Identifying Key Genes Involved in Axillary Lymph Node Metastasis in Breast Cancer Using Advanced RNA-Seq Analysis: A Methodological Approach with GLMQL and MAS
Abstract
1. Introduction
- Implementation of Generalized Linear Models with Quasi-Likelihood F-tests and Magnitude Altitude Scoring (GLMQL-MAS) [31,32]: Our application of GLMQL-MAS addresses the challenges posed by the discrete and overdispersed nature of RNA-Seq data. This approach enhances our capability to accurately identify and analyze key genes associated with ALNM, surpassing the limitations of traditional statistical methods.
- Utilization of Trimmed Mean of M-values (TMM) for Normalization [33]: We adopt TMM normalization to correct for library-specific compositional differences, significantly improving the accuracy of gene expression measurements compared to conventional methods such as TPM [34] or FPKM [21] normalizations.
- Focus on Untreated Patient Cohort: To the best of our knowledge, this is the first study that analyzes RNA-Seq data from a cohort of untreated breast cancer patients specifically for ALNM research. This unique focus allows us to observe the natural tumor environment without the confounding effects of prior treatments, enhancing the reliability of our findings in understanding the genetic underpinnings of lymph node metastasis.
- Exclusive Analysis of Protein-Coding Genes: By concentrating on protein-coding genes, our research targets those genomic elements most directly involved in cellular functions and disease mechanisms, ensuring that our findings are biologically significant and directly applicable to potential therapeutic interventions. Although this approach leaves out non-coding regions, which may include fragments involved in gene translation (such as microRNAs) and potentially influence gene expression, focusing on protein-coding genes provides clearer insights into disease mechanisms and therapeutic targets.
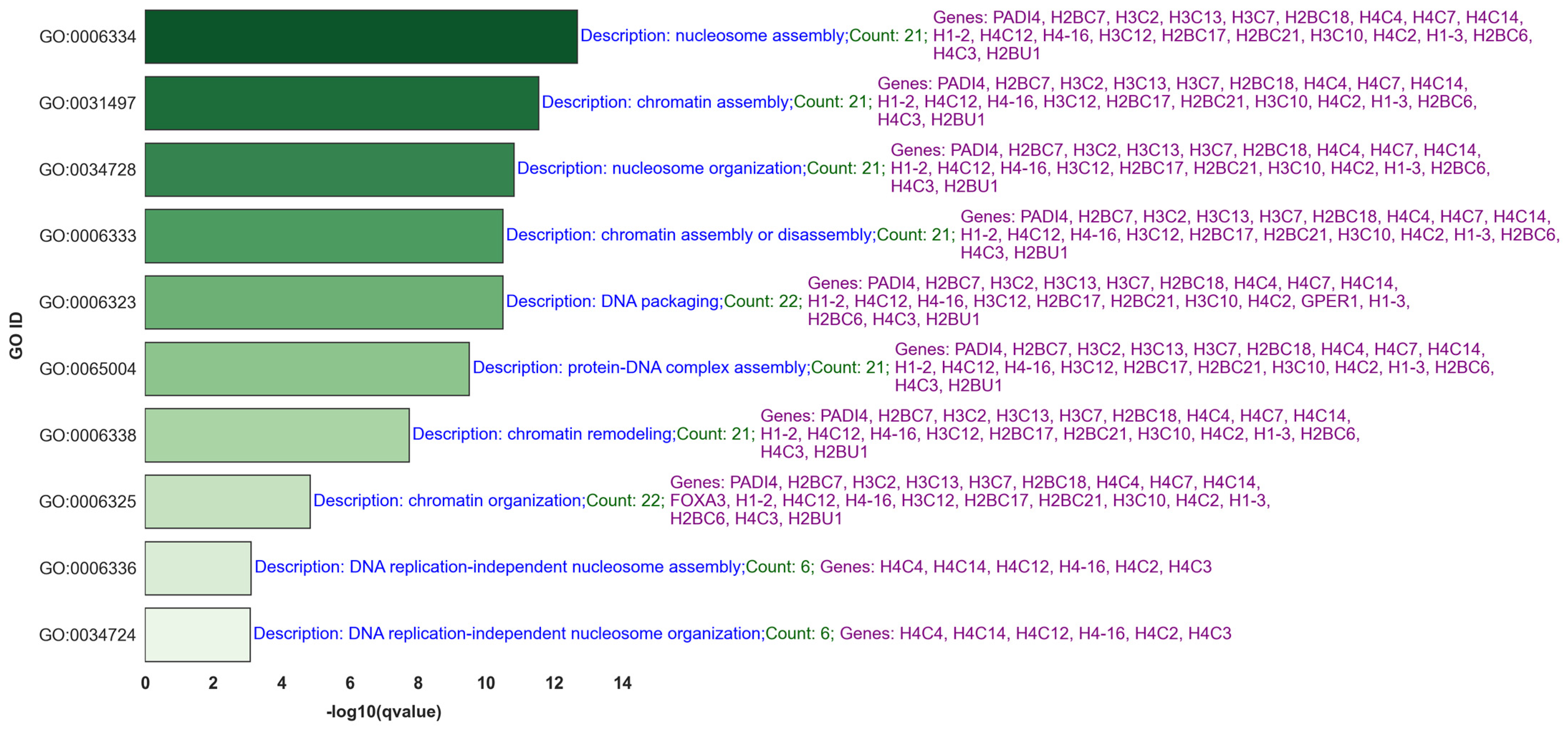
- Comprehensive Gene Ontology (GO) [35] and GSEA Hallmark Set [36,37] Analysis: Our study extends beyond traditional gene expression analysis by employing comprehensive GO and GSEA Hallmark set analyses. This approach allows us to categorize significant genes and identify disrupted biological pathways, providing actionable insights that could lead to the development of markers for early detection and targeted treatment strategies.
2. Results
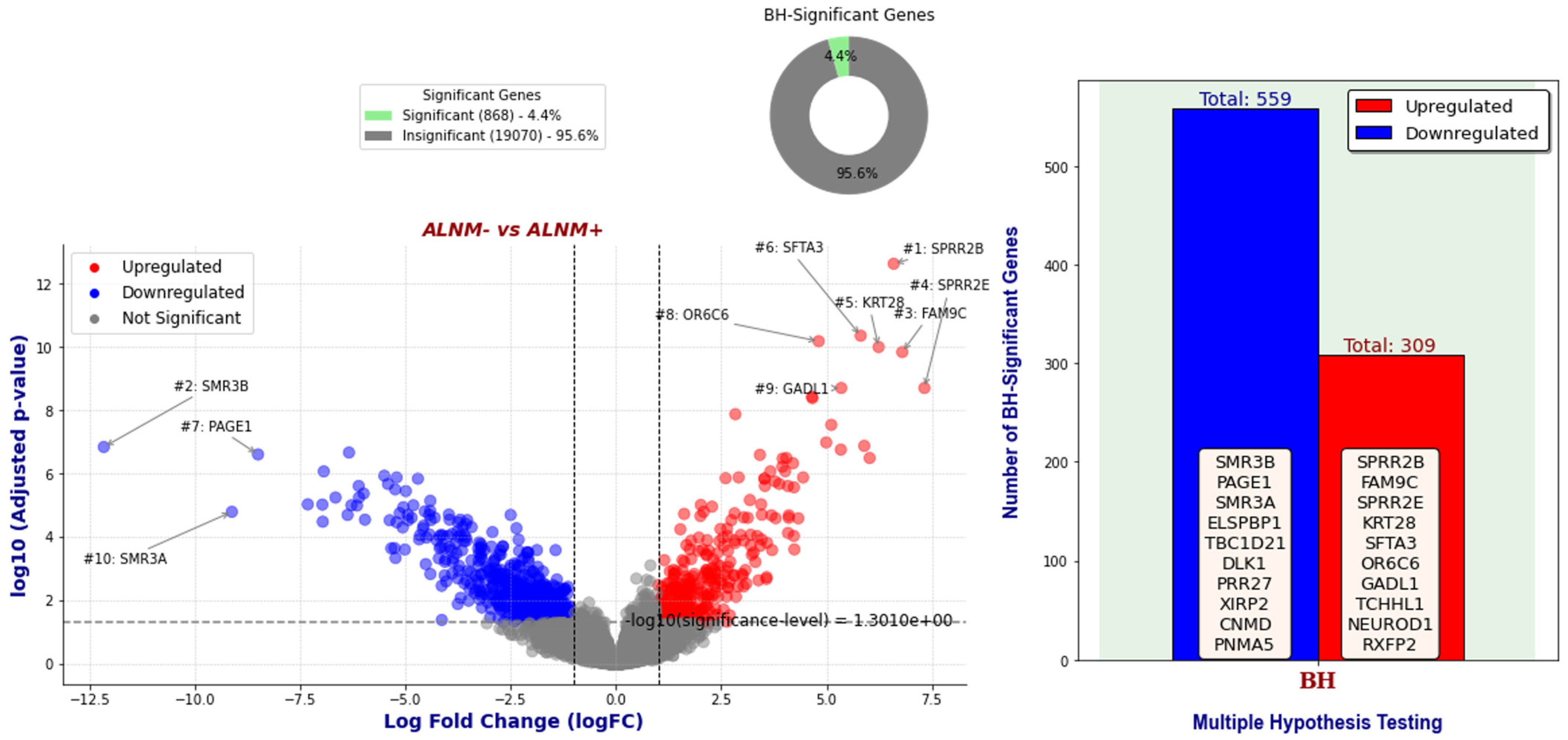
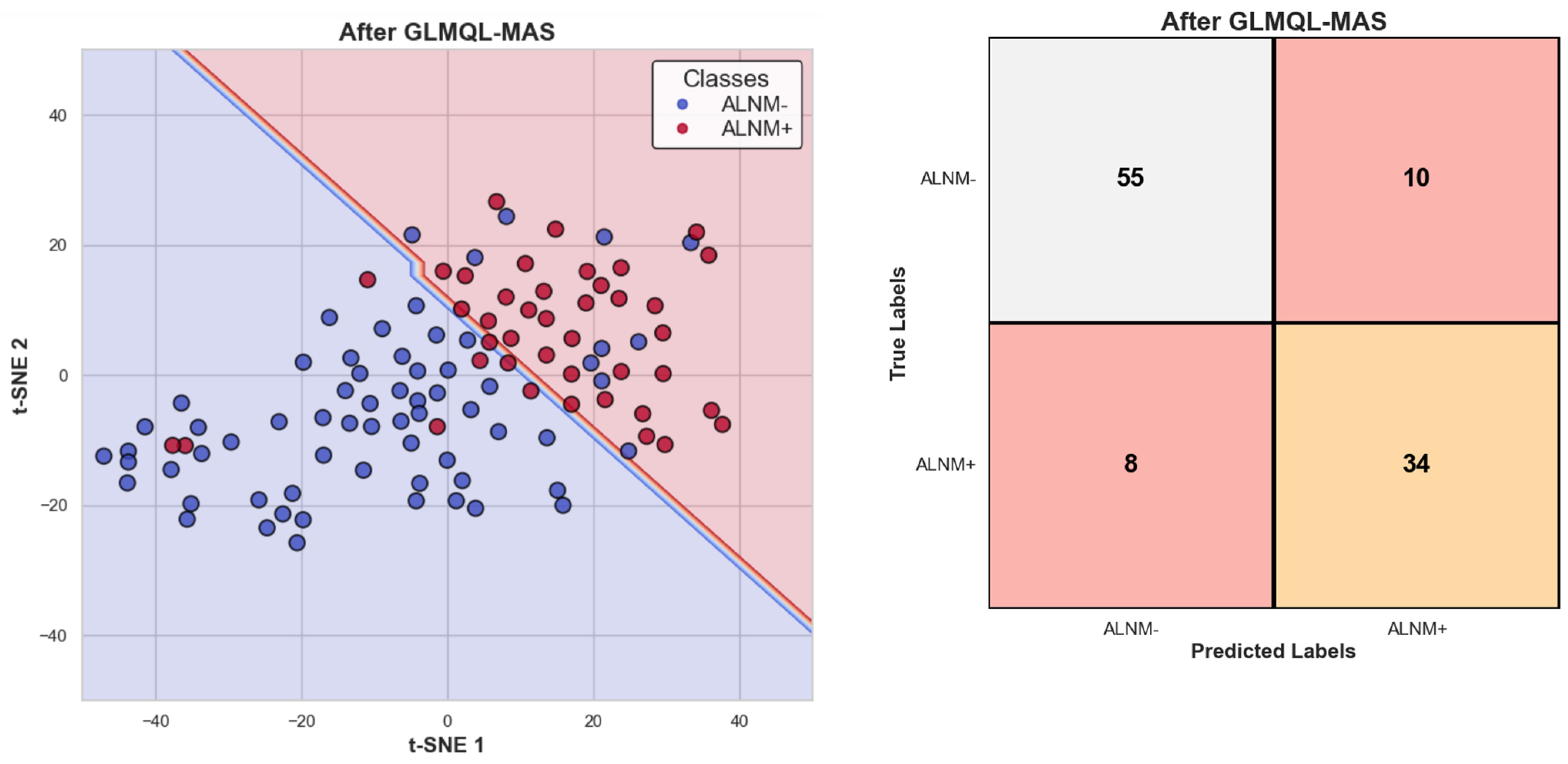
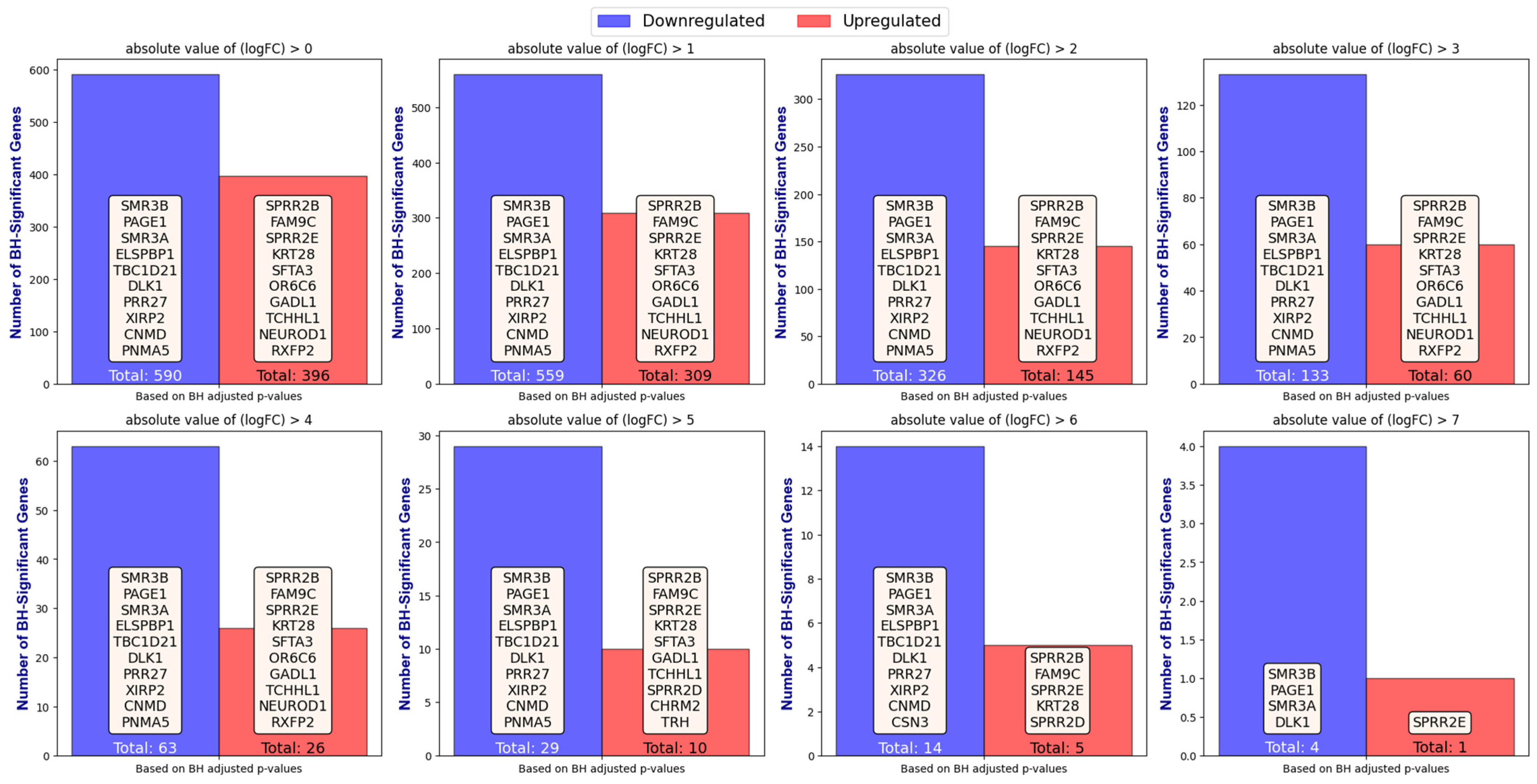
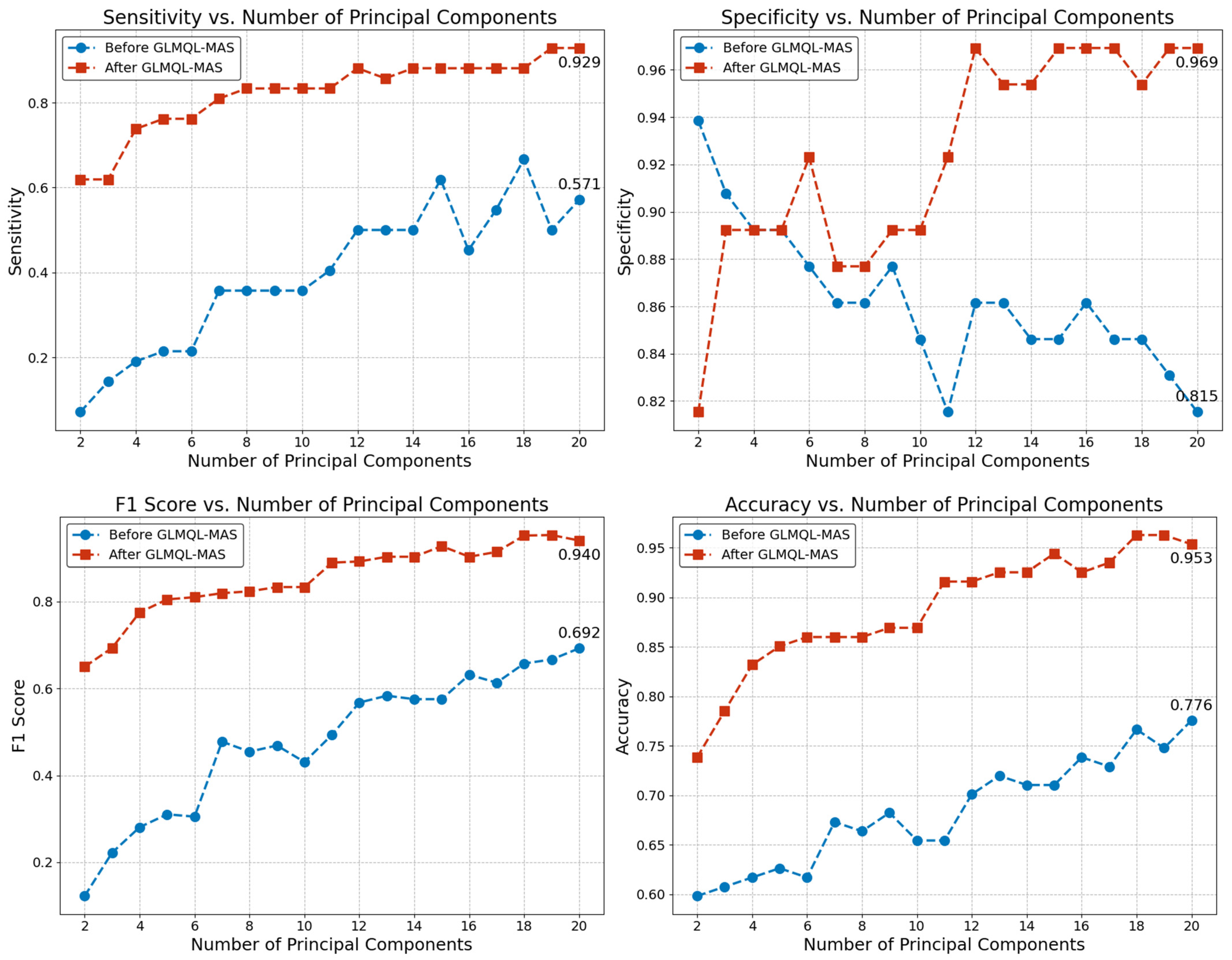
2.1. Applying GLMQL-MAS for the Analysis of Axillary Lymph Node Metastasis
Differential Expression Analysis
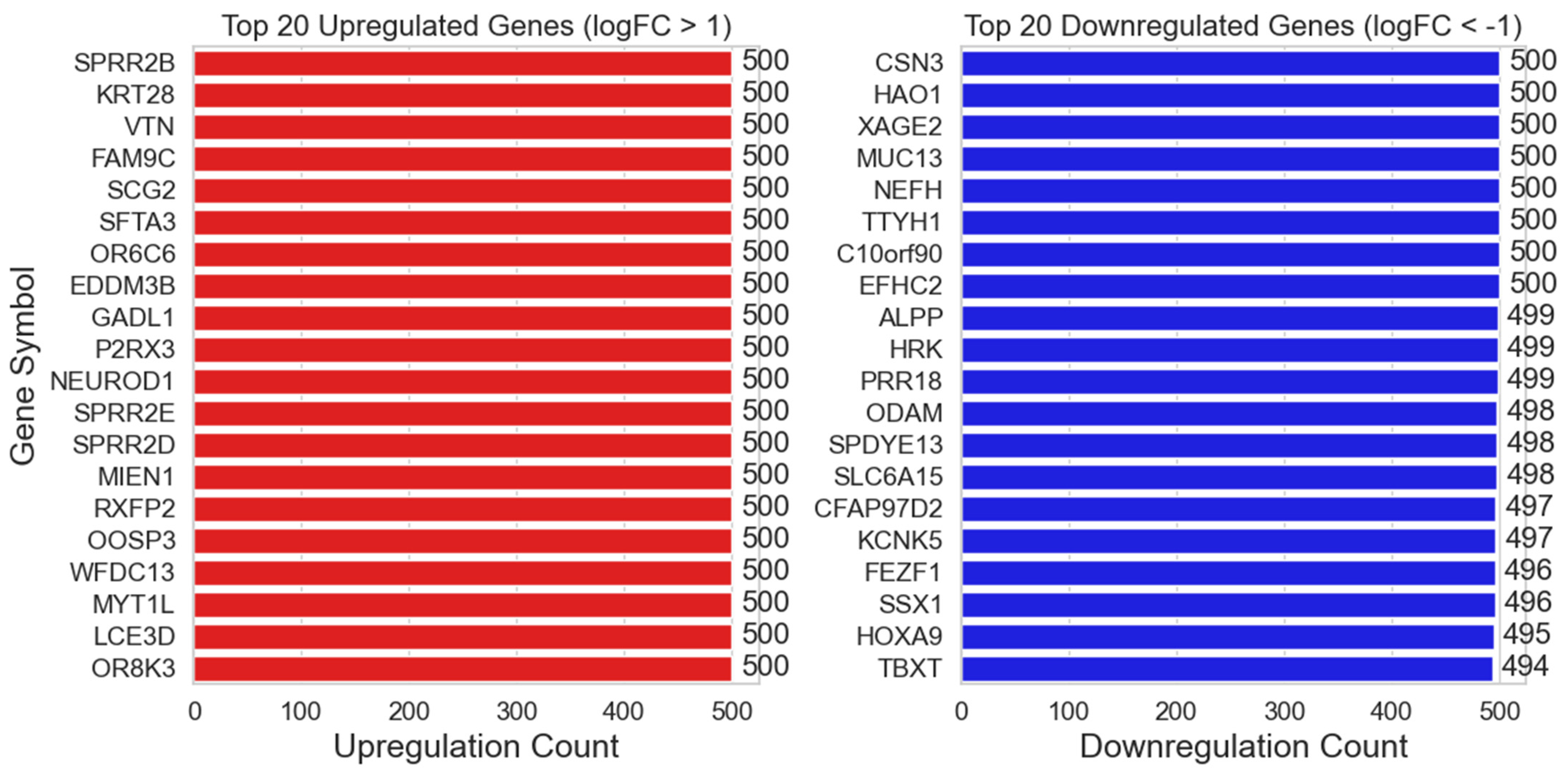
- Case 1: 65 ALNM− Versus 42 ALNM+:
- Case 2. Random Sampling of 42 ALNM− to Compare Against 42 ALNM+:
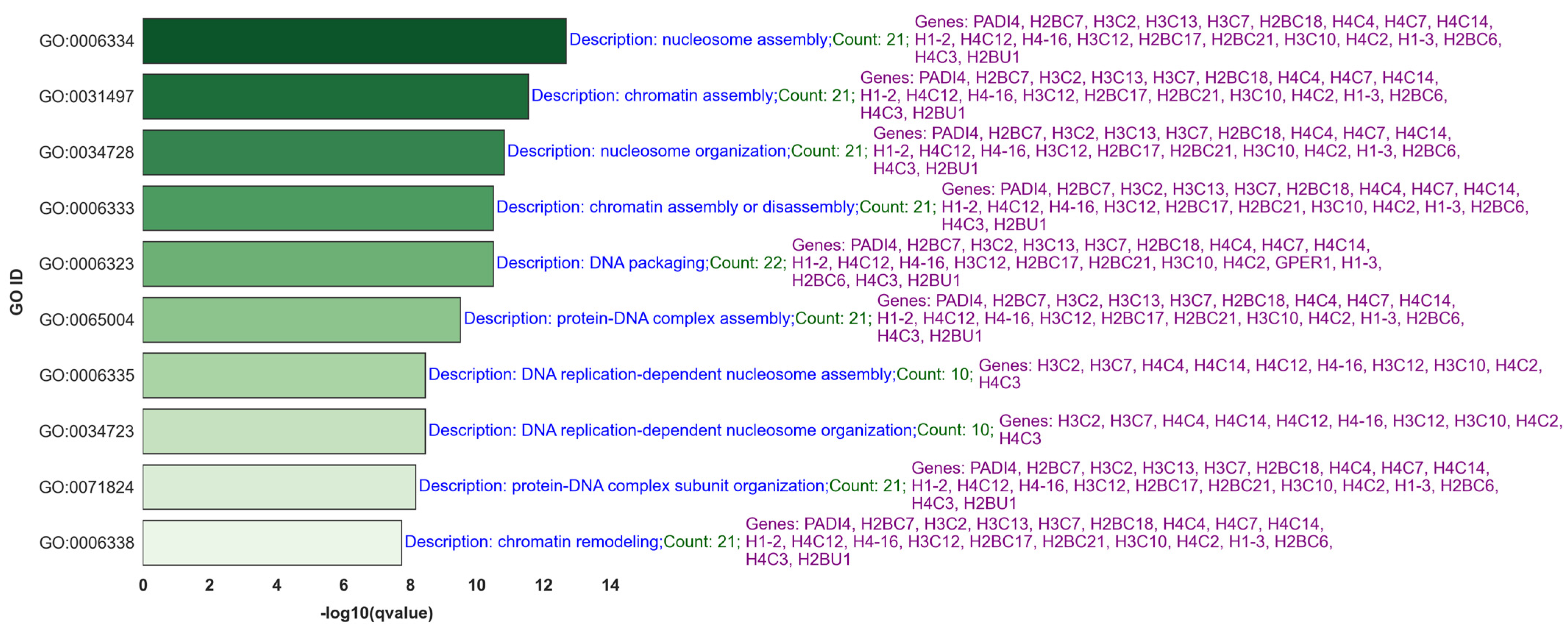
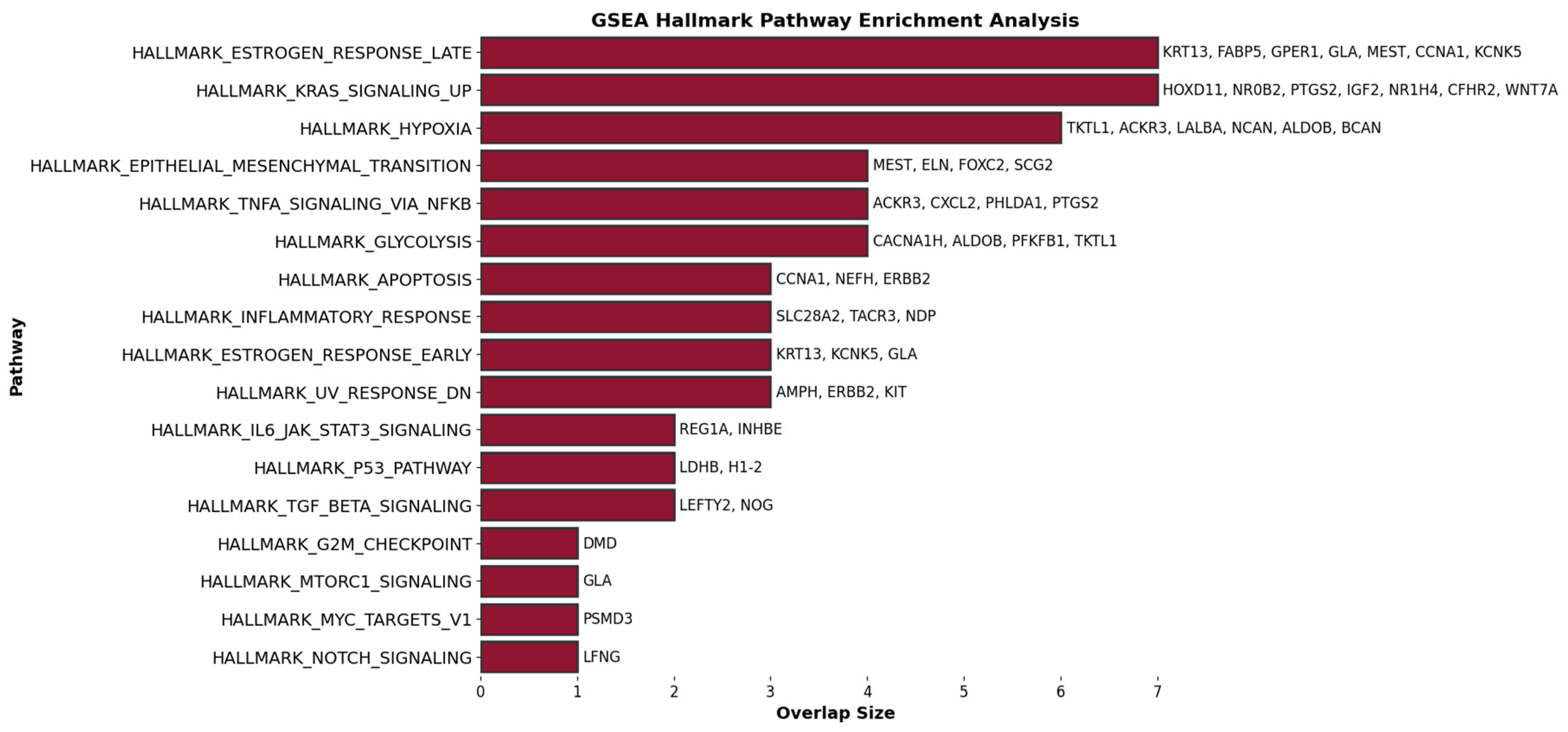
2.2. Comprehensive Gene Ontology (GO) and Gene Set Enrichment Analysis (GSEA) of Hallmark Gene Sets in Lymph Node Metastasis of Breast Cancer
- Case 1. 65 ALNM− Versus 42 ALNM+:
- Case 2. Random Sampling of 42 ALNM− to Compare Against 42 ALNM+:
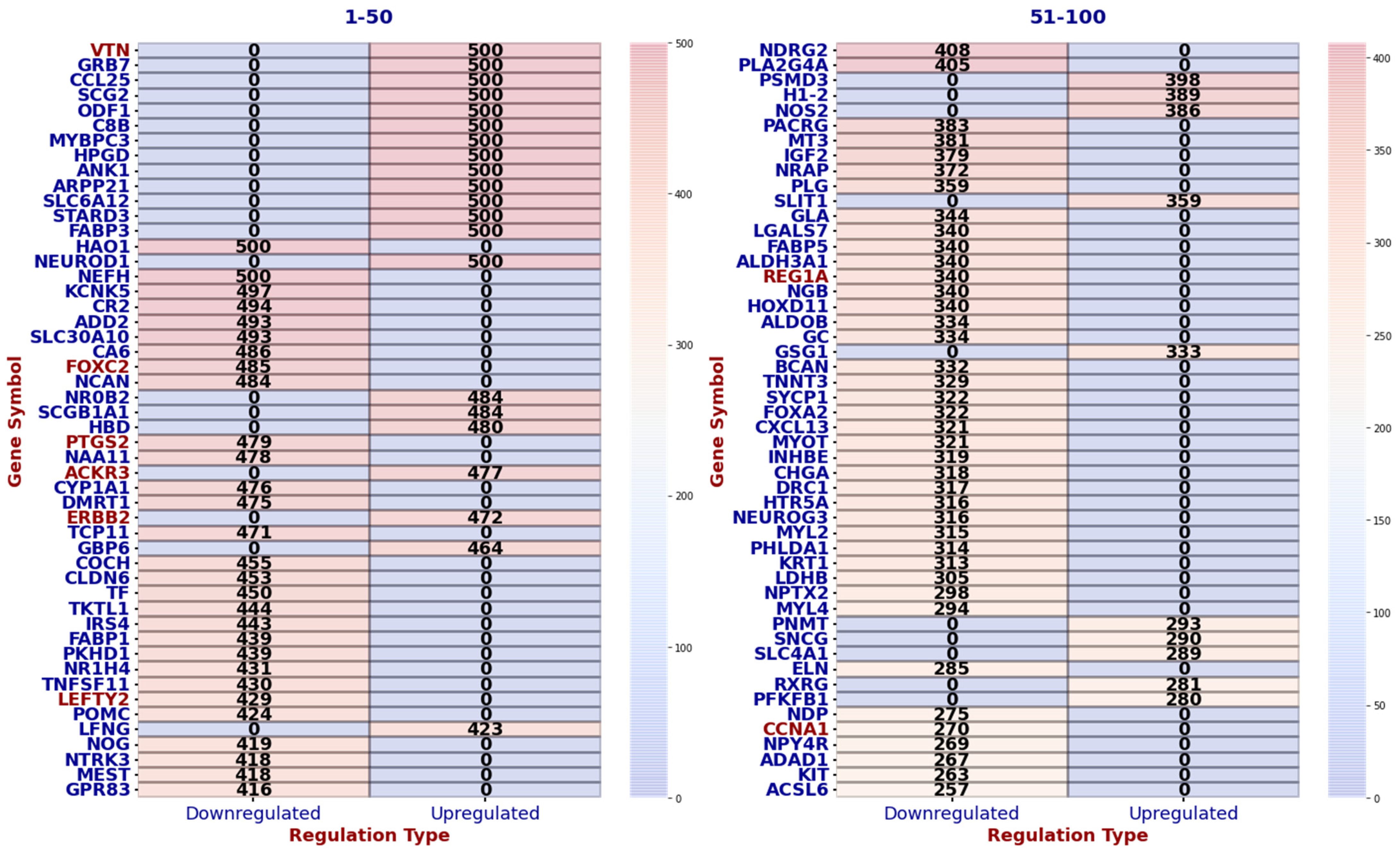
Common Genes and GO and GSEA Hallmark Processes in Cases 1 and 2
3. Discussion
3.1. Applying GLMQL-MAS for the Analysis of Axillary Lymph Node Metastasis
3.2. Comprehensive Gene Ontology (GO) and Gene Set Enrichment Analysis (GSEA) of Hallmark Gene Sets in Lymph Node Metastasis of Breast Cancer
4. Materials and Methods
4.1. Generalized Linear Models with Quasi-Likelihood F-Tests and Magnitude-Altitude Scoring (GLMQL-MAS)
- Formation of Design Matrix:
- A design matrix is constructed to reflect the experimental design, encapsulating all factors believed to influence gene expression (model.matrix()).
- It organizes samples in rows and experimental factors in columns, facilitating the modeling of gene expression influences.
- GLM Fitting:
- The TMM-normalized count data are subjected to a GLM fitting process, utilizing a link function appropriate for count data, typically the log link (glmQLFit()).
- The GLM quantifies the expected count values’ relationship to the linear predictors, with coefficients indicating log fold changes (LogFC) for each experimental factor.
- Application of Quasi-Likelihood F-test:
- Post model fitting, the Quasi-Likelihood F-test compares the full model against a reduced model to evaluate the impact of specific factors on gene expression (glmQLFTest()).
- It directly estimates data dispersion, addressing the overdispersion characteristic of RNA-Seq data.
- The test produces p-values to assess the significance of expression differences attributed to the experimental factors.
- Extraction of Significance and Log Fold Change (LogFC):
- Genes showing significant model deviations yield p-values.
- Concurrently, GLM calculates LogFC values, reflecting the magnitude of expression change between treatment and reference conditions.
4.2. Applying GLMQL-MAS for the Analysis of Axillary Lymph Node Metastasis
Differential Expression Analysis
- Case 1. 65 ALNM− Versus 42 ALNM+:
- Case 2. Random Sampling of 42 ALNM− to Compare Against 42 ALNM+:
4.3. Comprehensive Gene Ontology (GO) and Gene Set Enrichment Analysis (GSEA) of Hallmark Gene Sets in Lymph Node Metastasis of Breast Cancer
5. Conclusions
Limitations of Study
- The limited sample size significantly constrains the statistical power of our analyses, stemming from a focus on untreated patients within the TCGA Breast Invasive Carcinoma (BRCA) cohort. This decision was made to avoid the confounding effects of prior therapies, which could influence gene expression profiles.
- The reliance on a predefined dataset from untreated patients limits the diversity of the patient population and does not reflect the typical clinical scenario where patients may receive neoadjuvant therapies. These selection criteria restrict our findings’ generalizability to all breast cancer patients.
- The demographic and geographic homogeneity of the TCGA cohort might introduce selection bias, potentially influencing the study’s conclusions regarding populations that differ in ethnicity, age, or treatment history.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cronin, K.A.; Lake, A.J.; Scott, S.; Sherman, R.L.; Noone, A.M.; Howlader, N.; Henley, S.J.; Anderson, R.N.; Firth, A.U.; Ma, J.; et al. Annual Report to the Nation on the Status of Cancer, part I: National cancer statistics. Cancer 2018, 124, 2785–2800. [Google Scholar] [CrossRef]
- Negoita, S.; Chen, H.S.; Sanchez, P.V.; Sherman, R.L.; Henley, S.J.; Siegel, R.L.; Sung, H.; Scott, S.; Benard, V.B.; Kohler, B.A.; et al. Annual Report to the Nation on the Status of Cancer, part 2: Early assessment of the COVID-19 pandemic’s impact on cancer diagnosis. Cancer 2024, 130, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Cancer Stat Facts: Female Breast Cancer. Available online: https://seer.cancer.gov/statfacts/html/breast.html (accessed on 10 January 2024).
- DeSantis, C.E.; Bray, F.; Ferlay, J.; Lortet-Tieulent, J.; Anderson, B.O.; Jemal, A. International variation in female breast cancer incidence and mortality rates. Cancer Epidemiol. Biomark. Prev. 2015, 24, 1495–1506. [Google Scholar] [CrossRef] [PubMed]
- American Cancer Society. Available online: https://www.cancer.org/research/cancer-facts-statistics/all-cancer-facts-figures/2024-cancer-facts-figures.html (accessed on 10 January 2024).
- Humphrey, K.L.; Saksena, M.A.; Freer, P.E.; Smith, B.L.; Rafferty, E.A. To do or not to do: Axillary nodal evaluation after ACOSOG Z0011 trial. Radiographics 2014, 34, 1807–1816. [Google Scholar] [CrossRef] [PubMed]
- Carter, C.L.; Allen, C.; Henson, D.E. Relation of tumor size, lymph node status, and survival in 24,740 breast cancer cases. Cancer 1989, 63, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Razeq, H.; Mansour, A.; Edaily, S.; Dayyat, A. Delays in Initiating Anti-Cancer Therapy for Early-Stage Breast Cancer—How Slow Can We Go? J. Clin. Med. 2023, 12, 4502. [Google Scholar] [CrossRef]
- Hanna, T.P.; King, W.D.; Thibodeau, S.; Jalink, M.; Paulin, G.A.; Harvey-Jones, E.; O’Sullivan, D.E.; Booth, C.M.; Sullivan, R.; Aggarwal, A. Mortality due to cancer treatment delay: Systematic review and meta-analysis. BMJ 2020, 371, m4087. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Bhagabaty, S.M.; Tripathy, J.P.; Selvaraj, K.; Purkayastha, J.; Singh, R. Delays in diagnosis and treatment of breast cancer and the pathways of care: A mixed methods study from a tertiary cancer centre in North East India. Asian Pac. J. Cancer Prev. 2019, 20, 3711–3721. [Google Scholar] [CrossRef] [PubMed]
- Ginsburg, O.; Yip, C.; Brooks, A.; Cabanes, A.; Caleffi, M.; Yataco, J.A.D.; Gyawali, B.; McCormack, V.; de Anderson, M.M.; Mehrotra, R.; et al. Breast cancer early detection: A phased approach to implementation. Cancer 2020, 126, 2379–2393. [Google Scholar] [CrossRef]
- Biganzoli, L.; Cardoso, F.; Beishon, M.; Cameron, D.; Cataliotti, L.; Coles, C.E.; Bolton, R.C.D.; Trill, M.D.; Erdem, S.; Fjell, M.; et al. The requirements of a specialist breast centre. Breast 2020, 51, 65–84. [Google Scholar] [CrossRef]
- de Boniface, J.; Tvedskov, T.F.; Rydén, L.; Szulkin, R.; Reimer, T.; Kühn, T.; Kontos, M.; Gentilini, O.D.; Bagge, R.O.; Sund, M.; et al. Omitting axillary dissection in breast cancer with sentinel-node metastases. N. Engl. J. Med. 2024, 390, 1163–1175. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhang, C.; Du, J.; Zhang, R.; Yang, S.; Li, B.; Wang, P.; Deng, W. Prediction of lymph-node metastasis in cancers using differentially expressed mRNA and non-coding RNA signatures. Front. Cell Dev. Biol. 2021, 9, 605977. [Google Scholar] [CrossRef] [PubMed]
- Luo, N.; Wen, Y.; Zou, Q.; Ouyang, D.; Chen, Q.; Zeng, L.; He, H.; Anwar, M.; Qu, L.; Ji, J.; et al. Construction and validation of a risk prediction model for clinical axillary lymph node metastasis in T1–2 breast cancer. Sci. Rep. 2022, 12, 687. [Google Scholar] [CrossRef]
- Li, X.; Yang, L.; Jiao, X. Development and validation of a nomogram for predicting axillary lymph node metastasis in breast cancer. Clin. Breast Cancer 2023, 23, 538–545. [Google Scholar] [CrossRef]
- Dihge, L.; Vallon-Christersson, J.; Hegardt, C.; Saal, L.H.; Häkkinen, J.; Larsson, C.; Ehinger, A.; Loman, N.; Malmberg, M.; Bendahl, P.O.; et al. Prediction of lymph node metastasis in breast cancer by gene expression and clinicopathological models: Development and validation within a population-based cohort. Clin. Cancer Res. 2019, 25, 6368–6381. [Google Scholar] [CrossRef]
- Shiino, S.; Matsuzaki, J.; Shimomura, A.; Kawauchi, J.; Takizawa, S.; Sakamoto, H.; Aoki, Y.; Yoshida, M.; Tamura, K.; Kato, K.; et al. Serum miRNA–based prediction of axillary lymph node metastasis in breast cancer. Clin. Cancer Res. 2019, 25, 1817–1827. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, S. Prediction of tumor lymph node metastasis using wasserstein distance-based generative adversarial networks combing with neural architecture search for predicting. Mathematics 2023, 11, 729. [Google Scholar] [CrossRef]
- Li, B.; Tian, Y.; Tian, Y.; Zhang, S.; Zhang, X. Predicting cancer lymph-node metastasis from LncRNA expression profiles using local linear reconstruction guided distance metric learning. IEEE/ACM Trans. Comput. Biol. Bioinform. 2022, 19, 3179–3189. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Li, M.-C.; Konaté, M.M.; Chen, L.; Das, B.; Karlovich, C.; Williams, P.M.; Evrard, Y.A.; Doroshow, J.H.; McShane, L.M. TPM, FPKM, or normalized counts? A comparative study of quantification measures for the analysis of RNA-seq data from the NCI patient-derived models repository. J. Transl. Med. 2021, 19, 269. [Google Scholar] [CrossRef]
- Kim, G.-E.; Kim, N.I.; Lee, J.S.; Park, M.H.; Kang, K. Differentially expressed genes in matched normal, cancer, and lymph node metastases predict clinical outcomes in patients with breast cancer. Appl. Immunohistochem. Mol. Morphol. 2020, 28, 111–122. [Google Scholar] [CrossRef]
- Trapnell, C.; Pachter, L.; Salzberg, S.L. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [PubMed]
- Liang, F.; Qu, H.; Lin, Q.; Yang, Y.; Ruan, X.; Zhang, B.; Liu, Y.; Yu, C.; Zhang, H.; Fang, X.; et al. Molecular biomarkers screened by next-generation RNA sequencing for non-sentinel lymph node status prediction in breast cancer patients with metastatic sentinel lymph nodes. World J. Surg. Oncol. 2015, 13, 258. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Hendrickson, D.G.; Sauvageau, M.; Goff, L.; Rinn, J.L.; Pachter, L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 2013, 31, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Lyman, G.H.; Temin, S.; Edge, S.B.; Newman, L.A.; Turner, R.R.; Weaver, D.L.; Benson, A.B., III; Bosserman, L.D.; Burstein, H.J.; Cody, H., III; et al. Sentinel lymph node biopsy for patients with early-stage breast cancer: American Society of Clinical Oncology clinical practice guideline update. J. Clin. Oncol. 2014, 32, 1365–1383. [Google Scholar] [CrossRef] [PubMed]
- Weaver, D.L.; Ashikaga, T.; Krag, D.N.; Skelly, J.M.; Anderson, S.J.; Harlow, S.P.; Julian, T.B.; Mamounas, E.P.; Wolmark, N. Effect of occult metastases on survival in node-negative breast cancer. N. Engl. J. Med. 2011, 364, 412–421. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Rezapour, M.; Walker, S.J.; Ornelles, D.A.; McNutt, P.M.; Atala, A.; Gurcan, M.N. Analysis of gene expression dynamics and differential expression in viral infections using generalized linear models and quasi-likelihood methods. Front. Microbiol. 2024, 15, 1342328. [Google Scholar] [CrossRef]
- Rezapour, M.; Walker, S.J.; Ornelles, D.A.; Niazi, M.K.K.; McNutt, P.M.; Atala, A.; Gurcan, M.N. A Comparative Analysis of RNA-Seq and NanoString Technologies in Deciphering Viral Infection Response in Upper Airway Lung Organoids. Front. Genet. 2024, 15, 1327984. [Google Scholar] [CrossRef]
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010, 11, R25. [Google Scholar] [CrossRef] [PubMed]
- Wagner, G.P.; Kin, K.; Lynch, V.J. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 2012, 131, 281–285. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Liberzon, A.; Subramanian, A.; Pinchback, R.; Thorvaldsdóttir, H.; Tamayo, P.; Mesirov, J.P. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011, 27, 1739–1740. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Van der Maaten, L.; Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 2008, 9, 2579–2605. [Google Scholar]
- Balakrishnama, S.; Ganapathiraju, A. Linear discriminant analysis—A brief tutorial. Inst. Signal Inf. Process. 1998, 18, 1–8. [Google Scholar]
- Carregaro, F.; Stefanini, A.C.B.; Henrique, T.; Tajara, E.H. Study of small proline-rich proteins (SPRRs) in health and disease: A review of the literature. Arch. Dermatol. Res. 2013, 305, 857–866. [Google Scholar] [CrossRef] [PubMed]
- Yao, L.; Yan, J.; Cheng, F.; Gan, L.; Huang, Y.; Zheng, L.; Fang, N. Small proline-rich protein 2B Facilitates gastric adenocarcinoma proliferation via MDM2-p53/p21 signaling pathway. OncoTargets Ther. 2021, 14, 1453–1463. [Google Scholar] [CrossRef]
- Zhang, Y.-Y.; Mao, M.-H.; Han, Z.-X. Identification of a gene prognostic signature for oral squamous cell carcinoma by RNA sequencing and bioinformatics. BioMed Res. Int. 2021, 2021, 6657767. [Google Scholar] [CrossRef]
- Hao, S.-S. Gene Expression Profile of Early Prostate Cancer Cells. Doctoral Dissertation, UC San Diego, La Jolla, CA, USA, 2016. [Google Scholar]
- Liao, Z.; Zhou, J.; Xia, R.; Zhu, J.; Jia, Y.; Deng, Y. Construction and Evaluation of a Prognostic Model Based on Metastasis-Associated Genes in Breast Cancer. 2023. Available online: https://ssrn.com/abstract=4674392 (accessed on 20 April 2024).
- Cilek, E.E.; Ozturk, H.; Gur Dedeoglu, B. Construction of miRNA-miRNA networks revealing the complexity of miRNA-mediated mechanisms in trastuzumab treated breast cancer cell lines. PLoS ONE 2017, 12, e0185558. [Google Scholar] [CrossRef]
- Takan, I.; Karakülah, G.; Louka, A.; Pavlopoulou, A. “In the light of evolution:” keratins as exceptional tumor biomarkers. PeerJ 2023, 11, e15099. [Google Scholar] [CrossRef] [PubMed]
- Larson, M.H.; Pan, W.; Kim, H.J.; Mauntz, R.E.; Stuart, S.M.; Pimentel, M.; Zhou, Y.; Knudsgaard, P.; Demas, V.; Aravanis, A.M.; et al. A comprehensive characterization of the cell-free transcriptome reveals tissue- and subtype-specific biomarkers for cancer detection. Nat. Commun. 2021, 12, 2357. [Google Scholar] [CrossRef]
- Zhan, C.; Yan, L.; Wang, L.; Sun, Y.; Wang, X.; Lin, Z.; Zhang, Y.; Shi, Y.; Jiang, W.; Wang, Q. Identification of immunohistochemical markers for distinguishing lung adenocarcinoma from squamous cell carcinoma. J. Thorac. Dis. 2015, 7, 1398–1405. [Google Scholar] [CrossRef] [PubMed]
- Hirata, A.H.d.L.; Camargo, L.A.d.J.R.; da Silva, V.A.; de Almeida, R.J.; Bacigalupo, L.d.S.; Albejante, M.C.; Curi, F.S.D.; Varela, P.; Martins, L.; Pesquero, J.B.; et al. Exploring the Potential of Olfactory Receptor Circulating RNA Measurement for Preeclampsia Prediction and Its Linkage to Mild Gestational Hypothyroidism. Int. J. Mol. Sci. 2023, 24, 16681. [Google Scholar] [CrossRef]
- Wang, W.-Y.; Cao, Y.-X.; Zhou, X.; Wei, B.; Zhan, L.; Sun, S.-Y. Stimulative role of ST6GALNAC1 in proliferation, migration and invasion of ovarian cancer stem cells via the Akt signaling pathway. Cancer Cell Int. 2019, 19, 86. [Google Scholar] [CrossRef]
- Fiegl, H.; Jones, A.; Hauser-Kronberger, C.; Hutarew, G.; Reitsamer, R.; Jones, R.L.; Dowsett, M.; Mueller-Holzner, E.; Windbichler, G.; Daxenbichler, G.; et al. Methylated NEUROD1 promoter is a marker for chemosensitivity in breast cancer. Clin. Cancer Res. 2008, 14, 3494–3502. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ikematsu, Y.; Tanaka, K.; Toyokawa, G.; Ijichi, K.; Ando, N.; Yoneshima, Y.; Iwama, E.; Inoue, H.; Tagawa, T.; Nakanishi, Y.; et al. NEUROD1 is highly expressed in extensive-disease small cell lung cancer and promotes tumor cell migration. Lung Cancer 2020, 146, 97–104. [Google Scholar] [CrossRef]
- Sheau, Y.H.; Nakabayashi, K.; Nishi, S.; Kumagai, J.; Kudo, M.; Sherwood, O.D.; Hsueh, A.J.W. Activation of orphan receptors by the hormone relaxin. Science 2002, 295, 671–674. [Google Scholar] [CrossRef]
- Bathgate, R.A.D.; Halls, M.L.; van der Westhuizen, E.T.; Callander, G.E.; Kocan, M.; Summers, R.J. Relaxin family peptides and their receptors. Physiol. Rev. 2013, 93, 405–480. [Google Scholar] [CrossRef]
- Halls, M.L.; Bathgate, R.A.; Summers, R.J. Comparison of signaling pathways activated by the relaxin family peptide receptors, RXFP1 and RXFP2, using reporter genes. J. Pharmacol. Exp. Ther. 2007, 320, 281–290. [Google Scholar] [CrossRef]
- Chen, D.; Bao, X.; Zhang, R.; Ding, Y.; Zhang, M.; Li, B.; Zhang, H.; Li, X.; Tong, Z.; Liu, L.; et al. Depiction of the genomic and genetic landscape identifies CCL5 as a protective factor in colorectal neuroendocrine carcinoma. Br. J. Cancer 2021, 125, 994–1002. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Zhang, J.; Xia, R.; Wang, X.; Yang, J.; Xie, F.; Zhou, Q.; Li, J.; Zhang, T.; Chen, Q.; et al. The role of histone H1.2 in pancreatic cancer metastasis and chemoresistance. Drug Resist. Updat. 2024, 73, 101027. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Qu, Y.; Teng, X.; Xing, Y.; Li, D.; Li, C.; Cai, L. PADI4-mediated epithelial-mesenchymal transition in lung cancer cells. Mol. Med. Rep. 2019, 19, 3087–3094. [Google Scholar] [CrossRef] [PubMed]
- Bonner, E.R.; Dawood, A.; Gordish-Dressman, H.; Eze, A.; Bhattacharya, S.; Yadavilli, S.; Mueller, S.; Waszak, S.M.; Nazarian, J. Pan-cancer atlas of somatic core and linker histone mutations. NPJ Genom. Med. 2023, 8, 23. [Google Scholar] [CrossRef]
- Hannan, S.; Hami, I.; Dey, R.K.; Das Gupta, S. A Systematic Exploration of Key Candidate Genes and Pathways in the Biogenesis of Human Gastric Cancer: A Comprehensive Bioinformatics Investigation. J. Transl. Gastroenterol. 2024, 2, 9–20. [Google Scholar] [CrossRef]
- Jafari, S.; Ravan, M.; Karimi-Sani, I.; Aria, H.; Hasan-Abad, A.M.; Banasaz, B.; Atapour, A.; Sarab, G.A. Screening and identification of potential biomarkers for pancreatic cancer: An integrated bioinformatics analysis. Pathol.-Res. Pract. 2023, 249, 154726. [Google Scholar] [CrossRef]
- Jia, J.; Han, Z.; Wang, X.; Zheng, X.; Wang, S.; Cui, Y. H2B gene family: A prognostic biomarker and correlates with immune infiltration in glioma. Front. Oncol. 2022, 12, 966817. [Google Scholar] [CrossRef]
- Tang, W.; Hao, X.; He, F.; Li, L.; Xu, L. Abstract 565: Anti-CD44 antibody treatment inhibits pancreatic cancer metastasis and post-radiotherapy recurrence. Cancer Res. 2011, 71, 565. [Google Scholar] [CrossRef]
- Espiritu, D.; Gribkova, A.K.; Gupta, S.; Shaytan, A.K.; Panchenko, A.R. Molecular mechanisms of oncogenesis through the lens of nucleosomes and histones. J. Phys. Chem. B 2021, 125, 3963–3976. [Google Scholar] [CrossRef]
- Medrzycki, M.; Zhang, Y.; Zhang, W.; Cao, K.; Pan, C.; Lailler, N.; McDonald, J.F.; Bouhassira, E.E.; Fan, Y. Histone h1. 3 suppresses h19 noncoding RNA expression and cell growth of ovarian cancer cells. Cancer Res. 2014, 74, 6463–6473. [Google Scholar] [CrossRef] [PubMed]
- Rashid, M.; Shah, S.G.; Verma, T.; Chaudhary, N.; Rauniyar, S.; Patel, V.B.; Gera, P.B.; Smoot, D.; Ashaktorab, H.; Dalal, S.N.; et al. Tumor-specific overexpression of histone gene, H3C14 in gastric cancer is mediated through EGFR-FOXC1 axis. Biochim. Biophys. Acta BBA-Gene Regul. Mech. 2021, 1864, 194703. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Li, L.; Kang, Z.; Luo, H.; Lin, X.; Zhao, S.; Zhang, Q.; Li, Q.; Liu, H.; Li, M. Prognostic Model Associated with Necroptosis in Colorectal Cancer based on Transcriptomic Analysis and Experimental Validation. Front. Biosci. 2024, 29, 98. [Google Scholar] [CrossRef] [PubMed]
- Labi, V.; Erlacher, M. How cell death shapes cancer. Cell Death Dis. 2015, 6, e1675. [Google Scholar] [CrossRef]
- Ménard, S.; Tagliabue, E.; Campiglio, M.; Pupa, S.M. Role of HER2 gene overexpression in breast carcinoma. J. Cell. Physiol. 2000, 182, 150–162. [Google Scholar] [CrossRef]
- Bourova-Flin, E.; Derakhshan, S.; Goudarzi, A.; Wang, T.; Vitte, A.L.; Chuffart, F.; Khochbin, S.; Rousseaux, S.; Aminishakib, P. The combined detection of Amphiregulin, Cyclin A1 and DDX20/Gemin3 expression predicts aggressive forms of oral squamous cell carcinoma. Br. J. Cancer 2021, 125, 1122–1134. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Cao, Y.; Si, C.; Shao, P.; Su, G.; Wang, K.; Bao, J.; Yang, L. Emerging role of ERBB2 in targeted therapy for metastatic colorectal cancer: Signaling pathways to therapeutic strategies. Cancers 2022, 14, 5160. [Google Scholar] [CrossRef] [PubMed]
- Appert-Collin, A.; Hubert, P.; Crémel, G.; Bennasroune, A. Role of ErbB receptors in cancer cell migration and invasion. Front. Pharmacol. 2015, 6, 283. [Google Scholar] [CrossRef] [PubMed]
- Révillion, F.; Bonneterre, J.; Peyrat, J. ERBB2 oncogene in human breast cancer and its clinical significance. Eur. J. Cancer 1998, 34, 791–808. [Google Scholar] [CrossRef]
- Strickler, J.H.; Yoshino, T.; Graham, R.P.; Siena, S.; Bekaii-Saab, T. Diagnosis and treatment of ERBB2-positive metastatic colorectal cancer: A review. JAMA Oncol. 2022, 8, 760–769. [Google Scholar] [CrossRef]
- Wang, Y.; Zhou, B.P. Epithelial-mesenchymal transition—A hallmark of breast cancer metastasis. Cancer Hallm. 2013, 1, 38–49. [Google Scholar] [CrossRef] [PubMed]
- Hollier, B.G.; Tinnirello, A.A.; Werden, S.J.; Evans, K.W.; Taube, J.H.; Sarkar, T.R.; Sphyris, N.; Shariati, M.; Kumar, S.V.; Battula, V.L.; et al. FOXC2 expression links epithelial–mesenchymal transition and stem cell properties in breast cancer. Cancer Res. 2013, 73, 1981–1992. [Google Scholar] [CrossRef] [PubMed]
- Pham, T.N.D.; White, B.E.P.; Zhao, H.; Mortazavi, F.; Tonetti, D.A. Protein kinase C α enhances migration of breast cancer cells through FOXC2-mediated repression of p120-catenin. BMC Cancer 2017, 17, 832. [Google Scholar] [CrossRef] [PubMed]
- Mani, S.A.; Yang, J.; Brooks, M.; Schwaninger, G.; Zhou, A.; Miura, N.; Kutok, J.L.; Hartwell, K.; Richardson, A.L.; Weinberg, R.A. Mesenchyme Forkhead 1 (FOXC2) plays a key role in metastasis and is associated with aggressive basal-like breast cancers. Proc. Natl. Acad. Sci. USA 2007, 104, 10069–10074. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Zheng, L.; Wang, Q.; Hu, Y.-W. Emerging roles and mechanisms of FOXC2 in cancer. Clin. Chim. Acta 2018, 479, 84–93. [Google Scholar] [CrossRef]
- Meulmeester, E.; Ten Dijke, P. The dynamic roles of TGF-β in cancer. J. Pathol. 2011, 223, 206–219. [Google Scholar] [CrossRef] [PubMed]
- Alowayed, N.; Salker, M.S.; Zeng, N.; Singh, Y.; Lang, F. LEFTY2 controls migration of human endometrial cancer cells via focal adhesion kinase activity (FAK) and miRNA-200a. Cell. Physiol. Biochem. 2016, 39, 815–826. [Google Scholar] [CrossRef]
- Gao, X.; Cai, Y.; An, R. miR-215 promotes epithelial to mesenchymal transition and proliferation by regulating LEFTY2 in endometrial cancer. Int. J. Mol. Med. 2018, 42, 1229–1236. [Google Scholar] [CrossRef]
- Saaristo, A.; Karpanen, T.; Alitalo, K. Mechanisms of angiogenesis and their use in the inhibition of tumor growth and metastasis. Oncogene 2000, 19, 6122–6129. [Google Scholar] [CrossRef]
- Bera, A.; Subramanian, M.; Karaian, J.; Eklund, M.; Radhakrishnan, S.; Gana, N.; Rothwell, S.; Pollard, H.; Hu, H.; Shriver, C.D.; et al. Functional role of vitronectin in breast cancer. PLoS ONE 2020, 15, e0242141. [Google Scholar] [CrossRef]
- To, S.Q.; Dmello, R.S.; Richards, A.K.; Ernst, M.; Chand, A.L. STAT3 signaling in breast cancer: Multicellular actions and therapeutic potential. Cancers 2022, 14, 429. [Google Scholar] [CrossRef]
- Sasaki, Y.; Minamiya, Y.; Takahashi, N.; Nakagawa, T.; Katayose, Y.; Ito, A.; Saito, H.; Motoyama, S.; Ogawa, J.-I. REG1A expression is an independent factor predictive of poor prognosis in patients with breast cancer. Ann. Surg. Oncol. 2008, 15, 3244–3251. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wang, H.; Zogopoulos, G.; Shao, Q.; Dong, K.; Lv, F.; Nwilati, K.; Gui, X.-Y.; Cuggia, A.; Liu, J.-L.; et al. Reg proteins promote acinar-to-ductal metaplasia and act as novel diagnostic and prognostic markers in pancreatic ductal adenocarcinoma. Oncotarget 2016, 7, 77838–77853. [Google Scholar] [CrossRef] [PubMed]
- López-Lázaro, M. Role of oxygen in cancer: Looking beyond hypoxia. Anti-Cancer Agents Med. Chem. Former. Curr. Med. Chem.-Anti-Cancer Agents 2009, 9, 517–525. [Google Scholar] [CrossRef] [PubMed]
- Subarsky, P.; Hill, R.P. The hypoxic tumour microenvironment and metastatic progression. Clin. Exp. Metastasis 2003, 20, 237–250. [Google Scholar] [CrossRef] [PubMed]
- Wurth, R.; Tarn, K.; Jernigan, D.; Fernandez, S.V.; Cristofanilli, M.; Fatatis, A.; Meucci, O. A preclinical model of inflammatory breast cancer to study the involvement of CXCR4 and ACKR3 in the metastatic process. Transl. Oncol. 2015, 8, 358–367. [Google Scholar] [CrossRef] [PubMed]
- McCormick, F. KRAS as a therapeutic target. Clin. Cancer Res. 2015, 21, 1797–1801. [Google Scholar] [CrossRef] [PubMed]
- Venè, R.; Costa, D.; Augugliaro, R.; Carlone, S.; Scabini, S.; Pattacini, G.C.; Boggio, M.; Zupo, S.; Grillo, F.; Mastracci, L.; et al. Evaluation of glycosylated PTGS2 in colorectal cancer for NSAIDS-based adjuvant therapy. Cells 2020, 9, 683. [Google Scholar] [CrossRef]
- Kunzmann, A.T.; Murray, L.J.; Cardwell, C.R.; McShane, C.M.; McMenamin, U.C.; Cantwell, M.M. PTGS2 (Cyclooxygenase-2) expression and survival among colorectal cancer patients: A systematic review. Cancer Epidemiol. Biomark. Prev. 2013, 22, 1490–1497. [Google Scholar] [CrossRef]
- Li, J.; Kong, X.; Zhang, J.; Luo, Q.; Li, X.; Fang, L. MiRNA-26b inhibits proliferation by targeting PTGS2 in breast cancer. Cancer Cell Int. 2013, 13, 7. [Google Scholar] [CrossRef]
- Langsenlehner, U.; Yazdani-Biuki, B.; Eder, T.; Renner, W.; Wascher, T.C.; Paulweber, B.; Weitzer, W.; Samonigg, H.; Krippl, P. The cyclooxygenase-2 (PTGS2) 8473T> C polymorphism is associated with breast cancer risk. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2006, 12, 1392–1394. [Google Scholar] [CrossRef] [PubMed]
- Markosyan, N.; Li, J.; Sun, Y.H.; Richman, L.P.; Lin, J.H.; Yan, F.; Quinones, L.; Sela, Y.; Yamazoe, T.; Gordon, N.; et al. Tumor cell–intrinsic EPHA2 suppresses antitumor immunity by regulating PTGS2 (COX-2). J. Clin. Investig. 2019, 129, 3594–3609. [Google Scholar] [CrossRef]
- Zhou, Z.; Lu, C.; Meng, S.; Dun, L.; Yin, N.; An, H.; Xu, H.; Liu, G.; Cai, Y. Silencing of PTGS2 exerts promoting effects on angiogenesis endothelial progenitor cells in mice with ischemic stroke via repression of the NF-κB signaling pathway. J. Cell. Physiol. 2019, 234, 23448–23460. [Google Scholar] [CrossRef] [PubMed]
- Goos, J.A.; Coupé, V.M.; van de Wiel, M.A.; Diosdado, B.; Delis-Van Diemen, P.M.; Hiemstra, A.C.; de Cuba, E.M.; Beliën, J.A.; Menke-van der Houven, C.W.; Geldof, A.A.; et al. A prognostic classifier for patients with colorectal cancer liver metastasis, based on AURKA, PTGS2 and MMP9. Oncotarget 2023, 7, 2123–2134. [Google Scholar] [CrossRef] [PubMed]
- TCGA-BRCA. Available online: https://portal.gdc.cancer.gov/projects/TCGA-BRCA (accessed on 28 May 2024).
- Ding, L.; Ellis, M.J.; Li, S.; Larson, D.E.; Chen, K.; Wallis, J.W.; Harris, C.C.; McLellan, M.D.; Fulton, R.S.; Fulton, L.L.; et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature 2010, 464, 999–1005. [Google Scholar] [CrossRef] [PubMed]
- Amin, M.B.; Greene, F.L.; Edge, S.B.; Compton, C.C.; Gershenwald, J.E.; Brookland, R.K.; Meyer, L.; Gress, D.M.; Byrd, D.R.; Winchester, D.P. The eighth edition AJCC cancer staging manual: Continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA Cancer J. Clin. 2017, 67, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2018. CA Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef] [PubMed]
- de Boer, M.; van Dijck, J.A.; Bult, P.; Borm, G.F.; Tjan-Heijnen, V.C. Breast cancer prognosis and occult lymph node metastases, isolated tumor cells, and micrometastases. J. Natl. Cancer Inst. 2010, 102, 410–425. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Schneider, V.A.; Graves-Lindsay, T.; Howe, K.; Bouk, N.; Chen, H.C.; Kitts, P.A.; Murphy, T.D.; Pruitt, K.D.; Thibaud-Nissen, F.; Albracht, D.; et al. Evaluation of GRCh38 and de novo haploid genome assemblies demonstrates the enduring quality of the reference assembly. Genome Res. 2017, 27, 849–864. [Google Scholar] [CrossRef]
- Alberts, B.; Johnson, A.; Lewis, J.; Raff, M.; Roberts, K.; Walter, P. The molecular basis of cancer-cell behavior. In Molecular Biology of the Cell, 4th ed.; Garland Science: New York, NY, USA, 2002. [Google Scholar]
- Benjamini, Y.; Heller, R.; Yekutieli, D. Selective inference in complex research. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2009, 367, 4255–4271. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Dunn, O.J. Multiple comparisons among means. J. Am. Stat. Assoc. 1961, 56, 52–64. [Google Scholar] [CrossRef]
- Law, C.W.; Chen, Y.; Shi, W.; Smyth, G.K. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014, 15, R29. [Google Scholar] [CrossRef]
- Nelder, J.A.; Wedderburn, R.W.M. Generalized linear models. J. R. Stat. Soc. Ser. A Gen. 1972, 135, 370–384. [Google Scholar] [CrossRef]
- Wedderburn, R.W. Quasi-likelihood functions, generalized linear models, and the Gauss–Newton method. Biometrika 1974, 61, 439–447. [Google Scholar]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Abdi, H.; Williams, L.J. Principal component analysis. Wiley Interdiscip. Rev. Comput. Stat. 2010, 2, 433–459. [Google Scholar] [CrossRef]
- Menard, S. Applied Logistic Regression Analysis; Sage: Thousand Oaks, CA, USA, 2002. [Google Scholar]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Carlson, M.; Falcon, S.; Pages, H.; Li, N. org. Hs. eg. db: Genome wide annotation for Human. R Package Vers. 2019, 3, 3. [Google Scholar]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rezapour, M.; Wesolowski, R.; Gurcan, M.N. Identifying Key Genes Involved in Axillary Lymph Node Metastasis in Breast Cancer Using Advanced RNA-Seq Analysis: A Methodological Approach with GLMQL and MAS. Int. J. Mol. Sci. 2024, 25, 7306. https://doi.org/10.3390/ijms25137306
Rezapour M, Wesolowski R, Gurcan MN. Identifying Key Genes Involved in Axillary Lymph Node Metastasis in Breast Cancer Using Advanced RNA-Seq Analysis: A Methodological Approach with GLMQL and MAS. International Journal of Molecular Sciences. 2024; 25(13):7306. https://doi.org/10.3390/ijms25137306
Chicago/Turabian StyleRezapour, Mostafa, Robert Wesolowski, and Metin Nafi Gurcan. 2024. "Identifying Key Genes Involved in Axillary Lymph Node Metastasis in Breast Cancer Using Advanced RNA-Seq Analysis: A Methodological Approach with GLMQL and MAS" International Journal of Molecular Sciences 25, no. 13: 7306. https://doi.org/10.3390/ijms25137306
APA StyleRezapour, M., Wesolowski, R., & Gurcan, M. N. (2024). Identifying Key Genes Involved in Axillary Lymph Node Metastasis in Breast Cancer Using Advanced RNA-Seq Analysis: A Methodological Approach with GLMQL and MAS. International Journal of Molecular Sciences, 25(13), 7306. https://doi.org/10.3390/ijms25137306




