Selenium Discrepancies in Fetal Bovine Serum: Impact on Cellular Selenoprotein Expression
Abstract
:1. Introduction
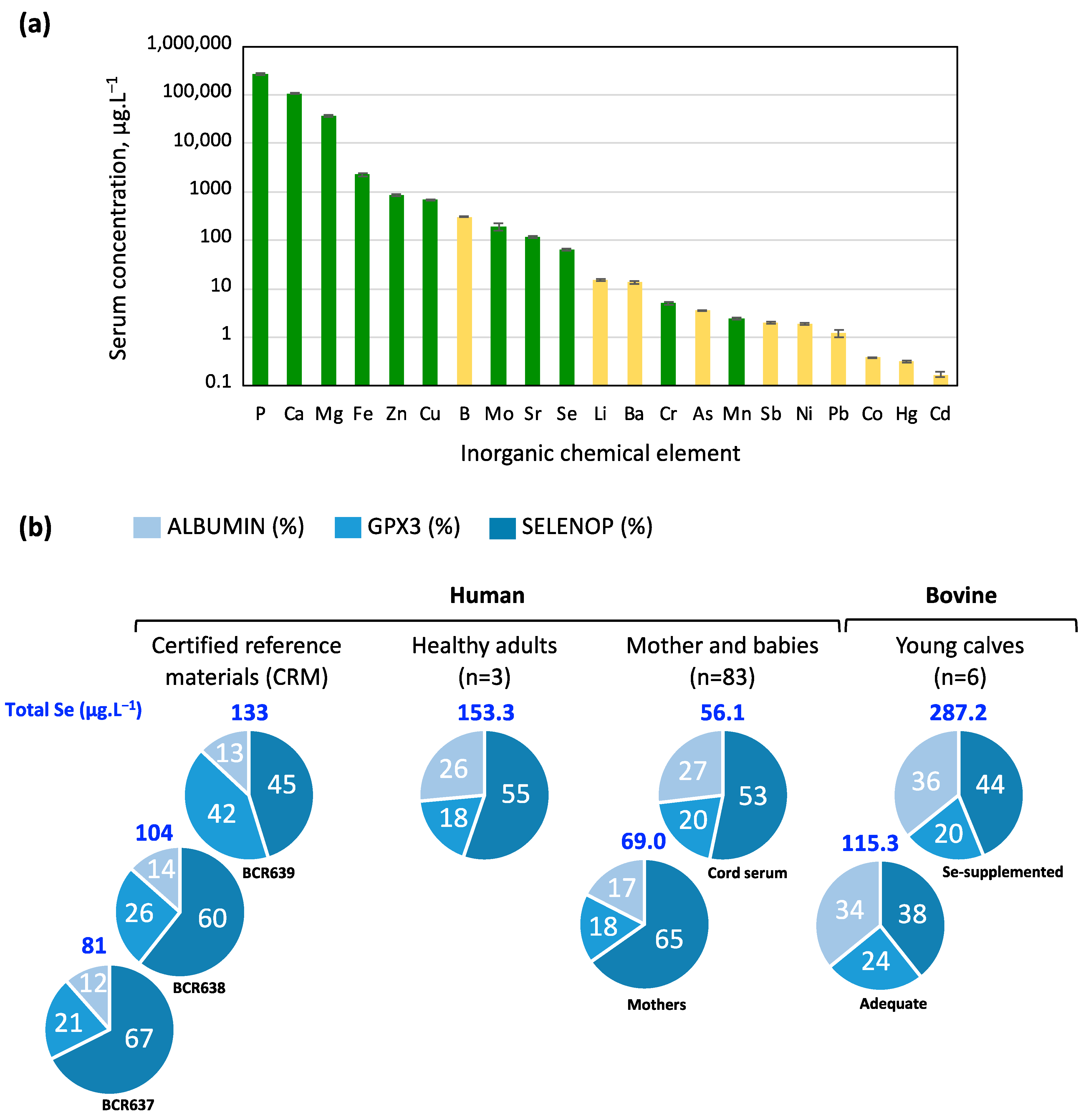
2. Results
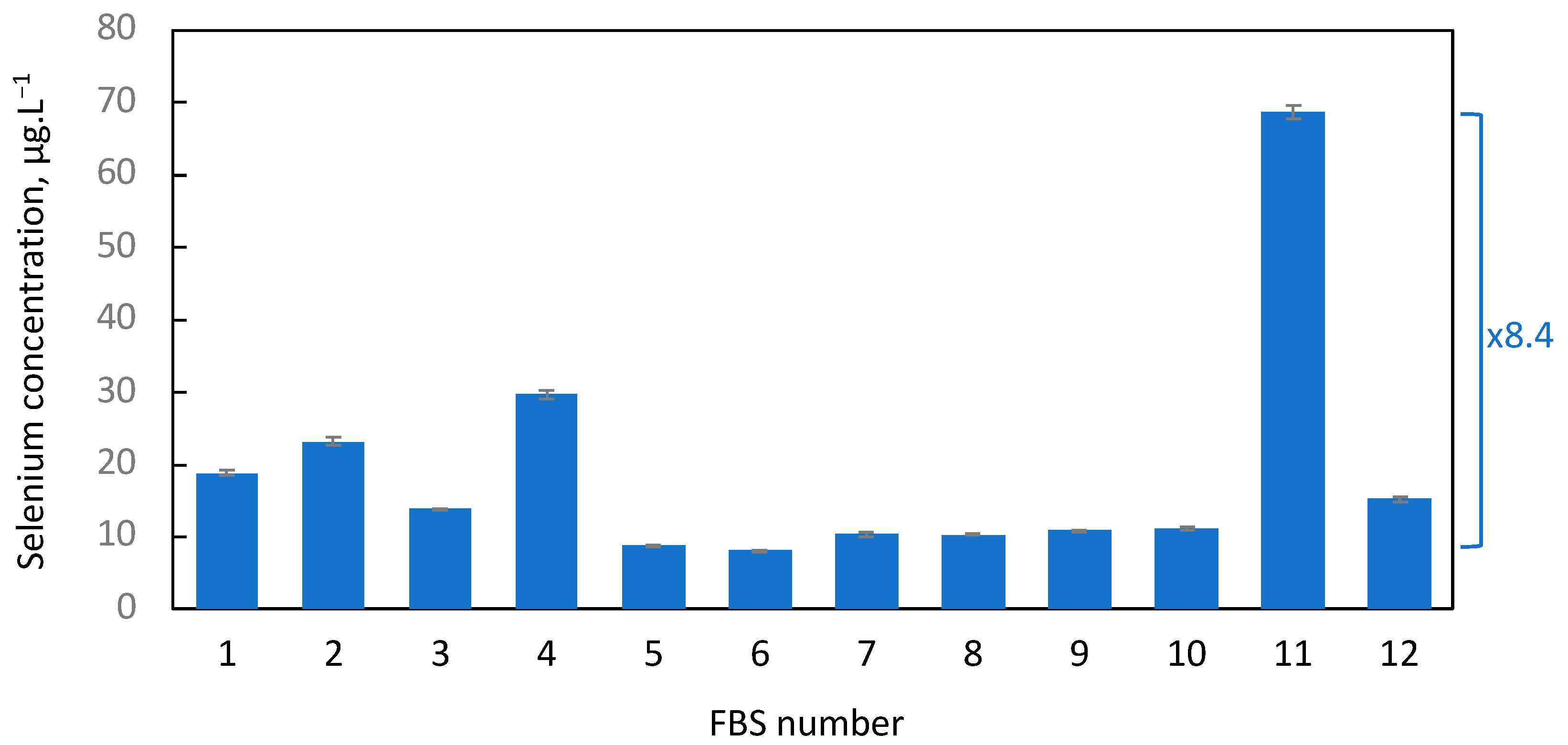
2.1. Selenium Levels in FBS Greatly Vary from Suppliers and Lot Number
2.2. Impact of FBS Selenium Levels on the Expression of GPX1, GPX4, and TXNRD1 in HEK293 Cells
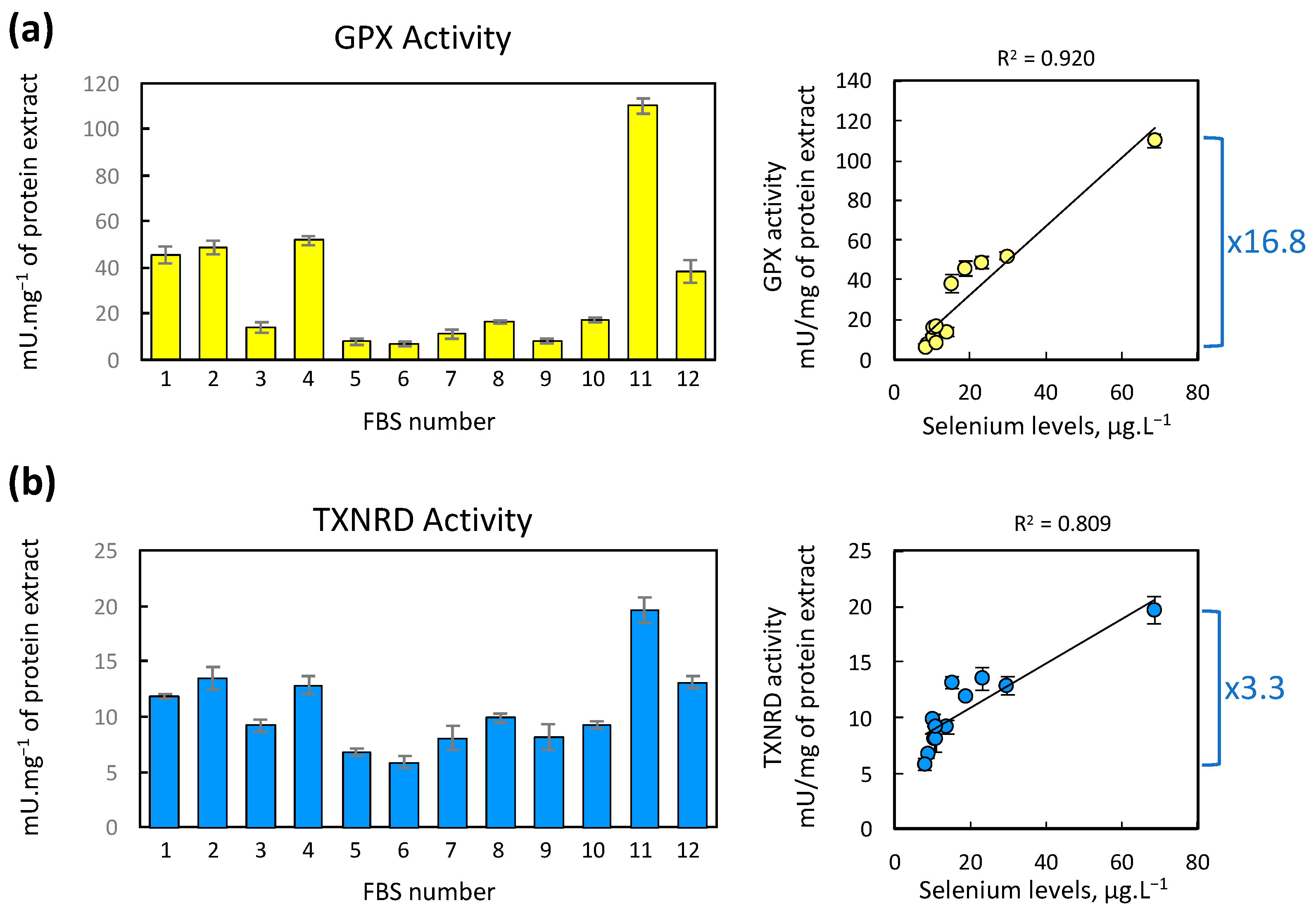
2.3. Impact of FBS Selenium Levels on the GPX and TXNRD Enzymatic Activities in HEK293 Cells
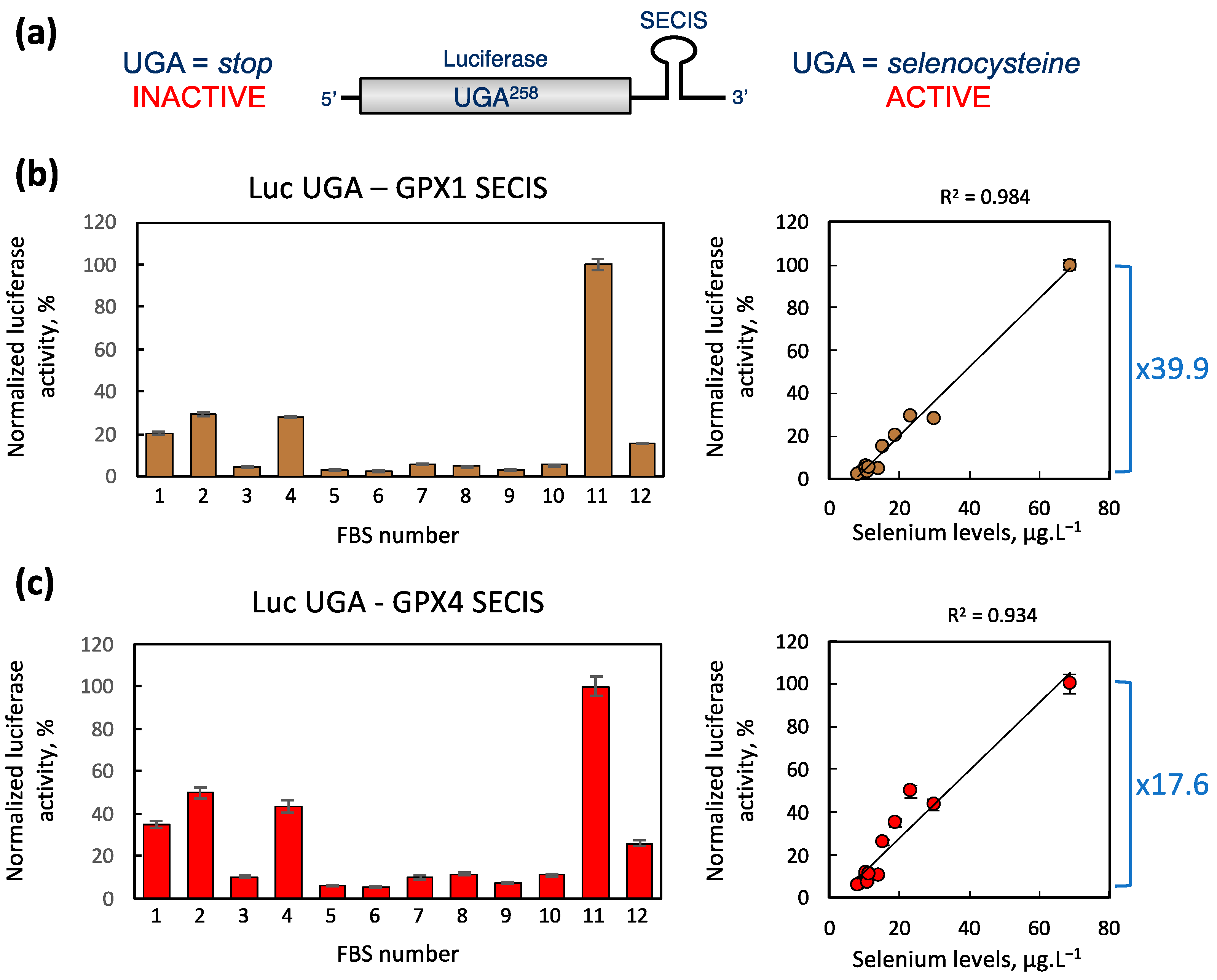
2.4. Impact of FBS Selenium Levels on the UGA Recoding Activities as Measured by Luciferase Reporter Assay
3. Discussion
3.1. Selenium Levels in FBS Greatly Vary from Suppliers and Lot Numbers
3.2. Selenium Levels in FBS Correlates with GPX1 and GPX4 Expression in Cells
3.3. Selenium Bioavailability from FBS Is Mostly Supported by SELENOP
4. Materials and Methods
4.1. Materials
4.2. Triple Quadrupole ICP-MS Analysis
4.3. Cell Culture
4.4. Protein Extraction and Analysis by Western Blot
4.5. GPX and TXNRD Enzymatic Assays
4.6. Luciferase Assays
4.7. Graphical Illustrations and Data Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Guillin, O.M.; Vindry, C.; Ohlmann, T.; Chavatte, L. Selenium, Selenoproteins and Viral Infection. Nutrients 2019, 11, 2101. [Google Scholar] [CrossRef] [PubMed]
- Papp, L.V.; Holmgren, A.; Khanna, K.K. Selenium and selenoproteins in health and disease. Antioxid. Redox Signal. 2010, 12, 793–795. [Google Scholar] [CrossRef]
- Rayman, M.P. Selenium and human health. Lancet 2012, 379, 1256–1268. [Google Scholar] [CrossRef] [PubMed]
- Hatfield, D.L.; Tsuji, P.A.; Carlson, B.A.; Gladyshev, V.N. Selenium and selenocysteine: Roles in cancer, health, and development. Trends Biochem. Sci. 2014, 39, 112–120. [Google Scholar] [CrossRef] [PubMed]
- Hatfield, D.L.; Carlson, B.A.; Xu, X.M.; Mix, H.; Gladyshev, V.N. Selenocysteine incorporation machinery and the role of selenoproteins in development and health. Prog. Nucleic Acid. Res. Mol. Biol. 2006, 81, 97–142. [Google Scholar] [PubMed]
- Schweizer, U.; Fabiano, M. Selenoproteins in brain development and function. Free Radic. Biol. Med. 2022, 190, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Vindry, C.; Ohlmann, T.; Chavatte, L. Translation regulation of mammalian selenoproteins. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 2480–2492. [Google Scholar] [CrossRef] [PubMed]
- Copeland, P.R.; Howard, M.T. Ribosome Fate during Decoding of UGA-Sec Codons. Int. J. Mol. Sci. 2021, 22, 13204. [Google Scholar] [CrossRef] [PubMed]
- Labunskyy, V.M.; Hatfield, D.L.; Gladyshev, V.N. Selenoproteins: Molecular pathways and physiological roles. Physiol. Rev. 2014, 94, 739–777. [Google Scholar] [CrossRef]
- Hatfield, D.L.; Gladyshev, V.N. How selenium has altered our understanding of the genetic code. Mol. Cell Biol. 2002, 22, 3565–3576. [Google Scholar] [CrossRef]
- Driscoll, D.M.; Copeland, P.R. Mechanism and regulation of selenoprotein synthesis. Annu. Rev. Nutr. 2003, 23, 17–40. [Google Scholar] [CrossRef] [PubMed]
- Shetty, S.P.; Copeland, P.R. Selenocysteine incorporation: A trump card in the game of mRNA decay. Biochimie 2015, 114, 97–101. [Google Scholar] [CrossRef]
- Squires, J.E.; Berry, M.J. Eukaryotic selenoprotein synthesis: Mechanistic insight incorporating new factors and new functions for old factors. IUBMB Life 2008, 60, 232–235. [Google Scholar] [CrossRef]
- Park, S.I.; Park, J.M.; Chittum, H.S.; Yang, E.S.; Carlson, B.A.; Lee, B.J.; Hatfield, D.L. Selenocysteine tRNAs as central components of selenoprotein biosynthesis in eukaryotes. Biomed. Environ. Sci. 1997, 10, 116–124. [Google Scholar] [PubMed]
- Hilal, T.; Killam, B.Y.; Grozdanovic, M.; Dobosz-Bartoszek, M.; Loerke, J.; Burger, J.; Mielke, T.; Copeland, P.R.; Simonovic, M.; Spahn, C.M.T. Structure of the mammalian ribosome as it decodes the selenocysteine UGA codon. Science 2022, 376, 1338–1343. [Google Scholar] [CrossRef] [PubMed]
- Berry, M.J.; Tujebajeva, R.M.; Copeland, P.R.; Xu, X.M.; Carlson, B.A.; Martin, G.W., 3rd; Low, S.C.; Mansell, J.B.; Grundner-Culemann, E.; Harney, J.W.; et al. Selenocysteine incorporation directed from the 3′UTR: Characterization of eukaryotic EFsec and mechanistic implications. Biofactors 2001, 14, 17–24. [Google Scholar] [CrossRef]
- Bermano, G.; Arthur, J.R.; Hesketh, J.E. Selective control of cytosolic glutathione peroxidase and phospholipid hydroperoxide glutathione peroxidase mRNA stability by selenium supply. FEBS Lett. 1996, 387, 157–160. [Google Scholar] [CrossRef] [PubMed]
- Bermano, G.; Arthur, J.R.; Hesketh, J.E. Role of the 3′ untranslated region in the regulation of cytosolic glutathione peroxidase and phospholipid-hydroperoxide glutathione peroxidase gene expression by selenium supply. Biochem. J. 1996, 320, 891–895. [Google Scholar] [CrossRef]
- Bermano, G.; Nicol, F.; Dyer, J.A.; Sunde, R.A.; Beckett, G.J.; Arthur, J.R.; Hesketh, J.E. Tissue-specific regulation of selenoenzyme gene expression during selenium deficiency in rats. Biochem. J. 1995, 311, 425–430. [Google Scholar] [CrossRef]
- Wingler, K.; Bocher, M.; Flohe, L.; Kollmus, H.; Brigelius-Flohe, R. mRNA stability and selenocysteine insertion sequence efficiency rank gastrointestinal glutathione peroxidase high in the hierarchy of selenoproteins. Eur. J. Biochem. 1999, 259, 149–157. [Google Scholar] [CrossRef]
- Weiss Sachdev, S.; Sunde, R.A. Selenium regulation of transcript abundance and translational efficiency of glutathione peroxidase-1 and -4 in rat liver. Biochem. J. 2001, 357, 851–858. [Google Scholar] [CrossRef] [PubMed]
- Lei, X.G.; Evenson, J.K.; Thompson, K.M.; Sunde, R.A. Glutathione peroxidase and phospholipid hydroperoxide glutathione peroxidase are differentially regulated in rats by dietary selenium. J. Nutr. 1995, 125, 1438–1446. [Google Scholar] [PubMed]
- Chittum, H.S.; Hill, K.E.; Carlson, B.A.; Lee, B.J.; Burk, R.F.; Hatfield, D.L. Replenishment of selenium deficient rats with selenium results in redistribution of the selenocysteine tRNA population in a tissue specific manner. Biochim. Biophys. Acta 1997, 1359, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Guillin, O.M.; Vindry, C.; Ohlmann, T.; Chavatte, L. Interplay between Selenium, Selenoproteins and HIV-1 Replication in Human CD4 T-Lymphocytes. Int. J. Mol. Sci. 2022, 23, 1394. [Google Scholar] [CrossRef] [PubMed]
- Sonet, J.; Bulteau, A.L.; Touat-Hamici, Z.; Mosca, M.; Bierla, K.; Mounicou, S.; Lobinski, R.; Chavatte, L. Selenoproteome Expression Studied by Non-Radioactive Isotopic Selenium-Labeling in Human Cell Lines. Int. J. Mol. Sci. 2021, 22, 7308. [Google Scholar] [CrossRef] [PubMed]
- Burk, R.F.; Hill, K.E. Regulation of Selenium Metabolism and Transport. Annu. Rev. Nutr. 2015, 35, 109–134. [Google Scholar] [CrossRef] [PubMed]
- Bellinger, F.P.; Raman, A.V.; Reeves, M.A.; Berry, M.J. Regulation and function of selenoproteins in human disease. Biochem. J. 2009, 422, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Copeland, P.R.; Driscoll, D.M. RNA binding proteins and selenocysteine. Biofactors 2001, 14, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Schomburg, L.; Schweizer, U. Hierarchical regulation of selenoprotein expression and sex-specific effects of selenium. Biochim. Biophys. Acta 2009, 1790, 1453–1462. [Google Scholar] [CrossRef]
- Santesmasses, D.; Gladyshev, V.N. Pathogenic Variants in Selenoproteins and Selenocysteine Biosynthesis Machinery. Int. J. Mol. Sci. 2021, 22, 11593. [Google Scholar] [CrossRef]
- Brigelius-Flohe, R.; Maiorino, M. Glutathione peroxidases. Biochim. Biophys. Acta 2012, 1830, 3289–3303. [Google Scholar] [CrossRef] [PubMed]
- Touat-Hamici, Z.; Bulteau, A.L.; Bianga, J.; Jean-Jacques, H.; Szpunar, J.; Lobinski, R.; Chavatte, L. Selenium-regulated hierarchy of human selenoproteome in cancerous and immortalized cells lines. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 2493–2505. [Google Scholar] [CrossRef] [PubMed]
- Arner, E.S. Focus on mammalian thioredoxin reductases—Important selenoproteins with versatile functions. Biochim. Biophys. Acta 2009, 1790, 495–526. [Google Scholar] [PubMed]
- Arner, E.S.; Holmgren, A. Physiological functions of thioredoxin and thioredoxin reductase. Eur. J. Biochem. 2000, 267, 6102–6109. [Google Scholar] [CrossRef] [PubMed]
- Sonet, J.; Mounicou, S.; Chavatte, L. Nonradioactive Isotopic Labeling and Tracing of Selenoproteins in Cultured Cell Lines. Methods Mol. Biol. 2018, 1661, 193–203. [Google Scholar]
- Luna, D.; Lopez-Alonso, M.; Cedeno, Y.; Rigueira, L.; Pereira, V.; Miranda, M. Determination of Essential and Toxic Elements in Cattle Blood: Serum vs Plasma. Animals 2019, 9, 465. [Google Scholar] [CrossRef] [PubMed]
- Karlenius, T.C.; Shah, F.; Yu, W.C.; Hawkes, H.J.; Tinggi, U.; Clarke, F.M.; Tonissen, K.F. The selenium content of cell culture serum influences redox-regulated gene expression. Biotechniques 2011, 50, 295–301. [Google Scholar] [CrossRef] [PubMed]
- Bishop, D.P.; Hare, D.J.; Fryer, F.; Taudte, R.V.; Cardoso, B.R.; Cole, N.; Doble, P.A. Determination of selenium in serum in the presence of gadolinium with ICP-QQQ-MS. Analyst 2015, 140, 2842–2846. [Google Scholar] [CrossRef]
- Suzuki, K.T.; Ogra, Y. Metabolic pathway for selenium in the body: Speciation by HPLC-ICP MS with enriched Se. Food Addit. Contam. 2002, 19, 974–983. [Google Scholar] [CrossRef]
- Guo, L.; Yang, W.; Huang, Q.; Qiang, J.; Hart, J.R.; Wang, W.; Hu, J.; Zhu, J.; Liu, N.; Zhang, Y. Selenocysteine-Specific Mass Spectrometry Reveals Tissue-Distinct Selenoproteomes and Candidate Selenoproteins. Cell Chem. Biol. 2018, 25, 1380–1388.e4. [Google Scholar] [CrossRef]
- Koyama, H.; Omura, K.; Ejima, A.; Kasanuma, Y.; Watanabe, C.; Satoh, H. Separation of selenium-containing proteins in human and mouse plasma using tandem high-performance liquid chromatography columns coupled with inductively coupled plasma-mass spectrometry. Anal. Biochem. 1999, 267, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Awadeh, F.T.; Abdelrahman, M.M.; Kincaid, R.L.; Finley, J.W. Effect of selenium supplements on the distribution of selenium among serum proteins in cattle. J. Dairy. Sci. 1998, 81, 1089–1094. [Google Scholar] [CrossRef] [PubMed]
- Letsiou, S.; Nomikos, T.; Panagiotakos, D.B.; Pergantis, S.A.; Fragopoulou, E.; Pitsavos, C.; Stefanadis, C.; Antonopoulou, S. Gender-specific distribution of selenium to serum selenoproteins: Associations with total selenium levels, age, smoking, body mass index, and physical activity. BioFactors 2014, 40, 524–535. [Google Scholar] [CrossRef] [PubMed]
- Santos, C.; Garcia-Fuentes, E.; Callejon-Leblic, B.; Garcia-Barrera, T.; Gomez-Ariza, J.L.; Rayman, M.P.; Velasco, I. Selenium, selenoproteins and selenometabolites in mothers and babies at the time of birth. Br. J. Nutr. 2017, 117, 1304–1311. [Google Scholar] [CrossRef] [PubMed]
- Lamarche, J.; Ronga, L.; Szpunar, J.; Lobinski, R. Characterization and Quantification of Selenoprotein P: Challenges to Mass Spectrometry. Int. J. Mol. Sci. 2021, 22, 6283. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, B.R.; Ganio, K.; Roberts, B.R. Expanding beyond ICP-MS to better understand selenium biochemistry. Metallomics 2019, 11, 1974–1983. [Google Scholar] [CrossRef] [PubMed]
- Sonet, J.; Bierla, K.; Bulteau, A.L.; Lobinski, R.; Chavatte, L. Comparison of analytical methods using enzymatic activity, immunoaffinity and selenium-specific mass spectrometric detection for the quantitation of glutathione peroxidase 1. Anal. Chim. Acta 2018, 1011, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Mounicou, S.; Szpunar, J.; Lobinski, R. Metallomics: The concept and methodology. Chem. Soc. Rev. 2009, 38, 1119–1138. [Google Scholar] [CrossRef] [PubMed]
- Latreche, L.; Duhieu, S.; Touat-Hamici, Z.; Jean-Jean, O.; Chavatte, L. The differential expression of glutathione peroxidase 1 and 4 depends on the nature of the SECIS element. RNA Biol. 2012, 9, 681–690. [Google Scholar] [CrossRef]
- Arnaud, J.; Bertrais, S.; Roussel, A.M.; Arnault, N.; Ruffieux, D.; Favier, A.; Berthelin, S.; Estaquio, C.; Galan, P.; Czernichow, S.; et al. Serum selenium determinants in French adults: The SU.VI.M.AX study. Br. J. Nutr. 2006, 95, 313–320. [Google Scholar] [CrossRef]
- Hurst, R.; Armah, C.N.; Dainty, J.R.; Hart, D.J.; Teucher, B.; Goldson, A.J.; Broadley, M.R.; Motley, A.K.; Fairweather-Tait, S.J. Establishing optimal selenium status: Results of a randomized, double-blind, placebo-controlled trial. Am. J. Clin. Nutr. 2010, 91, 923–931. [Google Scholar] [CrossRef] [PubMed]
- Combs, G.F., Jr. Selenium in global food systems. Br. J. Nutr. 2001, 85, 517–547. [Google Scholar] [CrossRef] [PubMed]
- Rayman, M.P. Food-chain selenium and human health: Emphasis on intake. Br. J. Nutr. 2008, 100, 254–268. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, M.; Loser, A.; Cheng, Q.; Wichmann-Costaganna, M.; Schadel, P.; Werz, O.; Arner, E.S.; Kipp, A.P. Side-by-side comparison of recombinant human glutathione peroxidases identifies overlapping substrate specificities for soluble hydroperoxides. Redox Biol. 2023, 59, 102593. [Google Scholar] [CrossRef] [PubMed]
- Saito, Y.; Takahashi, K. Characterization of selenoprotein P as a selenium supply protein. Eur. J. Biochem. 2002, 269, 5746–5751. [Google Scholar] [CrossRef]
- Saito, Y. Selenium Transport Mechanism via Selenoprotein P-Its Physiological Role and Related Diseases. Front. Nutr. 2021, 8, 685517. [Google Scholar] [CrossRef]

| FBS | Supplier (Distributor) | Supplier Location (City, State, Country) | Reference | Batch | Origin |
|---|---|---|---|---|---|
| #1 | Gibco (Thermo Fisher Scientific) | Waltham, MA, USA | 10-437-028 | 2230743RP | Mexico |
| #2 | Gibco (Thermo Fisher Scientific) | Waltham, MA, USA | 10-437-028 | 2286113RP | Mexico |
| #3 | Eurobio Scientific | Les Ulis, France | CVFSVF00-01 | S73136 | South America |
| #4 | Avantor (VWR) | Rosny-sous-Bois cedex, France | 97068-085 | 141K20 | USA |
| #5 | Biosera | Cholet, France | FB-1280/500 | S00AV10004 | EU |
| #6 | Biowest | Nuaillé, France | S1400-500 | S00F610003 | French |
| #7 | Biowest | Nuaillé, France | S1810-500 | S00H810001 | South America |
| #8 | Merck | Darmstadt, Germany | K7524 | BCBV8085 | Non-USA origin |
| #9 | Eurobio Scientific | Les Ulis, France | CVFSVF00-0U | S72121-1218 | South America |
| #10 | Cytiva Hyclone (Thermo Fisher Scientific) | Waltham, MA, USA | SV30160.02 | RF20200006 | South America |
| #11 | Seradigm (VWR) | Rosny-sous-Bois cedex, France | 3100-050 | 48B16 | USA |
| #12 | Gibco (Thermo Fisher Scientific) | Waltham, MA, USA | 10270-106 | 41Q5484K | Brazil |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parant, F.; Mure, F.; Maurin, J.; Beauvilliers, L.; Chorfa, C.; El Jamali, C.; Ohlmann, T.; Chavatte, L. Selenium Discrepancies in Fetal Bovine Serum: Impact on Cellular Selenoprotein Expression. Int. J. Mol. Sci. 2024, 25, 7261. https://doi.org/10.3390/ijms25137261
Parant F, Mure F, Maurin J, Beauvilliers L, Chorfa C, El Jamali C, Ohlmann T, Chavatte L. Selenium Discrepancies in Fetal Bovine Serum: Impact on Cellular Selenoprotein Expression. International Journal of Molecular Sciences. 2024; 25(13):7261. https://doi.org/10.3390/ijms25137261
Chicago/Turabian StyleParant, François, Fabrice Mure, Julien Maurin, Léana Beauvilliers, Chaïma Chorfa, Chaymae El Jamali, Théophile Ohlmann, and Laurent Chavatte. 2024. "Selenium Discrepancies in Fetal Bovine Serum: Impact on Cellular Selenoprotein Expression" International Journal of Molecular Sciences 25, no. 13: 7261. https://doi.org/10.3390/ijms25137261
APA StyleParant, F., Mure, F., Maurin, J., Beauvilliers, L., Chorfa, C., El Jamali, C., Ohlmann, T., & Chavatte, L. (2024). Selenium Discrepancies in Fetal Bovine Serum: Impact on Cellular Selenoprotein Expression. International Journal of Molecular Sciences, 25(13), 7261. https://doi.org/10.3390/ijms25137261







