract
1. Introduction
2. Results
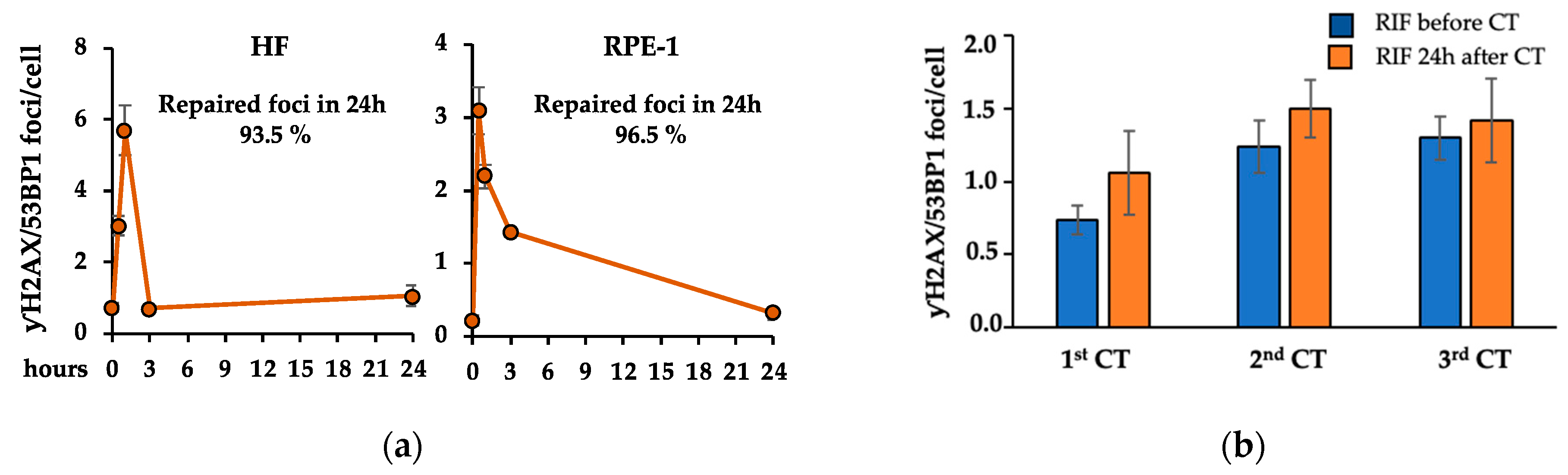
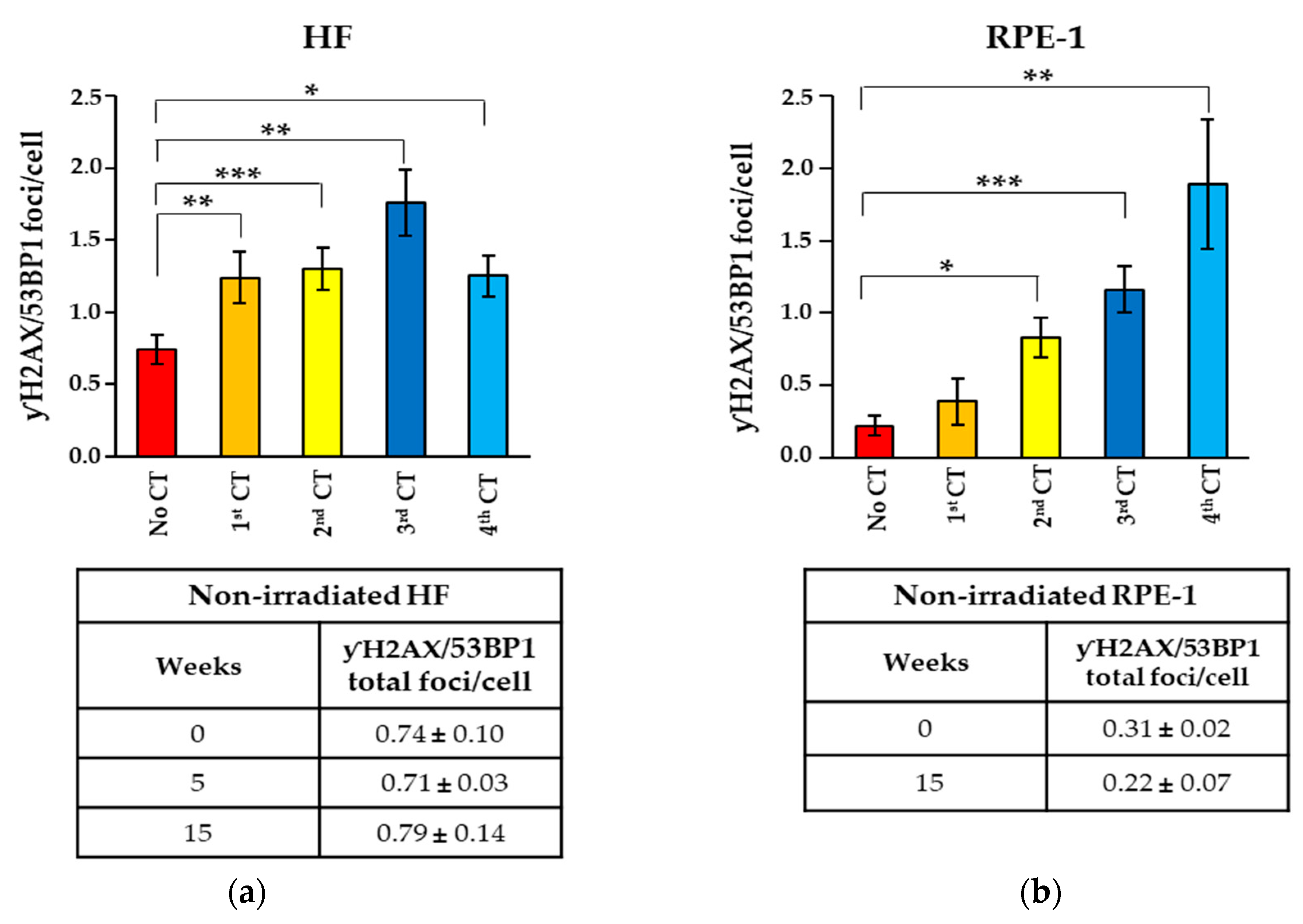
2.1. Recurrent CT Scans Increased the Number of Persistent γH2AX/53BP1 Foci
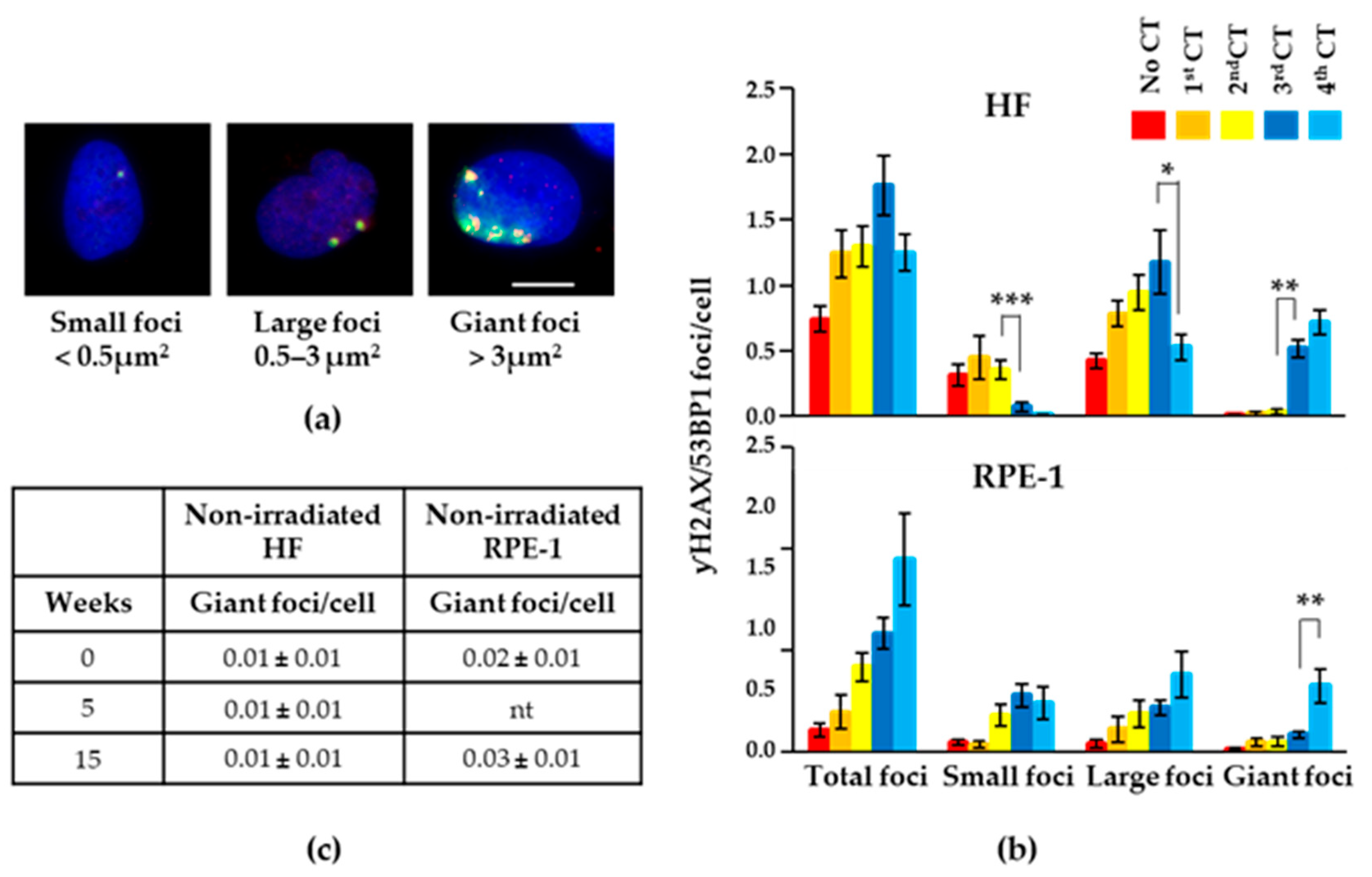
2.2. Recurrent CT Scans Increased the Size of Persistent γH2AX/53BP1 Foci
2.3. Persistent Foci Did Not Induce Cell Cycle Arrest
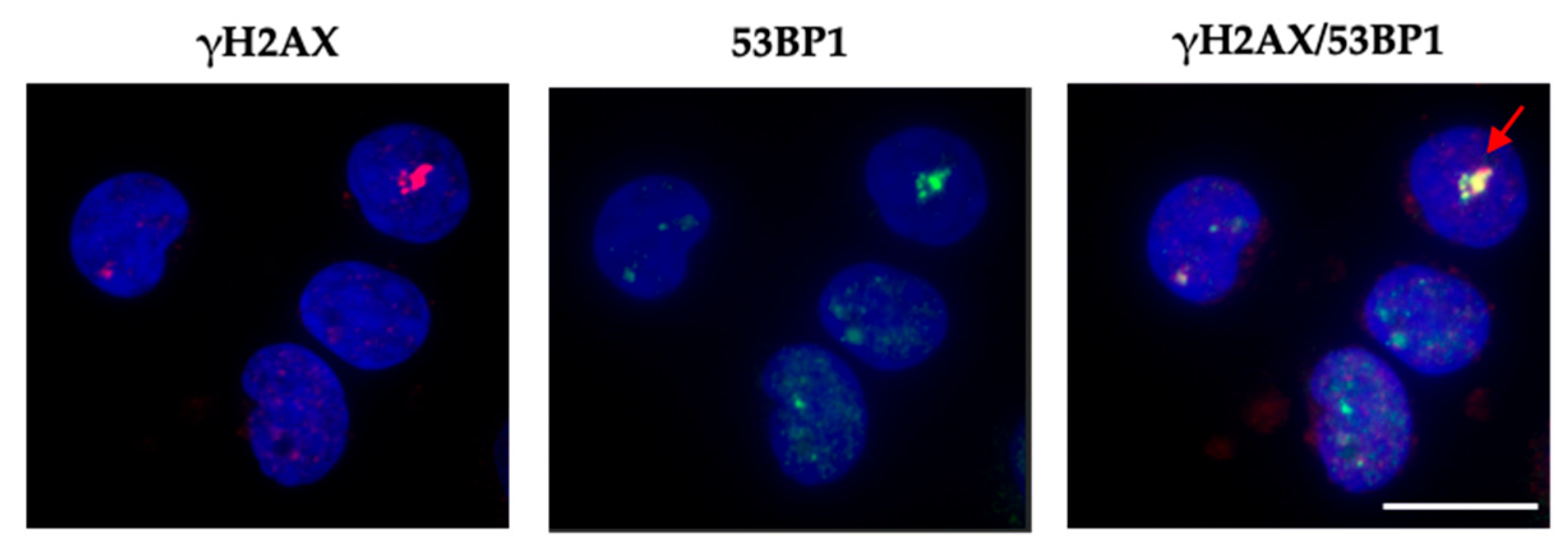
2.4. Giant Foci Were Observed In Vivo in Peripheral Blood Mononuclear Cells
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Culture
4.2. Irradiation (CT Scan)
4.3. Immunostaining/Immunofluorescence Assay
4.4. Foci Analysis
4.5. Cell Replication Assay
4.6. Peripheral Blood Mononuclear Cells (PBMCs) Isolation
4.7. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shao, Y.; Tsai, K.; Kim, S.; Wu, Y.; Demissie, K. Exposure to Tomographic Scans and Cancer Risks. JNCI Cancer Spectr. 2019, 4, pkz072. [Google Scholar] [CrossRef] [PubMed]
- Pola, A.; Corbella, D.; Righini, A.; Torresin, A.; Colombo, P.E.; Vismara, L.; Trombetta, L.; Maddalo, M.; Introini, M.V.; Tinelli, D.; et al. Computed Tomography use in a Large Italian Region: Trend Analysis 2004–2014 of Emergency and Outpatient CT Examinations in Children and Adults. Eur. Radiol. 2018, 28, 2308–2318. [Google Scholar] [CrossRef] [PubMed]
- Schauer, D.A.; Linton, O.W. NCRP Report no. 160, Ionizing Radiation Exposure of the Population of the United States, Medical Exposure—Are we Doing Less with More, and is there a Role for Health Physicists? Health Phys. 2009, 97, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Schauer, D.A.; Linton, O.W. National Council on Radiation Protection and Measurements Report shows Substantial Medical Exposure Increase. Radiology, 2009; 253, 293–296. [Google Scholar]
- Milano, M.T.; Mahesh, M.; Mettler, F.A.; Elee, J.; Vetter, R.J. Patient Radiation Exposure: Imaging during Radiation Oncology Procedures: Executive Summary of NCRP Report no. 184. J. Am. Coll. Radiol. 2020, 17, 1176–1182. [Google Scholar] [CrossRef] [PubMed]
- Pozzessere, C.; von Garnier, C.; Beigelman-Aubry, C. Radiation Exposure to Low-Dose Computed Tomography for Lung Cancer Screening: Should we be Concerned? Tomography 2023, 9, 166–177. [Google Scholar] [CrossRef] [PubMed]
- McCunney, R.J.; Li, J. Radiation Risks in Lung Cancer Screening Programs. Chest 2014, 145, 618–624. [Google Scholar] [CrossRef] [PubMed]
- Mettler, F.A., Jr.; Mahesh, M.; Bhargavan-Chatfield, M.; Chambers, C.E.; Elee, J.G.; Frush, D.P.; Miller, D.L.; Royal, H.D.; Milano, M.T.; Spelic, D.C.; et al. Patient Exposure from Radiologic and Nuclear Medicine Procedures in the United States: Procedure Volume and Effective Dose for the Period 2006–2016. Radiology 2020, 295, 418–427. [Google Scholar] [CrossRef] [PubMed]
- Frija, G.; Damilakis, J.; Paulo, G.; Loose, R.; Vano, E.; European Society of Radiology. Cumulative Effective Dose from Recurrent CT Examinations in Europe: Proposal for Clinical Guidance Based on an ESR EuroSafe Imaging Survey. Eur. Radiol. 2021, 31, 5514–5523. [Google Scholar] [CrossRef] [PubMed]
- Rehani, M.M.; Hauptmann, M. Estimates of the Number of Patients with High Cumulative Doses through Recurrent CT Exams in 35 OECD Countries. Phys. Med. 2020, 76, 173–176. [Google Scholar] [CrossRef] [PubMed]
- Brambilla, M.; Vassileva, J.; Kuchcinska, A.; Rehani, M.M. Multinational Data on Cumulative Radiation Exposure of Patients from Recurrent Radiological Procedures: Call for Action. Eur. Radiol. 2020, 30, 2493–2501. [Google Scholar] [CrossRef]
- Brambilla, M.; Cannillo, B.; D’Alessio, A.; Matheoud, R.; Agliata, M.F.; Carriero, A. Patients Undergoing Multiphase CT Scans and Receiving a Cumulative Effective Dose of ≥100 mSv in a Single Episode of Care. Eur. Radiol. 2021, 31, 4452–4458. [Google Scholar] [CrossRef] [PubMed]
- Westmark, S.; Hessellund, T.; Hoffmann, A.; Madsen, B.B.; Jensen, T.S.; Gielen, M.; Bøggild, H.; Leutscher, P.D.C. Increasing use of Computed Tomography Scans in the North Denmark Region Raises Patient Safety Concern. Eur. J. Radiol. 2023, 166, 110997. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.K.; Wang, M.; Staudt, C.; Iliakis, G. Post-Irradiation Chemical Processing of DNA Damage Generates Double-Strand Breaks in Cells Already Engaged in Repair. Nucleic Acids Res. 2011, 39, 8416–8429. [Google Scholar] [CrossRef] [PubMed]
- Jackson, S.P. Sensing and Repairing DNA Double-Strand Breaks. Carcinogenesis 2002, 23, 687–696. [Google Scholar] [CrossRef]
- Su, T.T. Cellular Responses to DNA Damage: One Signal, Multiple Choices. Anu. Rev. Genet. 2006, 40, 187–208. [Google Scholar] [CrossRef] [PubMed]
- Natale, F.; Rapp, A.; Yu, W.; Maiser, A.; Harz, H.; Scholl, A.; Grulich, S.; Anton, T.; Hörl, D.; Chen, W.; et al. Identification of the Elementary Structural Units of the DNA Damage Response. Nat. Commun. 2017, 8, 15760. [Google Scholar] [CrossRef] [PubMed]
- Willers, H.; Dahm-Daphi, J.; Powell, S.N. Repair of Radiation Damage to DNA. Br. J. Cancer 2004, 90, 1297–1301. [Google Scholar] [CrossRef] [PubMed]
- Jackson, S.P.; Bartek, J. The DNA-Damage Response in Human Biology and Disease. Nature 2009, 461, 1071–1078. [Google Scholar] [CrossRef] [PubMed]
- Ciccia, A.; Elledge, S.J. The DNA Damage Response: Making it Safe to Play with Knives. Mol. Cell 2010, 40, 179–204. [Google Scholar] [CrossRef] [PubMed]
- Polo, S.E.; Jackson, S.P. Dynamics of DNA Damage Response Proteins at DNA Breaks: A Focus on Protein Modifications. Genes Dev. 2011, 25, 409–433. [Google Scholar] [CrossRef] [PubMed]
- Ceccaldi, R.; Rondinelli, B.; D’Andrea, A.D. Repair Pathway Choices and Consequences at the Double-Strand Break. Trends Cell Biol. 2016, 26, 52–64. [Google Scholar] [CrossRef] [PubMed]
- Pinto, D.M.; Flaus, A. Structure and Function of Histone H2AX. Subcell Biochem. 2010, 50, 55–78. [Google Scholar] [PubMed]
- Modesti, M.; Kanaar, R. DNA Repair: Spot(Light)s on Chromatin. Curr. Biol. 2001, 11, 229. [Google Scholar] [CrossRef] [PubMed]
- Rogakou, E.P.; Boon, C.; Redon, C.; Bonner, W.M. Megabase Chromatin Domains Involved in DNA Double-Strand Breaks In Vivo. J. Cell Biol. 1999, 146, 905–916. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zou, L. Sensing, Signaling, and Responding to DNA Damage: Organization of the Checkpoint Pathways in Mammalian Cells. J. Cell Biochem. 2005, 94, 298–306. [Google Scholar] [CrossRef] [PubMed]
- Mirza-Aghazadeh-Attari, M.; Mohammadzadeh, A.; Yousefi, B.; Mihanfar, A.; Karimian, A.; Majidinia, M. 53BP1: A Key Player of DNA Damage Response with Critical Functions in Cancer. DNA Repair 2019, 73, 110–119. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Matsuoka, S.; Carpenter, P.B.; Elledge, S.J. 53BP1, a Mediator of the DNA Damage Checkpoint. Science 2002, 298, 1435–1438. [Google Scholar] [CrossRef] [PubMed]
- Lamarche, B.J.; Orazio, N.I.; Weitzman, M.D. The MRN Complex in Double-Strand Break Repair and Telomere Maintenance. FEBS Lett. 2010, 584, 3682–3695. [Google Scholar] [CrossRef] [PubMed]
- Shibata, A.; Jeggo, P.A. Roles for 53BP1 in the Repair of Radiation-Induced DNA Double Strand Breaks. DNA Repair 2020, 93, 102915. [Google Scholar] [CrossRef] [PubMed]
- Ström, L.; Lindroos, H.B.; Shirahige, K.; Sjögren, C. Postreplicative Recruitment of Cohesin to Double-Strand Breaks is Required for DNA Repair. Mol. Cell 2004, 16, 1003–1015. [Google Scholar] [CrossRef] [PubMed]
- Unal, E.; Arbel-Eden, A.; Sattler, U.; Shroff, R.; Lichten, M.; Haber, J.E.; Koshland, D. DNA Damage Response Pathway Uses Histone Modification to Assemble a Double-Strand Break-Specific Cohesin Domain. Mol. Cell 2004, 16, 991–1002. [Google Scholar] [CrossRef] [PubMed]
- Mognato, M.; Girardi, C.; Fabris, S.; Celotti, L. DNA Repair in Modeled Microgravity: Double Strand Break Rejoining Activity in Human Lymphocytes Irradiated with Gamma-Rays. Mutat. Res. 2009, 663, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Noda, A.; Hirai, Y.; Hamasaki, K.; Mitani, H.; Nakamura, N.; Kodama, Y. Unrepairable DNA Double-Strand Breaks that are Generated by Ionising Radiation Determine the Fate of Normal Human Cells. J. Cell Sci. 2012, 125, 5280–5287. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.M.; Ponnaiya, B.; Taveras, M.; Shuryak, I.; Turner, H.; Brenner, D.J. High Throughput Measurement of γH2AX DSB Repair Kinetics in a Healthy Human Population. PLoS ONE 2015, 10, e0121083. [Google Scholar] [CrossRef] [PubMed]
- Rossiello, F.; Herbig, U.; Longhese, M.P.; Fumagalli, M.; d’Adda di Fagagna, F. Irreparable Telomeric DNA Damage and Persistent DDR Signalling as a Shared Causative Mechanism of Cellular Senescence and Ageing. Curr. Opin. Genet. Dev. 2014, 26, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Antonelli, F.; Campa, A.; Esposito, G.; Giardullo, P.; Belli, M.; Dini, V.; Meschini, S.; Simone, G.; Sorrentino, E.; Gerardi, S.; et al. Induction and Repair of DNA DSB as Revealed by H2AX Phosphorylation Foci in Human Fibroblasts Exposed to Low- and High-LET Radiation: Relationship with Early and Delayed Reproductive Cell Death. Radiat. Res. 2015, 183, 417–431. [Google Scholar] [CrossRef] [PubMed]
- Costes, S.V.; Boissière, A.; Ravani, S.; Romano, R.; Parvin, B.; Barcellos-Hoff, M.H. Imaging Features that Discriminate between Foci Induced by High- and Low-LET Radiation in Human Fibroblasts. Radiat. Res. 2006, 165, 505–515. [Google Scholar] [CrossRef] [PubMed]
- Lee, U.; Lee, D.; Kim, E. Characterization of Γ-H2AX Foci Formation Under Alpha Particle and X-ray Exposures for Dose Estimation. Sci. Rep. 2022, 12, 3761. [Google Scholar] [CrossRef] [PubMed]
- Marnef, A.; Legube, G. Organizing DNA Repair in the Nucleus: DSBs Hit the Road. Curr. Opin. Cell Biol. 2017, 46, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Arnould, C.; Rocher, V.; Saur, F.; Bader, A.S.; Muzzopappa, F.; Collins, S.; Lesage, E.; Le Bozec, B.; Puget, N.; Clouaire, T.; et al. Chromatin Compartmentalization Regulates the Response to DNA Damage. Nature 2023, 623, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Durdik, M.; Kosik, P.; Gursky, J.; Vokalova, L.; Markova, E.; Belyaev, I. Imaging Flow Cytometry as a Sensitive Tool to Detect Low-Dose-Induced DNA Damage by Analyzing 53BP1 and γH2AX Foci in Human Lymphocytes. Cytom. A 2015, 87, 1070–1078. [Google Scholar] [CrossRef] [PubMed]
- Durdik, M.; Kosik, P.; Jakl, L.; Kozackova, M.; Markova, E.; Vigasova, K.; Beresova, K.; Jakubikova, J.; Horvathova, E.; Zastko, L.; et al. Imaging Flow Cytometry and Fluorescence Microscopy in Assessing Radiation Response in Lymphocytes from Umbilical Cord Blood and Cancer Patients. Cytom. A 2021, 99, 1198–1208. [Google Scholar] [CrossRef] [PubMed]
- Sedelnikova, O.A.; Horikawa, I.; Zimonjic, D.B.; Popescu, N.C.; Bonner, W.M.; Barrett, J.C. Senescing Human Cells and Ageing Mice Accumulate DNA Lesions with Unrepairable Double-Strand Breaks. Nat. Cell Biol. 2004, 6, 168–170. [Google Scholar] [CrossRef]
- Vaurijoux, A.; Voisin, P.; Freneau, A.; Barquinero, J.F.; Gruel, G. Transmission of Persistent Ionizing Radiation-Induced Foci through Cell Division in Human Primary Cells. Mutat. Res. 2017, 797–799, 15–25. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bogdanova, N.V.; Jguburia, N.; Ramachandran, D.; Nischik, N.; Stemwedel, K.; Stamm, G.; Werncke, T.; Wacker, F.; Dörk, T.; Christiansen, H. Persistent DNA Double-Strand Breaks After Repeated Diagnostic CT Scans in Breast Epithelial Cells and Lymphocytes. Front. Oncol. 2021, 11, 634389. [Google Scholar] [CrossRef]
- Rothkamm, K.; Barnard, S.; Moquet, J.; Ellender, M.; Rana, Z.; Burdak-Rothkamm, S. DNA Damage Foci: Meaning and Significance. Environ. Mol. Mutagen. 2015, 56, 491–504. [Google Scholar] [CrossRef] [PubMed]
- Hauptmann, M.; Byrnes, G.; Cardis, E.; Bernier, M.O.; Blettner, M.; Dabin, J.; Engels, H.; Istad, T.S.; Johansen, C.; Kaijser, M.; et al. Brain cancer after radiation exposure from CT examinations of children and young adults: Results from the EPI-CT cohort study. Lancet Oncol. 2023, 24, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Preston, D.L.; Ron, E.; Tokuoka, S.; Funamoto, S.; Nishi, N.; Soda, M.; Mabuchi, K.; Kodama, K. Solid Cancer Incidence in Atomic Bomb Survivors: 1958–1998. Radiat. Res. 2007, 168, 1–64. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Tashiro, S. Estimation of the Effects of Medical Diagnostic Radiation Exposure Based on DNA Damage. J. Radiat. Res. 2018, 59, ii121–ii129. [Google Scholar] [CrossRef] [PubMed]
- Pearce, M.S.; Salotti, J.A.; Little, M.P.; McHugh, K.; Lee, C.; Kim, K.P.; Howe, N.L.; Ronckers, C.M.; Rajaraman, P.; Sir Craft, A.W.; et al. Radiation Exposure from CT Scans in Childhood and Subsequent Risk of Leukaemia and Brain Tumours: A Retrospective Cohort Study. Lancet 2012, 380, 499–505. [Google Scholar] [CrossRef] [PubMed]
- National Research Council. Health Risks from Exposure to Low Levels of Ionizing Radiation: BEIR VII Phase 2; The National Academies Press: Washington, DC, USA, 2006. [Google Scholar]
- Lau, Y.S.; Chew, M.T.; Alqahtani, A.; Jones, B.; Hill, M.A.; Nisbet, A.; Bradley, D.A. Low Dose Ionising Radiation-Induced Hormesis: Therapeutic Implications to Human Health. Appl. Sci. 2021, 11, 8909. [Google Scholar] [CrossRef]
- Tang, H.; Cai, L.; He, X.; Niu, Z.; Huang, H.; Hu, W.; Bian, H.; Huang, H. Radiation-induced bystander effect and its clinical implications. Front. Oncol. 2023, 13, 1124412. [Google Scholar] [CrossRef]
- Kamiya, K.; Ozasa, K.; Akiba, S.; Niwa, O.; Kodama, K.; Takamura, N.; Zaharieva, E.K.; Kimura, Y.; Wakeford, R. Long-Term Effects of Radiation Exposure on Health. Lancet 2015, 386, 469–478. [Google Scholar] [CrossRef] [PubMed]
- McLean, A.R.; Adlen, E.K.; Cardis, E.; Elliott, A.; Goodhead, D.T.; Harms-Ringdahl, M.; Hendry, J.H.; Hoskin, P.; Jeggo, P.A.; Mackay, D.J.C.; et al. A Restatement of the Natural Science Evidence Base Concerning the Health Effects of Low-Level Ionizing Radiation. Proc. Biol. Sci. 2017, 284, 20171070. [Google Scholar] [CrossRef] [PubMed]
- Sampadi, B.; Vermeulen, S.; Mišovic, B.; Boei, J.J.; Batth, T.S.; Chang, J.; Paulsen, M.T.; Magnuson, B.; Schimmel, J.; Kool, H.; et al. Divergent Molecular and Cellular Responses to Low and High-Dose Ionizing Radiation. Cells 2022, 11, 3794. [Google Scholar] [CrossRef] [PubMed]
- Cao, C.; Ma, K.; Shan, H.; Liu, T.; Zhao, S.; Wan, Y.; Zhang, J.; Wang, H. CT Scans and Cancer Risks: A Systematic Review and Dose-Response Meta-Analysis. BMC Cancer 2022, 22, 1238. [Google Scholar] [CrossRef] [PubMed]
- Berrington de Gonzalez, A.; Pasqual, E.; Veiga, L. Epidemiological Studies of CT Scans and Cancer Risk: The State of the Science. Br. J. Radiol. 2021, 94, 20210471. [Google Scholar] [CrossRef] [PubMed]
- Ansari, A.; Kleinhans, K.; Boice, J.D. Potential Health Effects of Low Dose Radiation and what it Means to the Practice of Radiation Protection. J. Radiol. Prot. 2019, 39, E9–E13. [Google Scholar] [CrossRef] [PubMed]
- Prasad, A.; Visweswaran, S.; Kanagaraj, K.; Raavi, V.; Arunan, M.; Venkatachalapathy, E.; Paneerselvam, S.; Jose, M.T.; Ozhimuthu, A.; Perumal, V. 18F-FDG PET/CT Scanning: Biological Effects on Patients: Entrance Surface Dose, DNA Damage, and Chromosome Aberrations in Lymphocytes. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2019, 838, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Visweswaran, S.; Joseph, S.; Hegde, V.; Annalakshmi, O.; Jose, M.T.; Perumal, V. DNA Damage and Gene Expression Changes in Patients Exposed to Low-Dose X-Radiation during Neuro-Interventional Radiology Procedures. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2019, 844, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Bracalente, C.; Ibañez, I.L.; Molinari, B.; Palmieri, M.; Kreiner, A.; Valda, A.; Davidson, J.; Durán, H. Induction and Persistence of Large γH2AX Foci by High Linear Energy Transfer Radiation in DNA-Dependent Protein Kinase-Deficient Cells. Int. J. Radiat. Oncol. Biol. Phys. 2013, 87, 785–794. [Google Scholar] [CrossRef] [PubMed]
- Nickoloff, J.A.; Sharma, N.; Taylor, L. Clustered DNA Double-Strand Breaks: Biological Effects and Relevance to Cancer Radiotherapy. Genes 2020, 11, 99. [Google Scholar] [CrossRef] [PubMed]
- Ponomarev, A.L.; Costes, S.V.; Cucinotta, F.A. Stochastic Properties of Radiation-Induced DSB: DSB Distributions in Large Scale Chromatin Loops, the HPRT Gene and within the Visible Volumes of DNA Repair Foci. Int. J. Radiat. Biol. 2008, 84, 916–929. [Google Scholar] [CrossRef] [PubMed]
- Georgiades, E.; Crosetto, N.; Bienko, M. Compartmentalizing Damaged DNA: A Double-Edged Sword. Mol. Cell 2024, 84, 12–13. [Google Scholar] [CrossRef]
- Siepi, F.; Gatti, V.; Camerini, S.; Crescenzi, M.; Soddu, S. HIPK2 Catalytic Activity and Subcellular Localization are Regulated by Activation-Loop Y354 Autophosphorylation. Biochim. Biophys. Acta 2013, 1833, 1443–1453. [Google Scholar] [CrossRef] [PubMed]
- Stirling, D.R.; Swain-Bowden, M.J.; Lucas, A.M.; Carpenter, A.E.; Cimini, B.A.; Goodman, A. CellProfiler 4: Improvements in speed, utility and usability. BMC Bioinform. 2021, 22, 433. [Google Scholar] [CrossRef]
- Hanton, F.; Chaudhary, P.; Doria, D.; Gwynne, D.; Maiorino, C.; Scullion, C.; Ahmed, H.; Marshall, T.; Naughton, K.; Romagnani, L.; et al. DNA DSB Repair Dynamics Following Irradiation with Laser-Driven Protons at Ultra-High Dose Rates. Sci. Rep. 2019, 9, 4471–4476. [Google Scholar] [CrossRef] [PubMed]
- GraphPad Prism, version 9.0 for Windows; GraphPad Software: Boston, MA, USA.


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valente, D.; Gentileschi, M.P.; Valenti, A.; Burgio, M.; Soddu, S.; Bruzzaniti, V.; Guerrisi, A.; Verdina, A. Cumulative Dose from Recurrent CT Scans: Exploring the DNA Damage Response in Human Non-Transformed Cells. Int. J. Mol. Sci. 2024, 25, 7064. https://doi.org/10.3390/ijms25137064
Valente D, Gentileschi MP, Valenti A, Burgio M, Soddu S, Bruzzaniti V, Guerrisi A, Verdina A. Cumulative Dose from Recurrent CT Scans: Exploring the DNA Damage Response in Human Non-Transformed Cells. International Journal of Molecular Sciences. 2024; 25(13):7064. https://doi.org/10.3390/ijms25137064
Chicago/Turabian StyleValente, Davide, Maria Pia Gentileschi, Alessandro Valenti, Massimo Burgio, Silvia Soddu, Vicente Bruzzaniti, Antonino Guerrisi, and Alessandra Verdina. 2024. "Cumulative Dose from Recurrent CT Scans: Exploring the DNA Damage Response in Human Non-Transformed Cells" International Journal of Molecular Sciences 25, no. 13: 7064. https://doi.org/10.3390/ijms25137064
APA StyleValente, D., Gentileschi, M. P., Valenti, A., Burgio, M., Soddu, S., Bruzzaniti, V., Guerrisi, A., & Verdina, A. (2024). Cumulative Dose from Recurrent CT Scans: Exploring the DNA Damage Response in Human Non-Transformed Cells. International Journal of Molecular Sciences, 25(13), 7064. https://doi.org/10.3390/ijms25137064






