Characterization of Proteome Changes in Aged and Collagen VI-Deficient Human Pericyte Cultures
Abstract
:1. Introduction
2. Results
2.1. Ingenuity Pathway Analysis: Canonical Pathways
2.2. Ingenuity Pathway Analysis: Diseases and Biofunctions
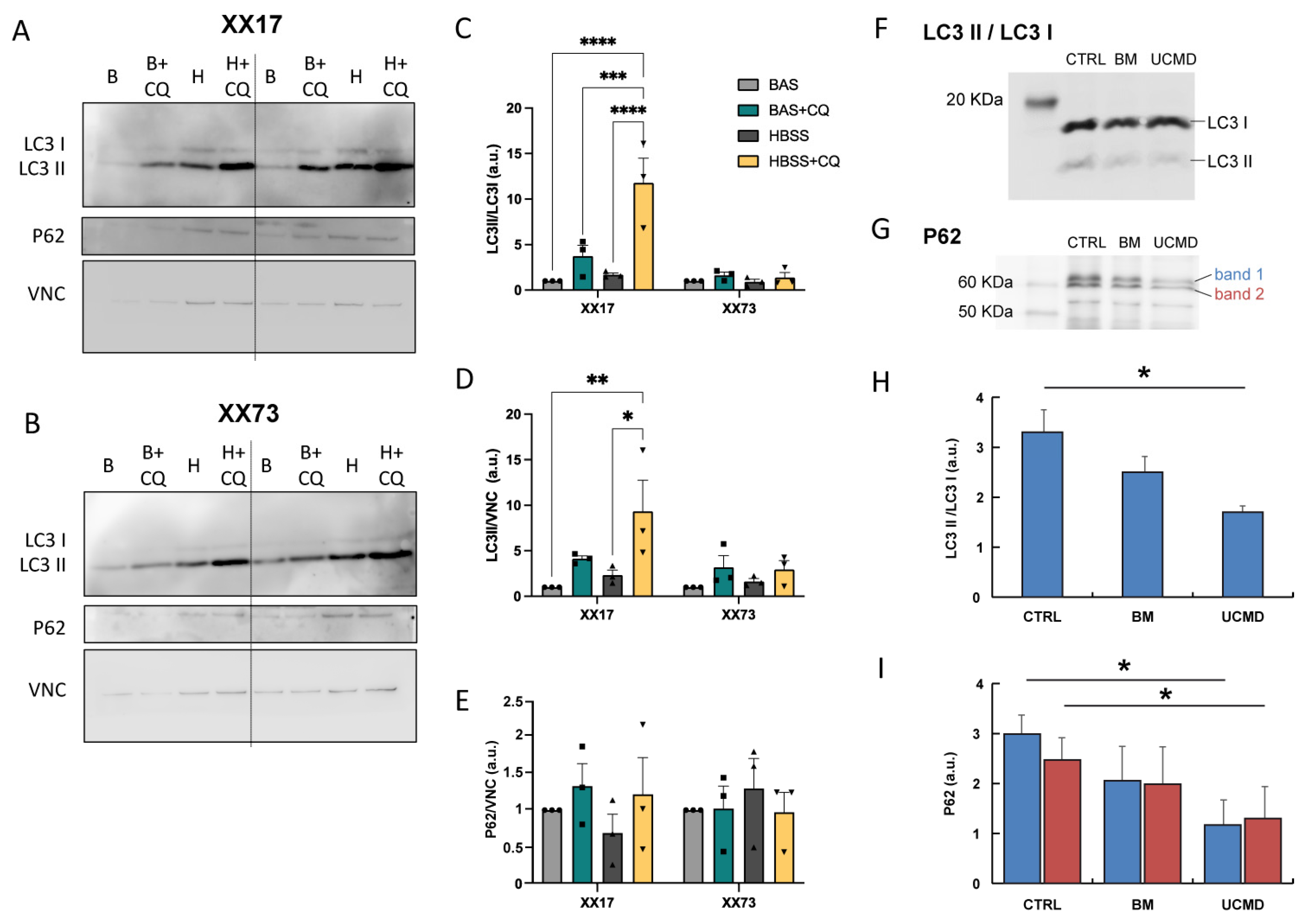
2.3. Autophagy Flux Analysis
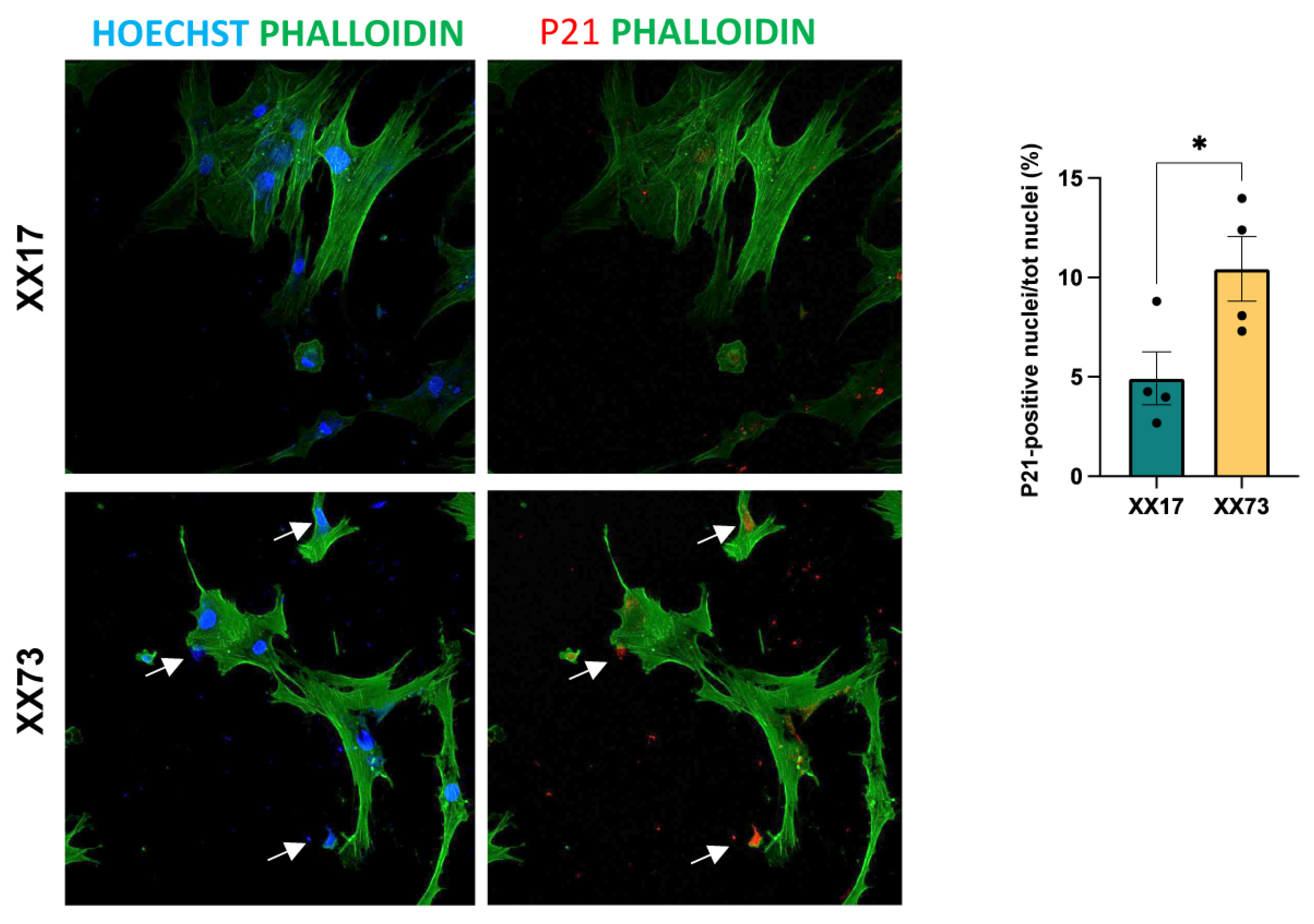
2.4. Analysis of Senescence
3. Discussion
4. Materials and Methods
4.1. Cell Cultures and Treatments
4.2. Protein Extraction for Label-Free LC–ESI–MS/MS Analysis
4.3. Label-Free Liquid Chromatography with Tandem Mass Spectrometry
4.4. Ingenuity Pathway Analysis
4.5. Western Blotting for the Analysis of Autophagic Flux
4.6. Immunofluorescence
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Armulik, A.; Genové, G.; Betsholtz, C. Pericytes: Developmental, Physiological, and Pathological Perspectives, Problems, and Promises. Dev. Cell 2011, 21, 193–215. [Google Scholar] [CrossRef]
- Attwell, D.; Mishra, A.; Hall, C.N.; O’Farrell, F.M.; Dalkara, T. What Is a Pericyte? J. Cereb. Blood Flow. Metab. 2016, 36, 451–455. [Google Scholar] [CrossRef]
- Birbrair, A. Pericyte Biology: Development, Homeostasis, and Disease. In Pericyte Biology-Novel Concepts. Advances in Experimental Medicine and Biology; Springer: Cham, Switzerland, 2018; pp. 1–3. [Google Scholar]
- Nakamura, K.; Kamouchi, M.; Arimura, K.; Nishimura, A.; Kuroda, J.; Ishitsuka, K.; Tokami, H.; Sugimori, H.; Ago, T.; Kitazono, T. Extracellular Acidification Activates CAMP Responsive Element Binding Protein via Na+/H+ Exchanger Isoform 1–Mediated Ca 2+ Oscillation in Central Nervous System Pericytes. Arter. Thromb. Vasc. Biol. 2012, 32, 2670–2677. [Google Scholar] [CrossRef]
- Sun, Z.; Gao, C.; Gao, D.; Sun, R.; Li, W.; Wang, F.; Wang, Y.; Cao, H.; Zhou, G.; Zhang, J.; et al. Reduction in Pericyte Coverage Leads to Blood–Brain Barrier Dysfunction via Endothelial Transcytosis Following Chronic Cerebral Hypoperfusion. Fluids Barriers CNS 2021, 18, 21. [Google Scholar] [CrossRef]
- Dohgu, S.; Takata, F.; Yamauchi, A.; Nakagawa, S.; Egawa, T.; Naito, M.; Tsuruo, T.; Sawada, Y.; Niwa, M.; Kataoka, Y. Brain Pericytes Contribute to the Induction and Up-Regulation of Blood–Brain Barrier Functions through Transforming Growth Factor-β Production. Brain Res. 2005, 1038, 208–215. [Google Scholar] [CrossRef]
- Proebstl, D.; Voisin, M.-B.; Woodfin, A.; Whiteford, J.; D’Acquisto, F.; Jones, G.E.; Rowe, D.; Nourshargh, S. Pericytes Support Neutrophil Subendothelial Cell Crawling and Breaching of Venular Walls in Vivo. J. Exp. Med. 2012, 209, 1219–1234. [Google Scholar] [CrossRef]
- Yamanaka, G.; Takata, F.; Kataoka, Y.; Kanou, K.; Morichi, S.; Dohgu, S.; Kawashima, H. The Neuroinflammatory Role of Pericytes in Epilepsy. Biomedicines 2021, 9, 759. [Google Scholar] [CrossRef]
- Smyth, L.C.D.; Rustenhoven, J.; Park, T.I.-H.; Schweder, P.; Jansson, D.; Heppner, P.A.; O’Carroll, S.J.; Mee, E.W.; Faull, R.L.M.; Curtis, M.; et al. Unique and Shared Inflammatory Profiles of Human Brain Endothelia and Pericytes. J. Neuroinflamm. 2018, 15, 138. [Google Scholar] [CrossRef]
- Shimizu, F.; Sano, Y.; Saito, K.; Abe, M.; Maeda, T.; Haruki, H.; Kanda, T. Pericyte-Derived Glial Cell Line-Derived Neurotrophic Factor Increase the Expression of Claudin-5 in the Blood–Brain Barrier and the Blood-Nerve Barrier. Neurochem. Res. 2012, 37, 401–409. [Google Scholar] [CrossRef]
- Polat, S.; Yazir, Y.; Duruksu, G.; Kiliç, K.C.; Mert, S.; Gacar, G.; Öncel Duman, B.; Halbutoğullari, Z.S. Investigation of the Differentiation Potential of Pericyte Cells as an Alternative Source of Mesenchymal Stem Cells. Acta Histochem. 2024, 126, 152145. [Google Scholar] [CrossRef]
- Dellavalle, A.; Maroli, G.; Covarello, D.; Azzoni, E.; Innocenzi, A.; Perani, L.; Antonini, S.; Sambasivan, R.; Brunelli, S.; Tajbakhsh, S.; et al. Pericytes Resident in Postnatal Skeletal Muscle Differentiate into Muscle Fibres and Generate Satellite Cells. Nat. Commun. 2011, 2, 499. [Google Scholar] [CrossRef] [PubMed]
- Carosio, S.; Berardinelli, M.G.; Aucello, M.; Musarò, A. Impact of Ageing on Muscle Cell Regeneration. Ageing Res. Rev. 2011, 10, 35–42. [Google Scholar] [CrossRef] [PubMed]
- García-Prat, L.; Sousa-Victor, P.; Muñoz-Cánoves, P. Functional Dysregulation of Stem Cells during Aging: A Focus on Skeletal Muscle Stem Cells. FEBS J. 2013, 280, 4051–4062. [Google Scholar] [CrossRef] [PubMed]
- Conboy, I.M.; Conboy, M.J.; Smythe, G.M.; Rando, T.A. Notch-Mediated Restoration of Regenerative Potential to Aged Muscle. Science 2003, 302, 1575–1577. [Google Scholar] [CrossRef] [PubMed]
- Baj, A.; Bettaccini, A.A.; Casalone, R.; Sala, A.; Cherubino, P.; Toniolo, A.Q. Culture of Skeletal Myoblasts from Human Donors Aged over 40 Years: Dynamics of Cell Growth and Expression of Differentiation Markers. J. Transl. Med. 2005, 3, 21. [Google Scholar] [CrossRef]
- Shefer, G.; Van de Mark, D.P.; Richardson, J.B.; Yablonka-Reuveni, Z. Satellite-Cell Pool Size Does Matter: Defining the Myogenic Potency of Aging Skeletal Muscle. Dev. Biol. 2006, 294, 50–66. [Google Scholar] [CrossRef] [PubMed]
- Day, K.; Shefer, G.; Shearer, A.; Yablonka-Reuveni, Z. The Depletion of Skeletal Muscle Satellite Cells with Age Is Concomitant with Reduced Capacity of Single Progenitors to Produce Reserve Progeny. Dev. Biol. 2010, 340, 330–343. [Google Scholar] [CrossRef] [PubMed]
- Shadrach, J.L.; Wagers, A.J. Stem Cells for Skeletal Muscle Repair. Philos. Trans. R. Soc. B Biol. Sci. 2011, 366, 2297–2306. [Google Scholar] [CrossRef] [PubMed]
- Carlson, B.M.; Faulkner, J.A. The Regeneration of Noninnervated Musele Grafts and Marcaine-Treated Muscles in Young and Old Rats. J. Gerontol. A Biol. Sci. Med. Sci. 1996, 51A, B43–B49. [Google Scholar] [CrossRef] [PubMed]
- Shavlakadze, T.; McGeachie, J.; Grounds, M.D. Delayed but Excellent Myogenic Stem Cell Response of Regenerating Geriatric Skeletal Muscles in Mice. Biogerontology 2010, 11, 363–376. [Google Scholar] [CrossRef]
- GROUNDS, M.D. Age-associated Changes in the Response of Skeletal Muscle Cells to Exercise and Regeneration a. Ann. N. Y. Acad. Sci. 1998, 854, 78–91. [Google Scholar] [CrossRef]
- García-Prat, L.; Muñoz-Cánoves, P.; Martinez-Vicente, M. Dysfunctional Autophagy Is a Driver of Muscle Stem Cell Functional Decline with Aging. Autophagy 2016, 12, 612–613. [Google Scholar] [CrossRef]
- García-Prat, L.; Martínez-Vicente, M.; Perdiguero, E.; Ortet, L.; Rodríguez-Ubreva, J.; Rebollo, E.; Ruiz-Bonilla, V.; Gutarra, S.; Ballestar, E.; Serrano, A.L.; et al. Autophagy Maintains Stemness by Preventing Senescence. Nature 2016, 529, 37–42. [Google Scholar] [CrossRef]
- Di Martino, A.; Cescon, M.; D’Agostino, C.; Schilardi, F.; Sabatelli, P.; Merlini, L.; Faldini, C. Collagen VI in the Musculoskeletal System. Int. J. Mol. Sci. 2023, 24, 5095. [Google Scholar] [CrossRef]
- Bushby, K.M.D.; Collins, J.; Hicks, D. Collagen Type VI Myopathies. In Progress in Heritable Soft Connective Tissue Diseases. Advances in Experimental Medicine and Biology; Springer: Dordrecht, The Netherlands, 2014; pp. 185–199. [Google Scholar]
- Urciuolo, A.; Quarta, M.; Morbidoni, V.; Gattazzo, F.; Molon, S.; Grumati, P.; Montemurro, F.; Tedesco, F.S.; Blaauw, B.; Cossu, G.; et al. Collagen VI Regulates Satellite Cell Self-Renewal and Muscle Regeneration. Nat. Commun. 2013, 4, 1964. [Google Scholar] [CrossRef]
- Cenni, V.; Sabatelli, P.; Di Martino, A.; Merlini, L.; Antoniel, M.; Squarzoni, S.; Neri, S.; Santi, S.; Metti, S.; Bonaldo, P.; et al. Collagen VI Deficiency Impairs Tendon Fibroblasts Mechanoresponse in Ullrich Congenital Muscular Dystrophy. Cells 2024, 13, 378. [Google Scholar] [CrossRef]
- Irwin, W.A.; Bergamin, N.; Sabatelli, P.; Reggiani, C.; Megighian, A.; Merlini, L.; Braghetta, P.; Columbaro, M.; Volpin, D.; Bressan, G.M.; et al. Mitochondrial Dysfunction and Apoptosis in Myopathic Mice with Collagen VI Deficiency. Nat. Genet. 2003, 35, 367–371. [Google Scholar] [CrossRef]
- Kawahara, G.; Okada, M.; Morone, N.; Ibarra, C.A.; Nonaka, I.; Noguchi, S.; Hayashi, Y.K.; Nishino, I. Reduced Cell Anchorage May Cause Sarcolemma-Specific Collagen VI Deficiency in Ullrich Disease. Neurology 2007, 69, 1043–1049. [Google Scholar] [CrossRef]
- Capitanio, D.; Moriggi, M.; de Palma, S.; Bizzotto, D.; Molon, S.; Torretta, E.; Fania, C.; Bonaldo, P.; Gelfi, C.; Braghetta, P. Collagen VI Null Mice as a Model for Early Onset Muscle Decline in Aging. Front. Mol. Neurosci. 2017, 10, 337. [Google Scholar] [CrossRef]
- Lawless, C.; Wang, C.; Jurk, D.; Merz, A.; von Zglinicki, T.; Passos, J.F. Quantitative Assessment of Markers for Cell Senescence. Exp. Gerontol. 2010, 45, 772–778. [Google Scholar] [CrossRef]
- Capitanio, D.; Vasso, M.; Fania, C.; Moriggi, M.; Viganò, A.; Procacci, P.; Magnaghi, V.; Gelfi, C. Comparative Proteomic Profile of Rat Sciatic Nerve and Gastrocnemius Muscle Tissues in Ageing by 2-D DIGE. Proteomics 2009, 9, 2004–2020. [Google Scholar] [CrossRef] [PubMed]
- Capitanio, D.; Vasso, M.; De Palma, S.; Fania, C.; Torretta, E.; Cammarata, F.P.; Magnaghi, V.; Procacci, P.; Gelfi, C. Specific Protein Changes Contribute to the Differential Muscle Mass Loss during Ageing. Proteomics 2016, 16, 645–656. [Google Scholar] [CrossRef] [PubMed]
- De Palma, S.; Leone, R.; Grumati, P.; Vasso, M.; Polishchuk, R.; Capitanio, D.; Braghetta, P.; Bernardi, P.; Bonaldo, P.; Gelfi, C. Changes in Muscle Cell Metabolism and Mechanotransduction Are Associated with Myopathic Phenotype in a Mouse Model of Collagen VI Deficiency. PLoS ONE 2013, 8, e56716. [Google Scholar] [CrossRef] [PubMed]
- Menaceur, C.; Hachani, J.; Dib, S.; Duban-Deweer, S.; Karamanos, Y.; Shimizu, F.; Kanda, T.; Gosselet, F.; Fenart, L.; Saint-Pol, J. Highlighting In Vitro the Role of Brain-like Endothelial Cells on the Maturation and Metabolism of Brain Pericytes by SWATH Proteomics. Cells 2023, 12, 1010. [Google Scholar] [CrossRef] [PubMed]
- Kolyada, A.Y.; Riley, K.N.; Herman, I.M. Rho GTPase Signaling Modulates Cell Shape and Contractile Phenotype in an Isoactin-Specific Manner. Am. J. Physiol.-Cell Physiol. 2003, 285, C1116–C1121. [Google Scholar] [CrossRef] [PubMed]
- Durham, J.T.; Surks, H.K.; Dulmovits, B.M.; Herman, I.M. Pericyte Contractility Controls Endothelial Cell Cycle Progression and Sprouting: Insights into Angiogenic Switch Mechanics. Am. J. Physiol.-Cell Physiol. 2014, 307, C878–C892. [Google Scholar] [CrossRef] [PubMed]
- Kemp, S.S.; Lin, P.K.; Sun, Z.; Castaño, M.A.; Yrigoin, K.; Penn, M.R.; Davis, G.E. Molecular Basis for Pericyte-Induced Capillary Tube Network Assembly and Maturation. Front. Cell Dev. Biol. 2022, 10, 943533. [Google Scholar] [CrossRef] [PubMed]
- Pellowe, A.S.; Wu, M.J.; Kang, T.-Y.; Chung, T.D.; Ledesma-Mendoza, A.; Herzog, E.; Levchenko, A.; Odell, I.; Varga, J.; Gonzalez, A.L. TGF-Β1 Drives Integrin-Dependent Pericyte Migration and Microvascular Destabilization in Fibrotic Disease. Am. J. Pathol. 2024, 194, 1171–1184. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Zhang, F.; Ikonomovic, M.; Yang, T. The Role of NRF2 in Cerebrovascular Protection: Implications for Vascular Cognitive Impairment and Dementia (VCID). Int. J. Mol. Sci. 2024, 25, 3833. [Google Scholar] [CrossRef]
- Pieper, C.; Galla, H.-J. Ultra Structure Analysis of Cell–Cell Interactions between Pericytes and Neutrophils in Vitro. Biochem. Biophys. Res. Commun. 2014, 445, 180–183. [Google Scholar] [CrossRef]
- Palmieri, M.; Impey, S.; Kang, H.; di Ronza, A.; Pelz, C.; Sardiello, M.; Ballabio, A. Characterization of the CLEAR Network Reveals an Integrated Control of Cellular Clearance Pathways. Hum. Mol. Genet. 2011, 20, 3852–3866. [Google Scholar] [CrossRef] [PubMed]
- Aman, Y.; Schmauck-Medina, T.; Hansen, M.; Morimoto, R.I.; Simon, A.K.; Bjedov, I.; Palikaras, K.; Simonsen, A.; Johansen, T.; Tavernarakis, N.; et al. Autophagy in Healthy Aging and Disease. Nat. Aging 2021, 1, 634–650. [Google Scholar] [CrossRef] [PubMed]
- Milani, S.Z.; Rezabakhsh, A.; Karimipour, M.; Salimi, L.; Mardi, N.; Narmi, M.T.; Sadeghsoltani, F.; Valioglu, F.; Rahbarghazi, R. Role of Autophagy in Angiogenic Potential of Vascular Pericytes. Front. Cell Dev. Biol. 2024, 12, 1347857. [Google Scholar] [CrossRef]
- Potes, Y.; Bermejo-Millo, J.C.; Mendes, C.; Castelão-Baptista, J.P.; Díaz-Luis, A.; Pérez-Martínez, Z.; Solano, J.J.; Sardão, V.A.; Oliveira, P.J.; Caballero, B.; et al. P66Shc Signaling and Autophagy Impact on C2C12 Myoblast Differentiation during Senescence. Cell Death Dis. 2024, 15, 200. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.N.; Green, J.; Wang, Z.; Deng, Y.; Qiao, M.; Peabody, M.; Zhang, Q.; Ye, J.; Yan, Z.; Denduluri, S.; et al. Bone Morphogenetic Protein (BMP) Signaling in Development and Human Diseases. Genes. Dis. 2014, 1, 87–105. [Google Scholar] [CrossRef] [PubMed]
- Bautch, V.L. Bone Morphogenetic Protein and Blood Vessels: New Insights into Endothelial Cell Junction Regulation. Curr. Opin. Hematol. 2019, 26, 154–160. [Google Scholar] [CrossRef] [PubMed]
- Harrison, D.A. The JAK/STAT Pathway. Cold Spring Harb. Perspect. Biol. 2012, 4, a011205. [Google Scholar] [CrossRef] [PubMed]
- Takata, F.; Dohgu, S.; Sakaguchi, S.; Sakai, K.; Yamanaka, G.; Iwao, T.; Matsumoto, J.; Kimura, I.; Sezaki, Y.; Tanaka, Y.; et al. Oncostatin-M-Reactive Pericytes Aggravate Blood–Brain Barrier Dysfunction by Activating JAK/STAT3 Signaling In Vitro. Neuroscience 2019, 422, 12–20. [Google Scholar] [CrossRef]
- Zhu, K.; Meng, Q.; Zhang, Z.; Yi, T.; He, Y.; Zheng, J.; Lei, W. Aryl Hydrocarbon Receptor Pathway: Role, Regulation and Intervention in Atherosclerosis Therapy (Review). Mol. Med. Rep. 2019, 20, 4763–4773. [Google Scholar] [CrossRef] [PubMed]
- Kidoya, H.; Ueno, M.; Yamada, Y.; Mochizuki, N.; Nakata, M.; Yano, T.; Fujii, R.; Takakura, N. Spatial and Temporal Role of the Apelin/APJ System in the Caliber Size Regulation of Blood Vessels during Angiogenesis. EMBO J. 2008, 27, 522–534. [Google Scholar] [CrossRef]
- Cox, C.M.; D’Agostino, S.L.; Miller, M.K.; Heimark, R.L.; Krieg, P.A. Apelin, the Ligand for the Endothelial G-Protein-Coupled Receptor, APJ, Is a Potent Angiogenic Factor Required for Normal Vascular Development of the Frog Embryo. Dev. Biol. 2006, 296, 177–189. [Google Scholar] [CrossRef] [PubMed]
- Lee, U.; Stuelsatz, P.; Karaz, S.; McKellar, D.W.; Russeil, J.; Deak, M.; De Vlaminck, I.; Lepper, C.; Deplancke, B.; Cosgrove, B.D.; et al. A Tead1-Apelin Axis Directs Paracrine Communication from Myogenic to Endothelial Cells in Skeletal Muscle. iScience 2022, 25, 104589. [Google Scholar] [CrossRef] [PubMed]
- Xiang, D.; Feng, Y.; Wang, J.; Zhang, X.; Shen, J.; Zou, R.; Yuan, Y. Platelet-derived Growth Factor-BB Promotes Proliferation and Migration of Retinal Microvascular Pericytes by Up-regulating the Expression of C-X-C Chemokine Receptor Types 4. Exp. Ther. Med. 2019, 18, 4022–4030. [Google Scholar] [CrossRef] [PubMed]
- Castagnaro, S.; Chrisam, M.; Cescon, M.; Braghetta, P.; Grumati, P.; Bonaldo, P. Extracellular Collagen VI Has Prosurvival and Autophagy Instructive Properties in Mouse Fibroblasts. Front. Physiol. 2018, 9, 391697. [Google Scholar] [CrossRef] [PubMed]
- Grumati, P.; Coletto, L.; Sabatelli, P.; Cescon, M.; Angelin, A.; Bertaggia, E.; Blaauw, B.; Urciuolo, A.; Tiepolo, T.; Merlini, L.; et al. Autophagy Is Defective in Collagen VI Muscular Dystrophies, and Its Reactivation Rescues Myofiber Degeneration. Nat. Med. 2010, 16, 1313–1320. [Google Scholar] [CrossRef] [PubMed]
- Castagnaro, S.; Pellegrini, C.; Pellegrini, M.; Chrisam, M.; Sabatelli, P.; Toni, S.; Grumati, P.; Ripamonti, C.; Pratelli, L.; Maraldi, N.M.; et al. Autophagy Activation in COL6 Myopathic Patients by a Low-Protein-Diet Pilot Trial. Autophagy 2016, 12, 2484–2495. [Google Scholar] [CrossRef]
- Abbonante, V.; Malara, A.; Chrisam, M.; Metti, S.; Soprano, P.; Semplicini, C.; Bello, L.; Bozzi, V.; Battiston, M.; Pecci, A.; et al. Lack of COL6/Collagen VI Causes Megakaryocyte Dysfunction by Impairing Autophagy and Inducing Apoptosis. Autophagy 2023, 19, 984–999. [Google Scholar] [CrossRef] [PubMed]
- Metti, S.; Gambarotto, L.; Chrisam, M.; La Spina, M.; Baraldo, M.; Braghetta, P.; Blaauw, B.; Bonaldo, P. The Polyphenol Pterostilbene Ameliorates the Myopathic Phenotype of Collagen VI Deficient Mice via Autophagy Induction. Front. Cell Dev. Biol. 2020, 8, 580933. [Google Scholar] [CrossRef] [PubMed]
- Gambarotto, L.; Metti, S.; Corpetti, M.; Baraldo, M.; Sabatelli, P.; Castagnaro, S.; Cescon, M.; Blaauw, B.; Bonaldo, P. Sustained Oral Spermidine Supplementation Rescues Functional and Structural Defects in COL6-Deficient Myopathic Mice. Autophagy 2023, 19, 3221–3229. [Google Scholar] [CrossRef]
- Merlini, L.; Sabatelli, P.; Gualandi, F.; Redivo, E.; Di Martino, A.; Faldini, C. New Clinical and Immunofluorescence Data of Collagen VI-Related Myopathy: A Single Center Cohort of 69 Patients. Int. J. Mol. Sci. 2023, 24, 12474. [Google Scholar] [CrossRef]
- Vono, R.; Fuoco, C.; Testa, S.; Pirrò, S.; Maselli, D.; Ferland McCollough, D.; Sangalli, E.; Pintus, G.; Giordo, R.; Finzi, G.; et al. Activation of the Pro-Oxidant PKCβII-P66Shc Signaling Pathway Contributes to Pericyte Dysfunction in Skeletal Muscles of Patients With Diabetes With Critical Limb Ischemia. Diabetes 2016, 65, 3691–3704. [Google Scholar] [CrossRef]
- Sacchetti, B.; Funari, A.; Michienzi, S.; Di Cesare, S.; Piersanti, S.; Saggio, I.; Tagliafico, E.; Ferrari, S.; Robey, P.G.; Riminucci, M.; et al. Self-Renewing Osteoprogenitors in Bone Marrow Sinusoids Can Organize a Hematopoietic Microenvironment. Cell 2007, 131, 324–336. [Google Scholar] [CrossRef]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal Sample Preparation Method for Proteome Analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant Enables High Peptide Identification Rates, Individualized p.p.b.-Range Mass Accuracies and Proteome-Wide Protein Quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Cox, J.; Hein, M.Y.; Luber, C.A.; Paron, I.; Nagaraj, N.; Mann, M. Accurate Proteome-Wide Label-Free Quantification by Delayed Normalization and Maximal Peptide Ratio Extraction, Termed MaxLFQ. Mol. Cell. Proteom. 2014, 13, 2513–2526. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus Computational Platform for Comprehensive Analysis of (Prote)Omics Data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Krämer, A.; Green, J.; Pollard, J.; Tugendreich, S. Causal Analysis Approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef] [PubMed]





| Canonical Pathways | Aged/Control | BM/Control | UCMD/Control |
|---|---|---|---|
| Glycolysis I | −3.464 | 1.342 | 0 |
| Gluconeogenesis I | −2.333 | 2.236 | 0.378 |
| Fatty Acid β-oxidation I | 2.236 | N/A | N/A |
| Oxidative Phosphorylation | 2.646 | 1 | −1.732 |
| ILK Signaling | −3.273 | −2.183 | −2.183 |
| Signaling by Rho Family GTPases | −3 | −0.775 | 1 |
| Actin Cytoskeleton Signaling | −2.785 | −2.324 | −0.943 |
| Integrin Signaling | −2.746 | −0.535 | −0.447 |
| Regulation of Actin-based Motility by Rho | −2.714 | −1.414 | −0.333 |
| RHOA Signaling | −2.673 | −1 | 0.905 |
| CDC42 Signaling | −2.111 | −1.134 | 0.378 |
| RHOGDI Signaling | 2.5 | 1.732 | 0.577 |
| Pentose Phosphate Pathway | N/A | 2 | 2 |
| Aryl Hydrocarbon Receptor Signaling | 0 | 2.449 | 2 |
| Glutathione-mediated Detoxification | 1 | 2 | 2 |
| NRF2-mediated Oxidative Stress Response | 2.53 | 2.714 | 1.897 |
| Leukocyte Extravasation Signaling | −2.828 | −2.496 | −1.604 |
| EIF2 Signaling | −2.138 | −4.707 | −0.728 |
| CLEAR Signaling Pathway | −2.138 | N/A | N/A |
| CXCR4 Signaling | −0.707 | N/A | 2 |
| Apelin Adipocyte Signaling Pathway | 1.414 | 2.449 | 2.236 |
| BMP signaling pathway | 2 | N/A | N/A |
| JAK/STAT Signaling | 2.236 | N/A | N/A |
| Diseases and Biofunctions | Aged/Control | BM/Control | UCMD/Control |
|---|---|---|---|
| Synthesis of protein | −2.875 | −2.06 | −0.459 |
| Metabolism of protein | −2.589 | −1.764 | −0.797 |
| Attachment of cells | −3.065 | −0.288 | −1.16 |
| Binding of connective tissue cells | −2.717 | −0.694 | −0.548 |
| Adhesion of connective tissue cells | −2.531 | −0.141 | −0.423 |
| Cell movement of epithelial cell lines | −2.462 | −0.342 | 0.385 |
| Shape change of embryonic cell lines | −2.449 | N/A | −0.447 |
| Shape change of epithelial cell lines | −2.138 | −1.664 | −0.896 |
| Epithelial−mesenchymal transition | N/A | −0.116 | −2.029 |
| Angiogenesis | −1.723 | 1.421 | 2.29 |
| Organismal death | −1.257 | 1.069 | 2.673 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moriggi, M.; Torretta, E.; Cescon, M.; Russo, L.; Gregorio, I.; Braghetta, P.; Sabatelli, P.; Faldini, C.; Merlini, L.; Gargioli, C.; et al. Characterization of Proteome Changes in Aged and Collagen VI-Deficient Human Pericyte Cultures. Int. J. Mol. Sci. 2024, 25, 7118. https://doi.org/10.3390/ijms25137118
Moriggi M, Torretta E, Cescon M, Russo L, Gregorio I, Braghetta P, Sabatelli P, Faldini C, Merlini L, Gargioli C, et al. Characterization of Proteome Changes in Aged and Collagen VI-Deficient Human Pericyte Cultures. International Journal of Molecular Sciences. 2024; 25(13):7118. https://doi.org/10.3390/ijms25137118
Chicago/Turabian StyleMoriggi, Manuela, Enrica Torretta, Matilde Cescon, Loris Russo, Ilaria Gregorio, Paola Braghetta, Patrizia Sabatelli, Cesare Faldini, Luciano Merlini, Cesare Gargioli, and et al. 2024. "Characterization of Proteome Changes in Aged and Collagen VI-Deficient Human Pericyte Cultures" International Journal of Molecular Sciences 25, no. 13: 7118. https://doi.org/10.3390/ijms25137118





