Prokaryotic Expression and Functional Verification of Antimicrobial Peptide LRGG
Abstract
1. Introduction
2. Results
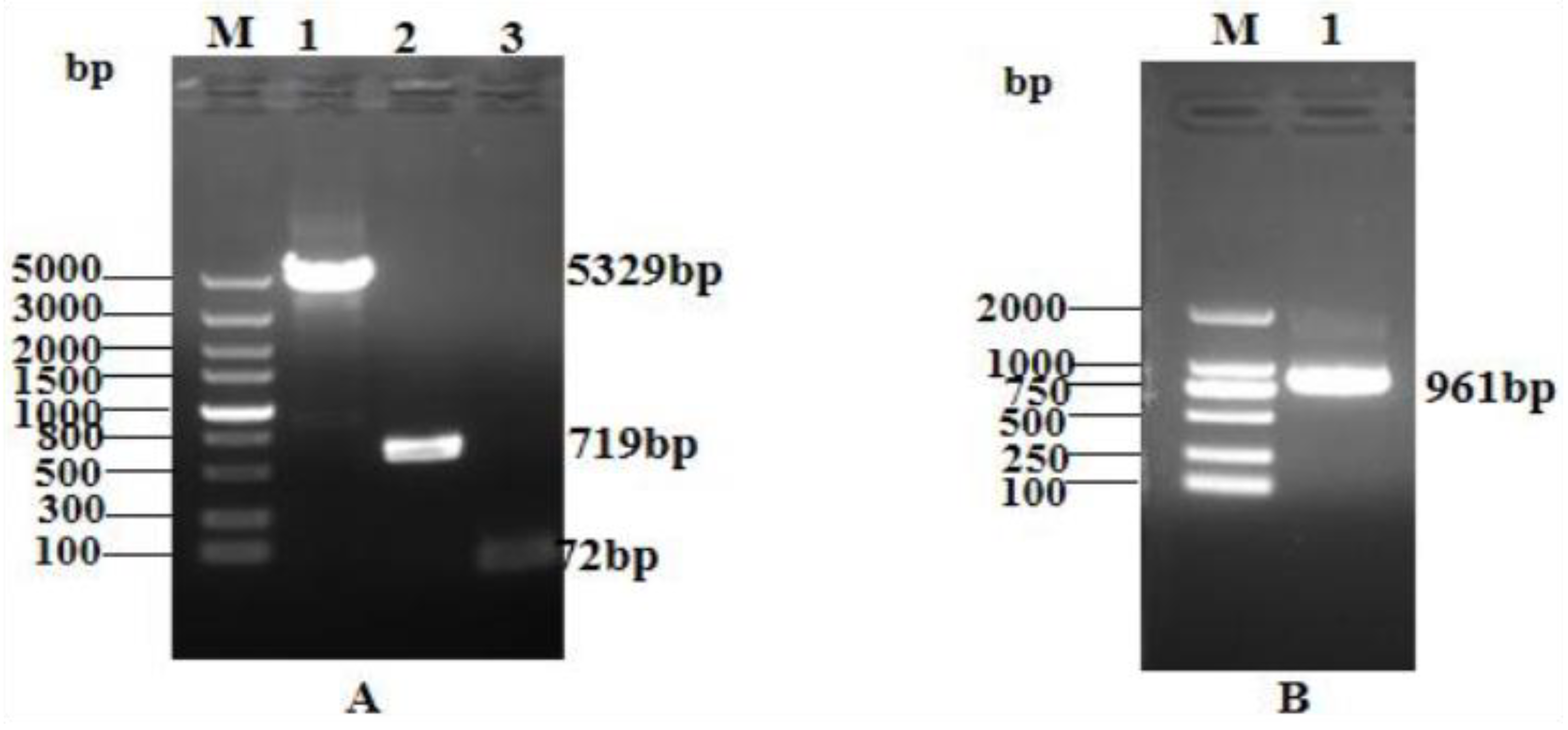
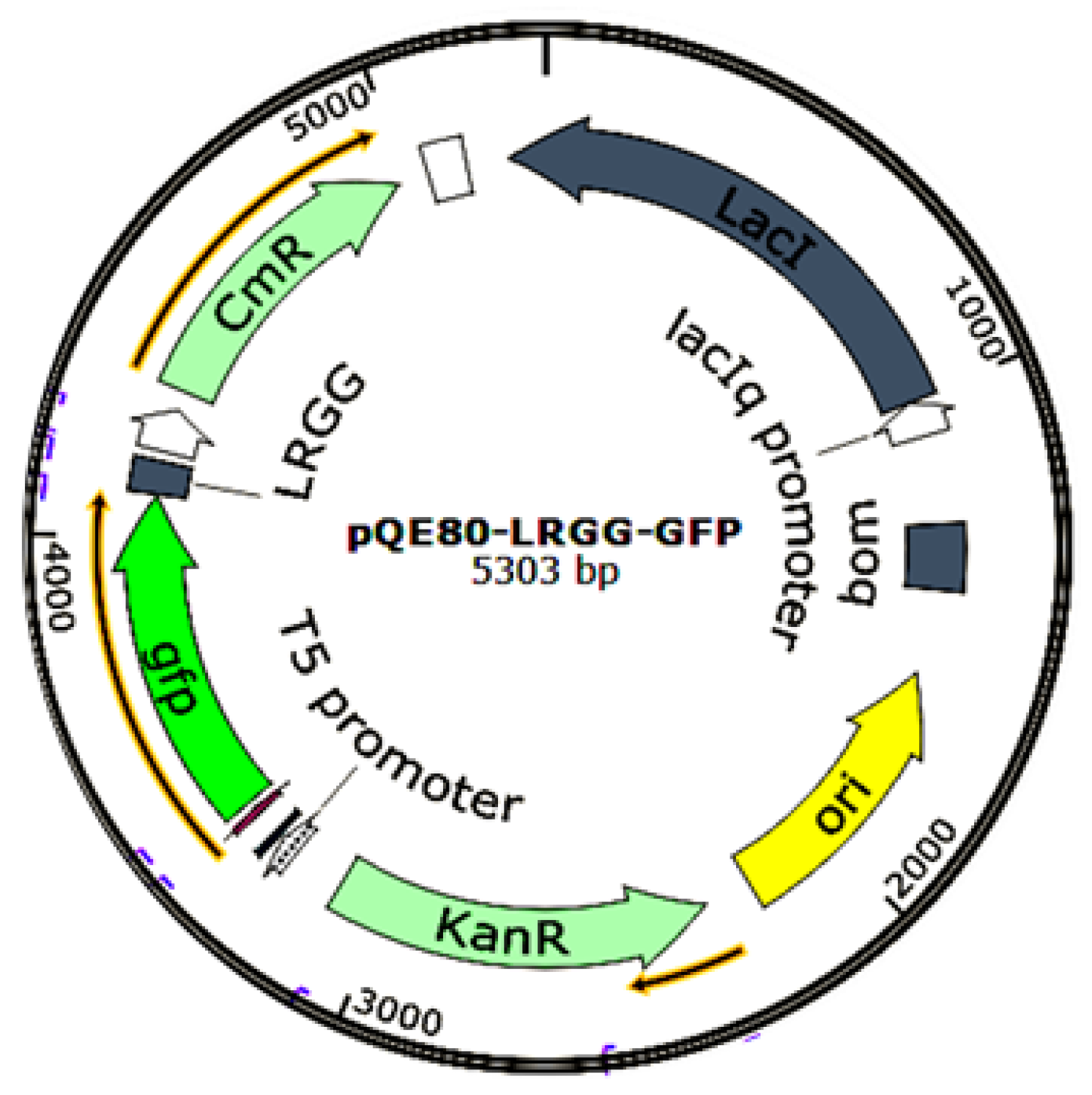
2.1. Construction of the pQE-GFP-LRGG Expression Vector
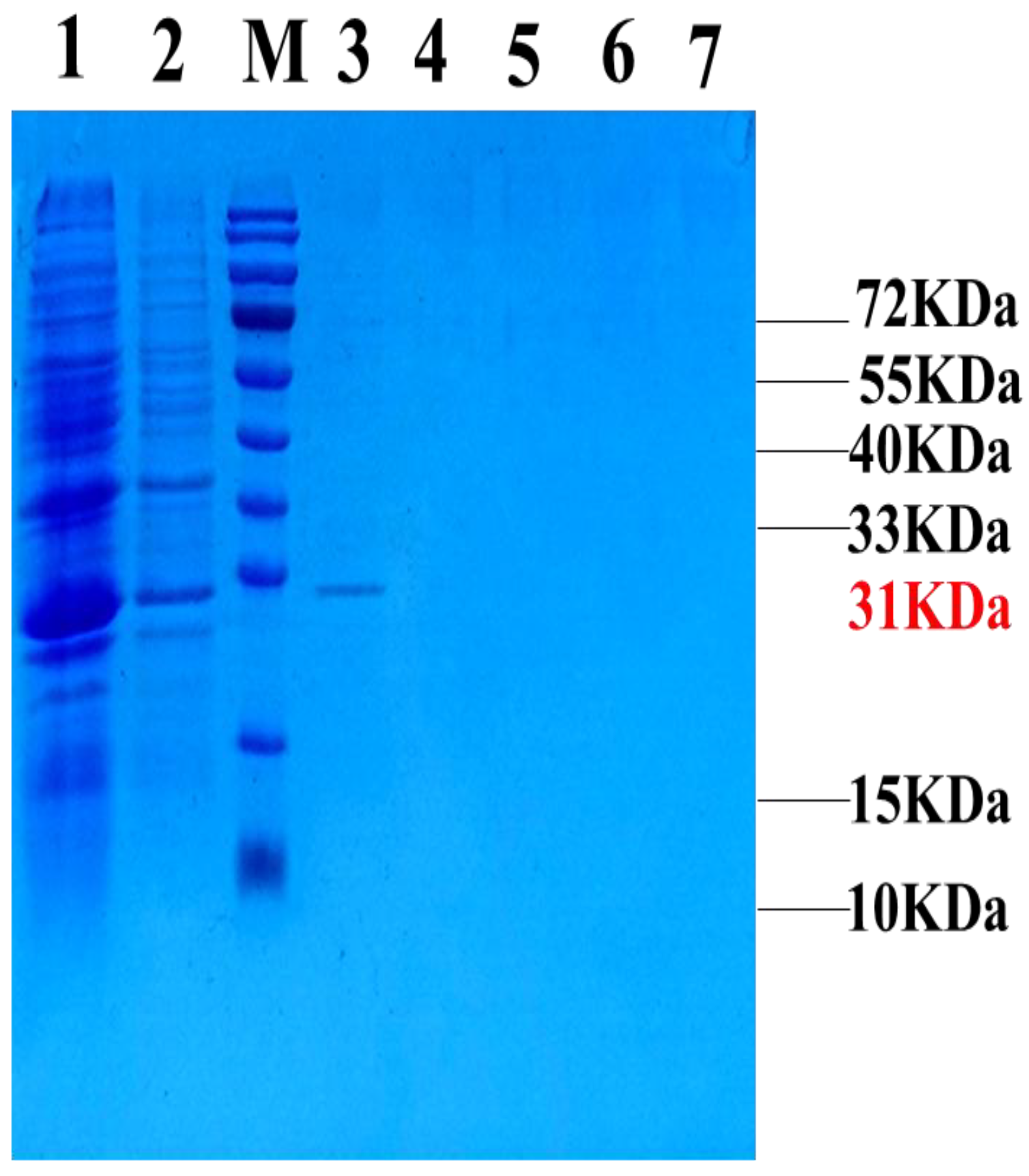
2.2. Expression and Purification of the Fusion Protein GFP-LRGG
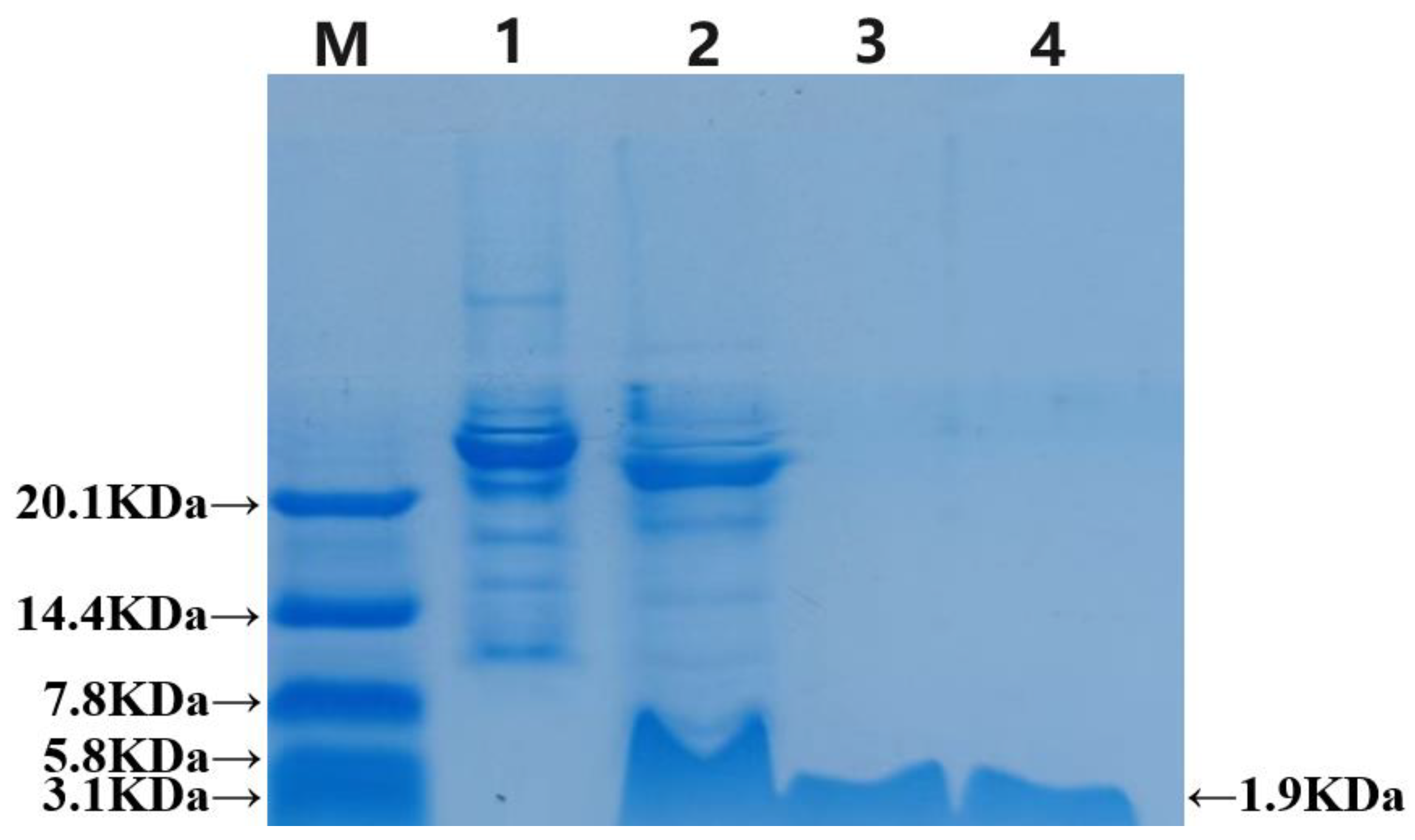
2.3. Cutting of the Fusion Protein and Purification of the Antibacterial Peptide LRGG
2.4. Determination of the Bacteriostatic Activity and Kinetics Curve of Fusion-Expressed Antimicrobial Peptide LRGG
2.5. Environmental Sensitivity of Fusion Expressed Peptide LRGG
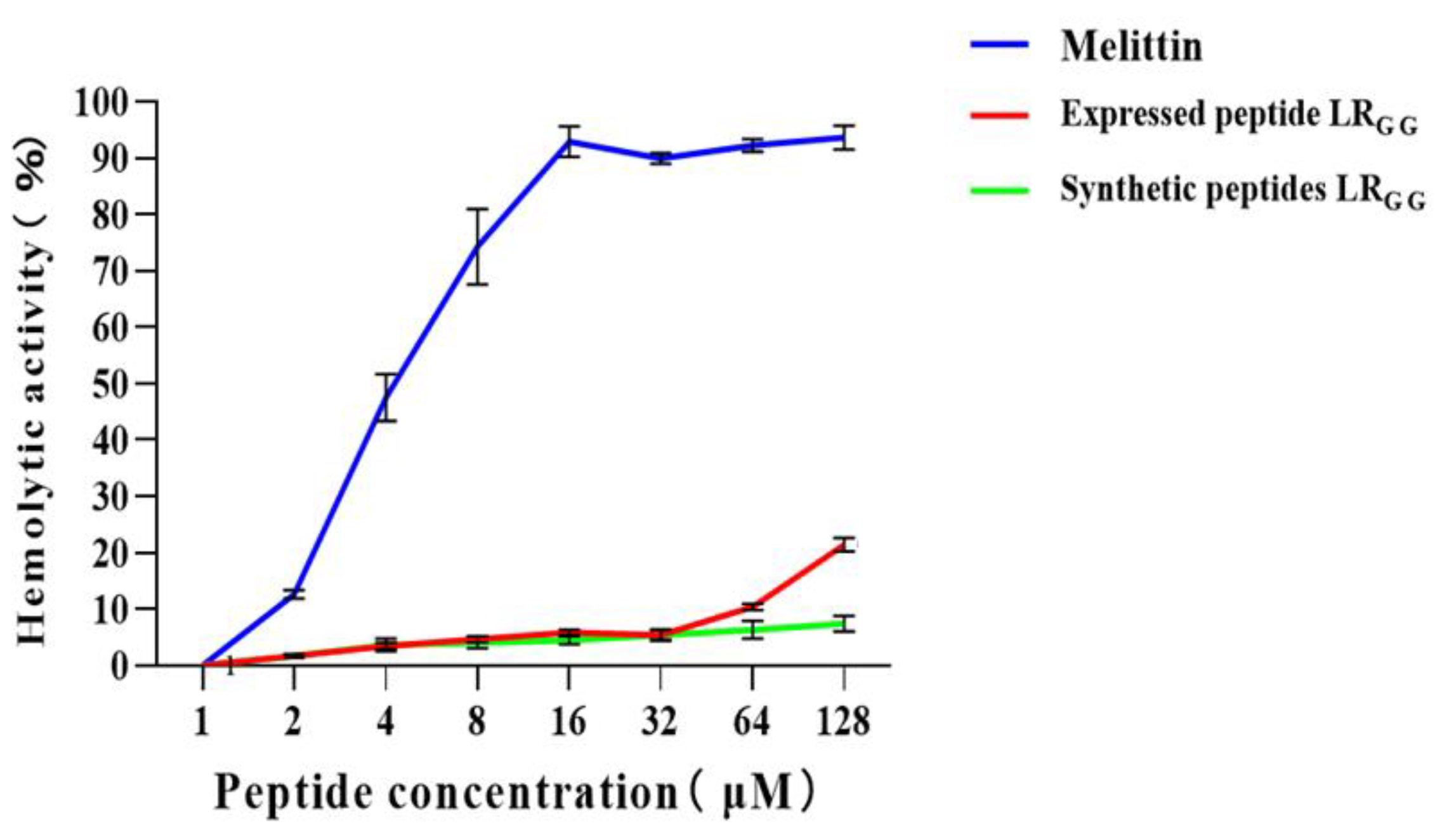
2.6. Cytotoxicity and Hemolytic Activity of Antimicrobial Peptide LRGG
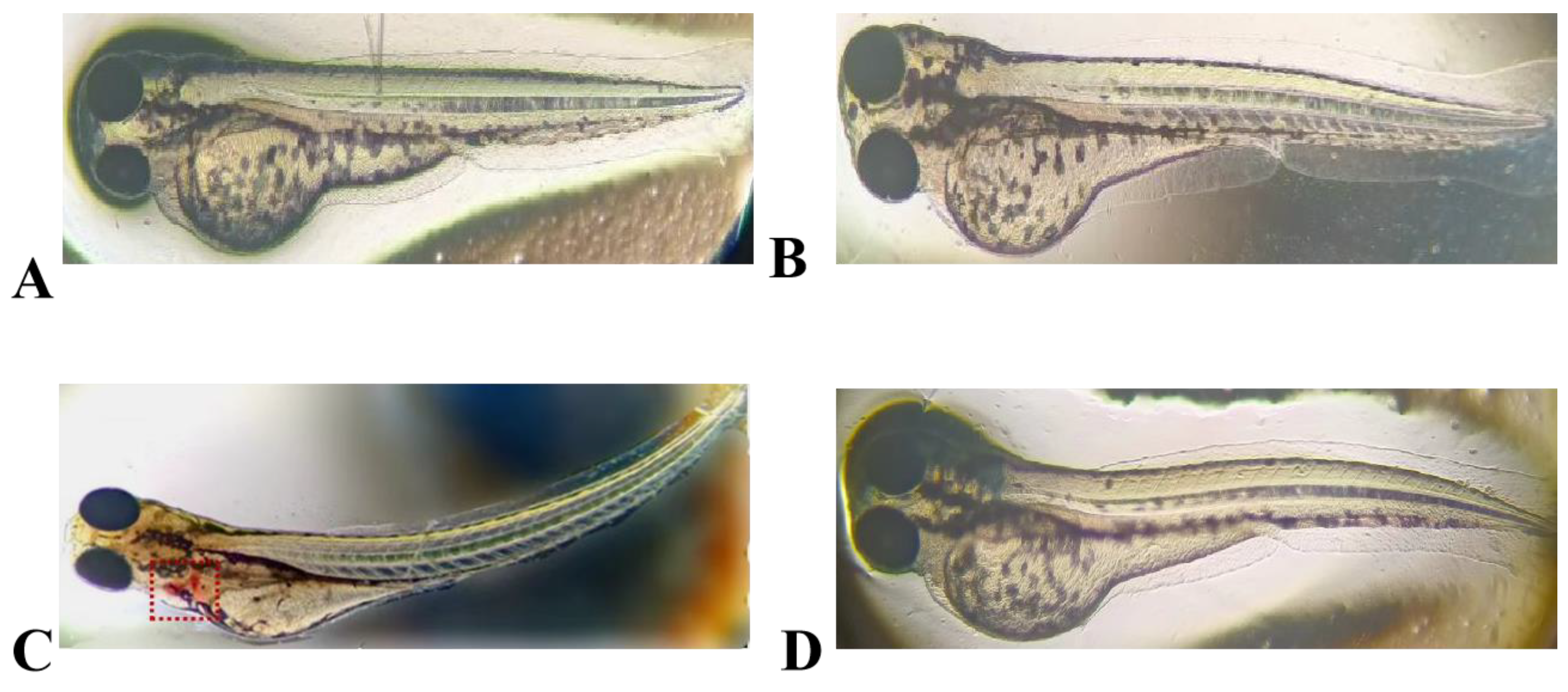
2.7. Embryotoxicity of the Antimicrobial Peptide LRGG in Zebrafish
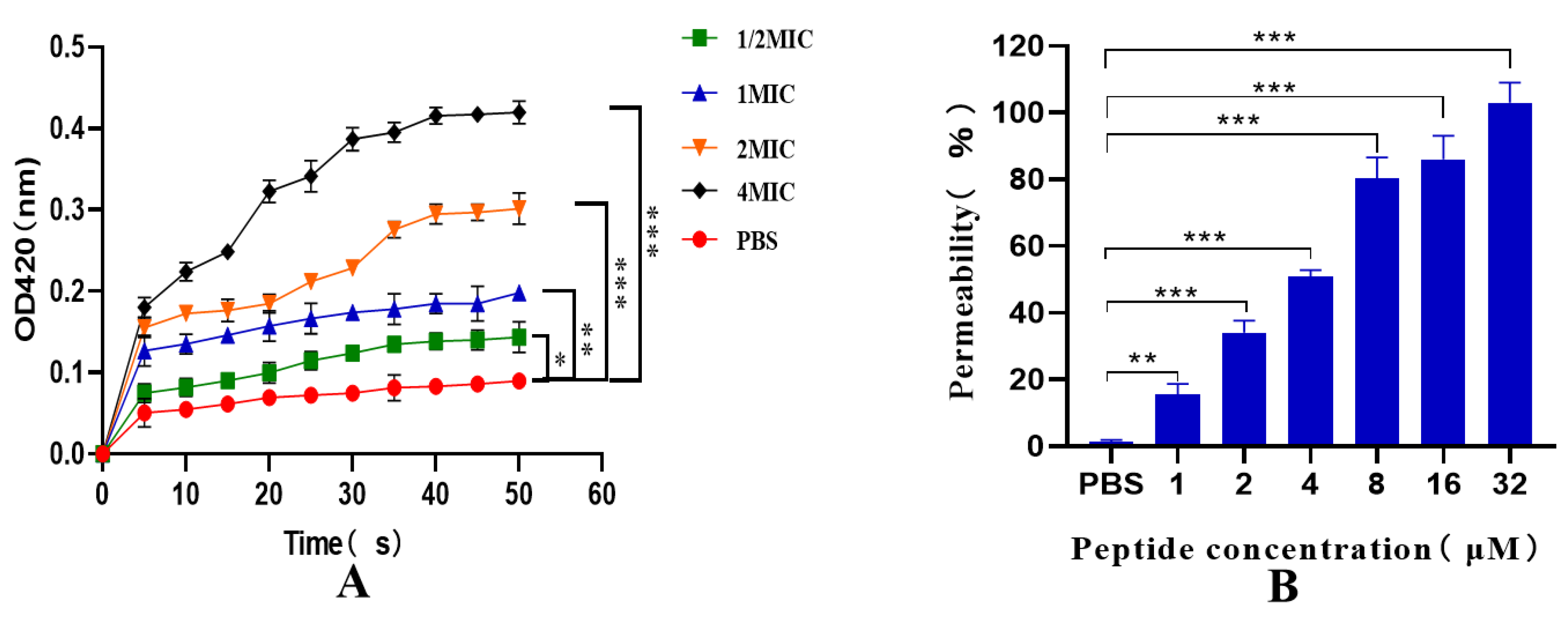
2.8. Inner and Outer Membrane Permeability Tests
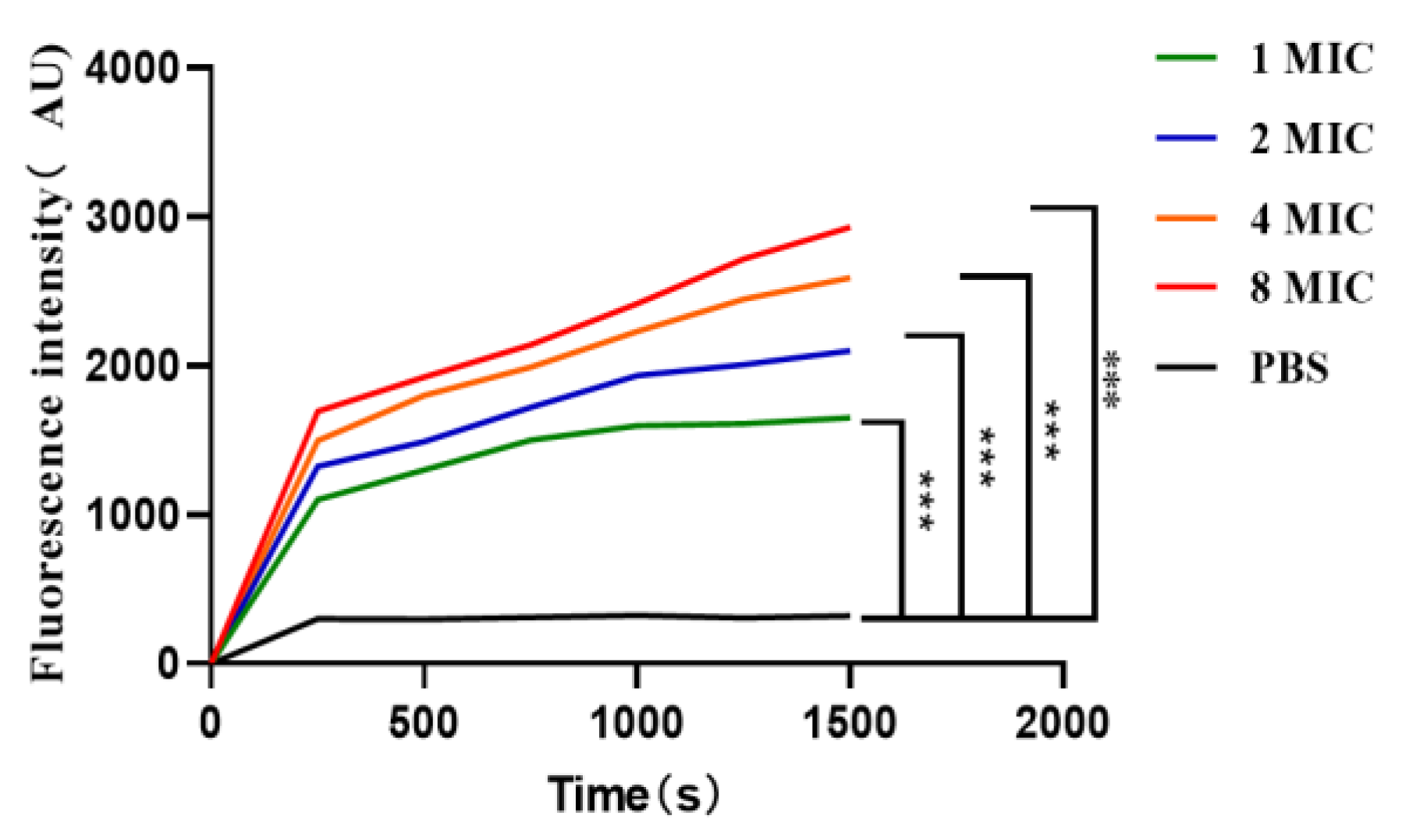
2.9. Effect of the Antimicrobial Peptide LRGG on the Bacterial Plasma Membrane Potential
2.10. DNA Gel Retardation Assay
3. Discussion
4. Materials and Methods
4.1. Strains and Plasmids
4.2. Acquisition of Target Genes
- (1)
- (2)
- To target the gene sequence of the antimicrobial peptide LRGG, the primers LRGG-F1 and LRGG-R1 were designed. The seamless cloning method were followed to construct the plasmids, as shown in Figure 10.
4.3. Construction of pQE-GFP-LRGG Expression Vector
- (1)
- The gfp gene was amplified by PCR reaction using the primers GFP-F and GFP-R, and the pTZ18U-GFP plasmid served as the template.
- (2)
- To synthesize the Tev site and LRGG sequence, two oligonucleotides (TEV-LRGG-F2 and TEV-LRGG-R2) were mixed at a 1:1 ratio in a PCR reaction buffer. The mixture was denatured at 95 °C and annealed by reducing the temperature by one degree per minute from 95 °C to 25 °C within 70 min. This process yielded a double-stranded LRGG DNA sequence containing the TEV target site.
- (3)
- The linearized pQE80 vector was amplified by PCR using the pQE-VT-F and pQE-VT-R primers and the pQE80-KAN plasmid as a template.
4.4. Expression and Purification of Fusion Protein GFP-LRGG
4.5. Cleavage of the Fusion Protein and Purification of the Antimicrobial Peptide LRGG
4.6. Determination of the Antibacterial Activity of Fusion Expressed Antimicrobial Peptide LRGG
4.7. Determination of the Bactericidal Kinetic Curve of Fusion Expressed Antimicrobial Peptide LRGG
4.8. Environmental Sensitivity of Fusion Expressed Antimicrobial Peptide LRGG
4.9. Cytotoxicity Assay of the Antimicrobial Peptide LRGG
4.10. Hemolytic Activity Assay of the Antimicrobial Peptide LRGG
4.11. Embryotoxicity of the Antimicrobial Peptide LRGG in Zebrafish
4.12. Inner and Outer Membrane Permeability Tests
4.13. Effect of the Antimicrobial Peptide LRGG on the Bacterial Plasma Membrane Potential
4.14. DNA Gel Retardation Assay
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bruce, J.; Oyedemi, B.; Parsons, N.; Harrison, F. Phase 1 safety trial of a natural product cocktail with antibacterial activity in human volunteers. Sci. Rep. 2022, 12, 19656. [Google Scholar] [CrossRef] [PubMed]
- Tsang, J. Bacterial plasmid addiction systems and their implications for antibiotic drug development. Postdoc J. 2017, 5, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Wang, W.; Xu, W. Effects of tetracycline antibiotics in chicken manure on soil microbes and antibiotic resistance genes (ARGs). Environ. Geochem. Health 2021, 44, 273–284. [Google Scholar] [CrossRef] [PubMed]
- Wiman, E.; Zattarin, E.; Aili, D.; Bengtsson, T.; Selegård, R.; Khalaf, H. Development of novel broad-spectrum antimicrobial lipopeptides derived from plantaricin NC8 β. Sci. Rep. 2023, 13, 4104. [Google Scholar] [CrossRef] [PubMed]
- Topalova, Y.; Belouhova, M.; Velkova, L.; Dolashki, A.; Zheleva, N.; Daskalova, E.; Kaynarov, D.; Voelter, W.; Dolashka, P. Effect and Mechanisms of Antibacterial Peptide Fraction from Mucus of C. aspersum against Escherichia coli NBIMCC 8785. Biomedicines 2022, 10, 672. [Google Scholar] [CrossRef] [PubMed]
- Tallet, L.; Frisch, E.; Bornerie, M.; Medemblik, C.; Frisch, B.; Lavalle, P.; Guichard, G.; Douat, C.; Kichler, A. Design of Oligourea-Based Foldamers with Antibacterial and Antifungal Activities. Molecules 2022, 27, 1749. [Google Scholar] [CrossRef] [PubMed]
- Mardirossian, M.; Rubini, M.; Adamo, M.F.A.; Scocchi, M.; Saviano, M.; Tossi, A.; Gennaro, R.; Caporale, A. Natural and Synthetic Halogenated Amino Acids—Structural and Bioactive Features in Antimicrobial Peptides and Peptidomimetics. Molecules 2021, 26, 7401. [Google Scholar] [CrossRef] [PubMed]
- Hanson, M.A.; Lemaitre, B.; Unckless, R.L. Dynamic Evolution of Antimicrobial Peptides Underscores Trade-Offs Between Immunity and Ecological Fitness. Front. Immunol. 2019, 10, 2620. [Google Scholar] [CrossRef]
- Lin, Y.; Jiang, Y.; Zhao, Z.; Lu, Y.; Xi, X.; Ma, C.; Chen, X.; Zhou, M.; Chen, T.; Shaw, C.; et al. Discovery of a Novel Antimicrobial Peptide, Temporin-PKE, from the Skin Secretion of Pelophylax kl. esculentus, and Evaluation of Its Structure-Activity Relationships. Biomolecules 2022, 12, 759. [Google Scholar] [CrossRef]
- Tomb, R.M.; Maclean, M.; Coia, J.E.; MacGregor, S.J.; Anderson, J.G. Assessment of the potential for resistance to antimicrobial violet-blue light in Staphylococcus aureus. Antimicrob. Resist. Infect. Control. 2017, 6, 100. [Google Scholar] [CrossRef]
- Czechowicz, P.; Neubauer, D.; Nowicka, J.; Kamysz, W.; Gościniak, G. Antifungal Activity of Linear and Disulfide-Cyclized Ultrashort Cationic Lipopeptides Alone and in Combination with Fluconazole against Vulvovaginal Candida spp. Pharmaceutics 2021, 13, 1589. [Google Scholar] [CrossRef]
- Popa, C.; Shi, X.; Ruiz, T.; Ferrer, P.; Coca, M. Biotechnological Production of the Cell Penetrating Antifungal PAF102 Peptide in Pichia pastoris. Front. Microbiol. 2019, 10, 1472. [Google Scholar] [CrossRef] [PubMed]
- Vinutha, A.S.; Rajasekaran, R. Insight on the mechanism of hexameric Pseudin-4 against bacterial membrane-mimetic environment. J. Comput. Mol. Des. 2023, 37, 419–434. [Google Scholar] [CrossRef]
- Won, T.; Mohid, S.A.; Choi, J.; Kim, M.; Krishnamoorthy, J.; Biswas, I.; Bhunia, A.; Lee, D. The role of hydrophobic patches of de novo designed MSI-78 and VG16KRKP antimicrobial peptides on fragmenting model bilayer membranes. Biophys. Chem. 2023, 296, 106981. [Google Scholar] [CrossRef]
- Kumar, P.; Kizhakkedathu, J.N.; Straus, S.K. Antimicrobial Peptides: Diversity, Mechanism of Action and Strategies to Improve the Activity and Biocompatibility In Vivo. Biomolecules 2018, 8, 4. [Google Scholar] [CrossRef] [PubMed]
- Mandel, S.; Michaeli, J.; Nur, N.; Erbetti, I.; Zazoun, J.; Ferrari, L.; Felici, A.; Cohen-Kutner, M.; Bachnoff, N. OMN6 a novel bioengineered peptide for the treatment of multidrug resistant Gram negative bacteria. Sci. Rep. 2021, 11, 6603. [Google Scholar] [CrossRef] [PubMed]
- Mumtaz, S.; Behera, S.; Mukhopadhyay, K. Lipidated Short Analogue of α-Melanocyte Stimulating Hormone Exerts Bactericidal Activity against the Stationary Phase of Methicillin-Resistant Staphylococcus aureus and Inhibits Biofilm Formation. ACS Omega 2020, 5, 28425–28440. [Google Scholar] [CrossRef]
- Panevska, A.; Hodnik, V.; Skočaj, M.; Novak, M.; Modic, Š.; Pavlic, I.; Podržaj, S.; Zarić, M.; Resnik, N.; Maček, P.; et al. Pore-forming protein complexes from Pleurotus mushrooms kill western corn rootworm and Colorado potato beetle through targeting membrane ceramide phosphoethanolamine. Sci. Rep. 2019, 9, 5073. [Google Scholar] [CrossRef]
- Panevska, A.; Čegovnik, N.; Fortuna, K.; Vukovič, A.; Grundner, M.; Modic, Š.; Bajc, G.; Skočaj, M.; Bohte, M.M.; Popošek, L.L.; et al. A single point mutation expands the applicability of ostreolysin A6 in biomedicine. Sci. Rep. 2023, 13, 2149. [Google Scholar] [CrossRef]
- Rončević, T.; Puizina, J.; Tossi, A. Antimicrobial Peptides as Anti-Infective Agents in Pre-Post-Antibiotic Era. Int. J. Mol. Sci. 2019, 20, 5713. [Google Scholar] [CrossRef]
- Rima, M.; Rima, M.; Fajloun, Z.; Sabatier, J.M.; Bechinger, B.; Naas, T. Antimicrobial Peptides: A Potent Alternative to Antibiotics. Antibiotics 2021, 10, 1095. [Google Scholar] [CrossRef]
- Lei, J.; Sun, L.C.; Huang, S.; Zhu, C.; Li, P.; He, J.; Mackey, V.; Coy, D.H.; He, Q.Y. The antimicrobial peptides and their potentialclinical applications. Am. J. Transl. Res. 2019, 11, 3919–3931. [Google Scholar]
- Mahlapuu, M.; Håkansson, J.; Ringstad, L.; Björn, C. Antimicrobial Peptides: An Emerging Category of Therapeutic Agents. Front. Cell. Infect. Microbiol. 2016, 6, 194. [Google Scholar] [CrossRef] [PubMed]
- Bin Hafeez, A.; Jiang, X.; Bergen, P.J.; Zhu, Y. Antimicrobial Peptides: An Update on Classifications and Databases. Int. J. Mol. Sci. 2021, 22, 11691. [Google Scholar] [CrossRef] [PubMed]
- Upton, M.; Cotter, P.; Tagg, J. Antimicrobial Peptides as Therapeutic Agents. Int. J. Microbiol. 2012, 2012, 326503–326512. [Google Scholar] [CrossRef] [PubMed]
- Jia, B.; Wang, Y.; Zhang, Y.; Wang, Z.; Wang, X.; Muhammad, I.; Kong, L.; Pei, Z.; Ma, H.; Jiang, X. High Cell Selectivity and Bactericidal Mechanism of Symmetric Peptides Centered on d-Pro–Gly Pairs. Int. J. Mol. Sci. 2020, 21, 1140. [Google Scholar] [CrossRef]
- Sampaio de Oliveira, K.B.; Leite, M.L.; Rodrigues, G.R.; Duque, H.M.; da Costa, R.A.; Cunha, V.A.; de Loiola Costa, L.S.; da Cunha, N.B.; Franco, O.L.; Dias, S.C. Strategies for recombinant production of antimicrobial peptides with pharmacological potential. Expert Rev. Clin. Pharmacol. 2020, 13, 367–390. [Google Scholar] [CrossRef] [PubMed]
- Mejía-Pitta, A.; Broset, E.; de la Fuente-Nunez, C. Probiotic engineering strategies for heterologous production of antimicrobial peptides. Adv. Drug Deliv. Rev. 2021, 176, 113863. [Google Scholar] [CrossRef] [PubMed]
- Bartolo-Aguilar, Y.; Chávez-Cabrera, C.; Flores-Cotera, L.B.; Badillo-Corona, J.A.; Oliver-Salvador, C.; Marsch, R. The potential of cold-shock promoters for the expression of recombinant proteins in microbes and mammalian cells. J. Genet. Eng. Biotechnol. 2022, 20, 173. [Google Scholar] [CrossRef]
- Ahmadi, Z.; Farajnia, S.; Farajzadeh, D.; Pouladi, N.; Pourvatan, N.; Karbalaeimahdi, M.; Shayegh, F.; Arya, M. Optimized Signal Peptide for Secretory Expression of Human Recombinant Somatropin in E. coli. Adv. Pharm. Bull. 2022, 13, 339–349. [Google Scholar] [CrossRef]
- Starr, C.G.; Wimley, W.C. Antimicrobial peptides are degraded by the cytosolic proteases of human erythrocytes. Biochim. Biophys. Acta (BBA)-Biomembr. 2017, 1859, 2319–2326. [Google Scholar] [CrossRef] [PubMed]
- Deo, S.; Turton, K.L.; Kainth, T.; Kumar, A.; Wieden, H.J. Strategies for improving antimicrobial peptide production. Biotechnol. Adv. 2022, 59, 107968. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.H.; Carpenter, T.S.; D’haeseleer, P.; Savage, D.F.; Yung, M.C. Encapsulin carrier proteins for enhanced expression of antimicrobial peptides. Biotechnol. Bioeng. 2020, 117, 603–613. [Google Scholar] [CrossRef] [PubMed]
- Chalfie, M. Green fluorescent protein as a marker for gene expression. Trends Genet. 1994, 10, 151. [Google Scholar] [CrossRef]
- Müller, H.; Salzig, D.; Czermak, P. Considerations for the process development of insect-derived antimicrobial peptide production. Biotechnol. Prog. 2015, 31, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Blommel, P.G.; Fox, B.G. A combined approach to improving large-scale production of tobacco etch virus protease. Protein Expr. Purif. 2007, 55, 53–68. [Google Scholar] [CrossRef] [PubMed]
- Blomstrand, E.; Posch, E.; Stepulane, A.; Rajasekharan, A.K.; Andersson, M. Antibacterial and Hemolytic Activity of Antimicrobial Hydrogels Utilizing Immobilized Antimicrobial Peptides. Int. J. Mol. Sci. 2024, 25, 4200. [Google Scholar] [CrossRef] [PubMed]
- Chou, S.; Shao, C.; Wang, J.; Shan, A.; Xu, L.; Dong, N.; Li, Z. Short, multiple-stranded β-hairpin peptides have antimicrobial potency with high selectivity and salt resistance. Acta Biomater. 2016, 30, 78–93. [Google Scholar] [CrossRef]
- Chen, X.; Shi, J.; Chen, R.; Wen, Y.; Shi, Y.; Zhu, Z.; Guo, S.; Li, L. Molecular chaperones (TrxA, SUMO, Intein, and GST) mediating expression, purification, and antimicrobial activity assays of plectasin in Escherichia coli. Biotechnol. Appl. Biochem. 2015, 62, 606–614. [Google Scholar] [CrossRef]
- Ali, M.P.; Yoshimatsu, K.; Suzuki, T.; Kato, T.; Park, E.Y. Expression and purification of cyto-insectotoxin (Cit1a) using silkworm larvae targeting for an antimicrobial therapeutic agent. Appl. Microbiol. Biotechnol. 2014, 98, 6973–6982. [Google Scholar] [CrossRef]
- Dale, B.A.; Fredericks, L.P. Antimicrobial peptides in the oral environment: Expression and function in health and disease. Curr. Issues Mol. Biol. 2005, 7, 119–133. [Google Scholar] [PubMed]
- Brancatisano, F.L.; Maisetta, G.; Barsotti, F.; Esin, S.; Miceli, M.; Gabriele, M.; Giuca, M.R.; Campa, M.; Batoni, G. Reduced Human Beta Defensin 3 in Individuals with Periodontal Disease. J. Dent. Res. 2011, 90, 241–245. [Google Scholar] [CrossRef] [PubMed]
- Raran-Kurussi, S.; Cherry, S.; Zhang, D.; Waugh, D.S. Removal of Affinity Tags with TEV Protease. Methods Mol. Biol. 2017, 1586, 221–230. [Google Scholar] [PubMed]
- Dong, N.; Zhu, X.; Chou, S.; Shan, A.; Li, W.; Jiang, J. Antimicrobial potency and selectivity of simplified symmetricend peptides. Biomaterials 2014, 35, 8028–8039. [Google Scholar] [CrossRef]


| Test Strains | MICs (μg/mL) | ||
|---|---|---|---|
| Chem. Syn. LRGG | Expressed LRGG | Fusion Protein GFP-LRGG | |
| Gram-negative | |||
| Eschericha coli ATCC25922 | 2 | 2 | ›512 |
| S. pullorum NCTC5776 | 4 | 4 | ›512 |
| K. pneumoniae ATCC46117 | 8 | 16 | ›512 |
| P.aeruginosa ATCC27853 | 8 | 8 | ›512 |
| S.flexneri CMCC51572 | 8 | 8 | ›512 |
| Gram-positive bacteria | |||
| Staphylococcus aureus ATCC25923 | 32 | 32 | ›512 |
| S. aureus ATCC29213 | 16 | 16 | ›512 |
| Enterococcus faecalis ATCC29212 | 32 | 32 | ›512 |
| MRSA | 256 | 128 | ›512 |
| AMPs | Control (pH 7) | Temperature | pH | |||||
|---|---|---|---|---|---|---|---|---|
| 0 °C | 37 °C | 100 °C | pH 4 | pH 6 | pH 8 | pH 10 | ||
| Chem. syn. LRGG | 2 | 2 | 2 | 8 | 2 | 2 | 4 | 4 |
| Expressed LRGG | 2 | 2 | 4 | 8 | 2 | 4 | 2 | 16 |
| Melittin | 1 | 1 | 1 | 2 | 2 | 1 | 1 | 2 |
| Peptide | Control | Proteinase (1 mg/mL) | |||
|---|---|---|---|---|---|
| Trypsin | Pepsin | Papain | Protease K | ||
| Chem. syn. LRGG | 2 | >128 | >128 | >64 | >64 |
| Exprssed LRGG | 4 | >128 | >128 | >64 | >64 |
| Melittin | 2 | 4 | 4 | 2 | 4 |
| Peptide | Control | Physical Salt Concentration | ||||
|---|---|---|---|---|---|---|
| CaCl2 | NaCl | KCl | NH4Cl | MgCl2 | ||
| Chem. Syn. LRGG | 2 | 4 | 8 | 2 | 2 | 4 |
| Expressed LRGG | 4 | 4 | 8 | 8 | 4 | 8 |
| Melittin | 2 | 4 | 4 | 2 | 2 | 2 |
| Peptide | Control | Serum | ||||
|---|---|---|---|---|---|---|
| 5% | 10% | 20% | 40% | 50% | ||
| Chem. syn. LRGG | 2 | 2 | 2 | 8 | 16 | 16 |
| Expressed LRGG | 2 | 2 | 4 | 32 | 32 | 32 |
| Melittin | 2 | 32 | 128 | 128 | 128 | 128 |
| Gene | Primer | Sequence (5′-3′) |
|---|---|---|
| linear pQE Vector | pQE-VT-F | GTAAAAGCTTAATTAGCTGAGCTTGGACTCC |
| pQE-VT-R | CATATCTCTAGAGGATCCGTGATGGTG | |
| GFP | GFP-F | CTAGAGATATGCGTAAAGGAGAAGAACTTTTCACTG |
| GFP-R | AAGATTCTCATACTTGTATAGTTCATCCATGCCATGTGTAATCCC | |
| LRGG | LRGG-F1 | CTTACAGCAGACGCAGCAGACGACGGCCGCCACGACGCAG |
| LRGG-R1 | GAGAATCTTTATTTTCAGGGCCTGCTGCGTCTGCTGCGTCGTGGCGGC | |
| TEV cleavage site + LRGG | TEV-LRGG-F2 | GAGAATCTTTATTTTCAGGGCCTGCTGCGTCTGCTGCGTCGTGGCGGC |
| TEV-LRGG-R2 | TTACAGCAGACGCAGCAGACGACGGCCGCCACGACGCAG | |
| validation primers | M13-F | AGGGTTTTCCCAGTCACG |
| M13-R | GAGCGGATAACAATTTCACAC | |
| pQE30+ | GTGAGCGGATAACAATTTCAC | |
| pQE30− | CTGAACAAATCCAGATGGAG |
| Strains | Source |
|---|---|
| Eschericha coli stellar component cell | Takara |
| Transetta component cell | Takara |
| Eschericha coli ATCC 25922 | Preserved by the Pharmacology and Toxicology Laboratory of Jilin Agricultural University |
| S. pullorum NCTC5776 | |
| K.Pneumoniae CMCC 46117 | |
| P.aeruginosa ATCC27853 | |
| S.flexneri CMCC51572 | |
| S.aureus ATCC 25923 | |
| S. faecalis ATCC 29212 | |
| Plasmids | |
| pQE-80-Kan | Qiagen |
| pMD-18T | Takara |
| pTZ18U-GFP | Takara |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, X.; Ding, Y.; Shen, Y.; Liu, S.; Liu, Y.; Wang, Y.; Wang, S.; Gualerzi, C.O.; Fabbretti, A.; Guan, L.; et al. Prokaryotic Expression and Functional Verification of Antimicrobial Peptide LRGG. Int. J. Mol. Sci. 2024, 25, 7072. https://doi.org/10.3390/ijms25137072
Liu X, Ding Y, Shen Y, Liu S, Liu Y, Wang Y, Wang S, Gualerzi CO, Fabbretti A, Guan L, et al. Prokaryotic Expression and Functional Verification of Antimicrobial Peptide LRGG. International Journal of Molecular Sciences. 2024; 25(13):7072. https://doi.org/10.3390/ijms25137072
Chicago/Turabian StyleLiu, Xiang, Yining Ding, Yuhan Shen, Sizhuo Liu, Yuehua Liu, Yuting Wang, Shikun Wang, Claudio Orlando Gualerzi, Attilio Fabbretti, Lili Guan, and et al. 2024. "Prokaryotic Expression and Functional Verification of Antimicrobial Peptide LRGG" International Journal of Molecular Sciences 25, no. 13: 7072. https://doi.org/10.3390/ijms25137072
APA StyleLiu, X., Ding, Y., Shen, Y., Liu, S., Liu, Y., Wang, Y., Wang, S., Gualerzi, C. O., Fabbretti, A., Guan, L., Kong, L., Zhang, H., Ma, H., & He, C. (2024). Prokaryotic Expression and Functional Verification of Antimicrobial Peptide LRGG. International Journal of Molecular Sciences, 25(13), 7072. https://doi.org/10.3390/ijms25137072







