Expression and Functional Analysis of the Metallothionein and Metal-Responsive Transcription Factor 1 in Phascolosoma esculenta under Zn Stress
Abstract
:1. Introduction
2. Results
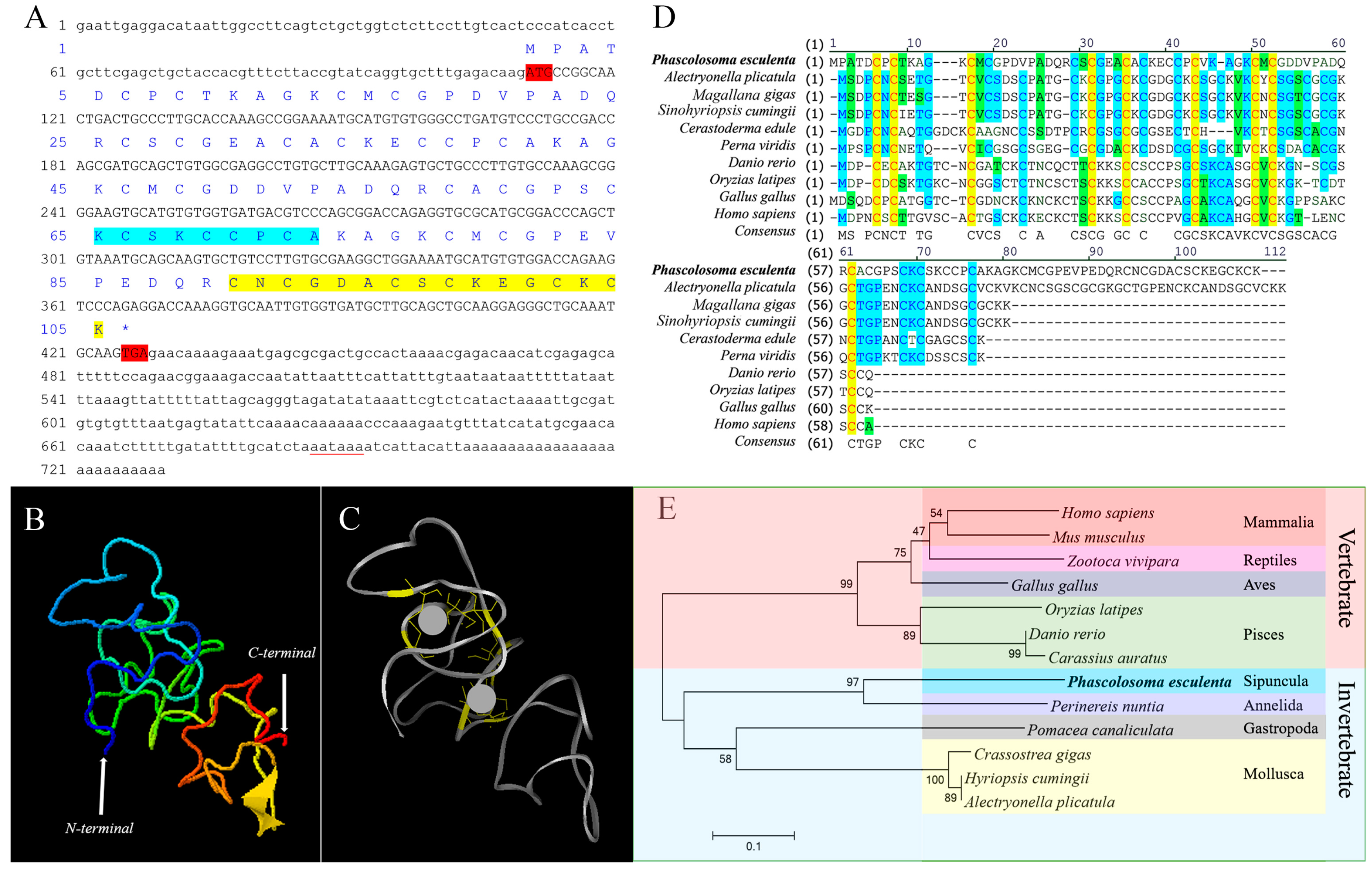
2.1. Sequence Features and Bioinformatics Analysis of PeMT
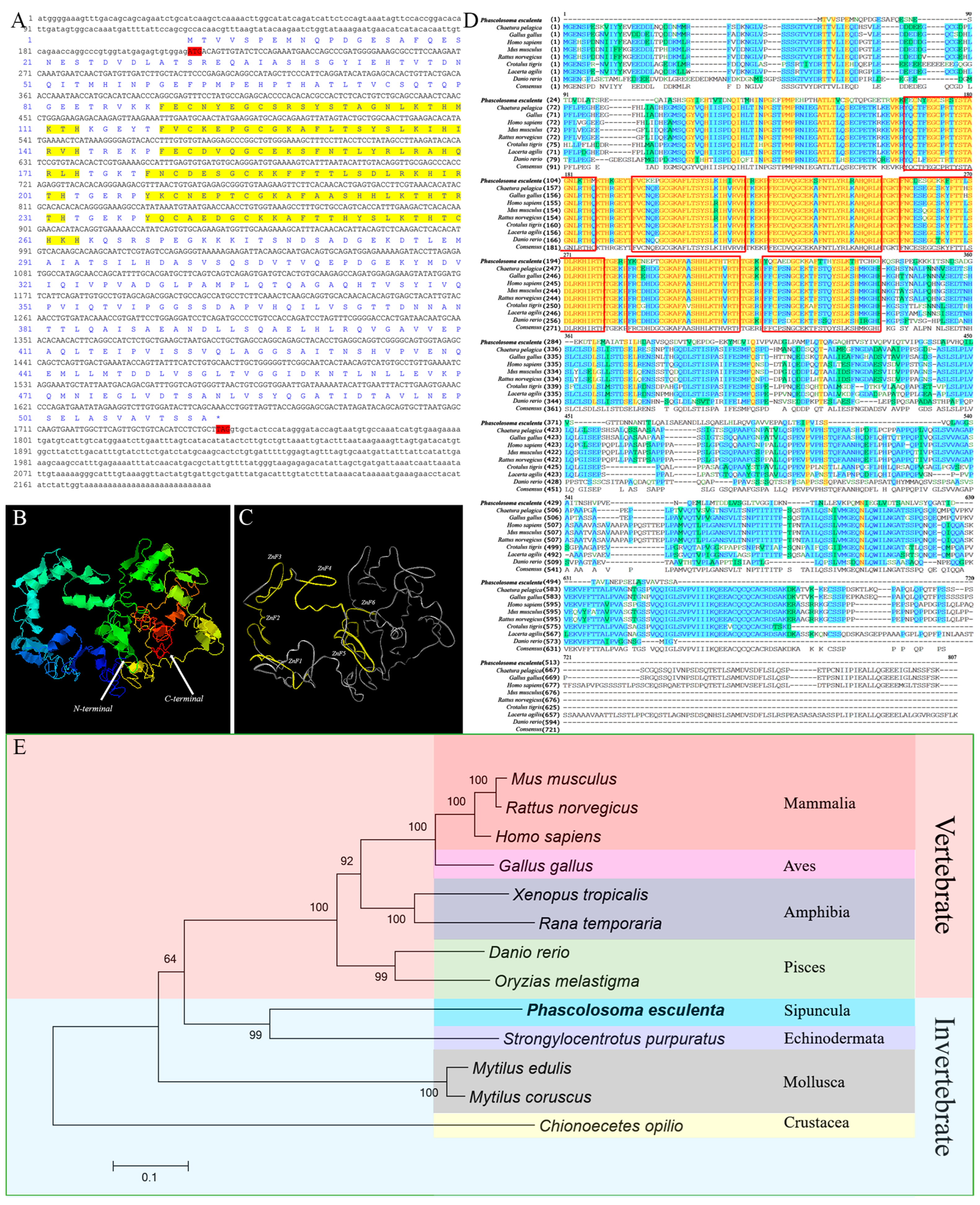
2.2. Sequence Features and Bioinformatics Analysis of PeMTF1
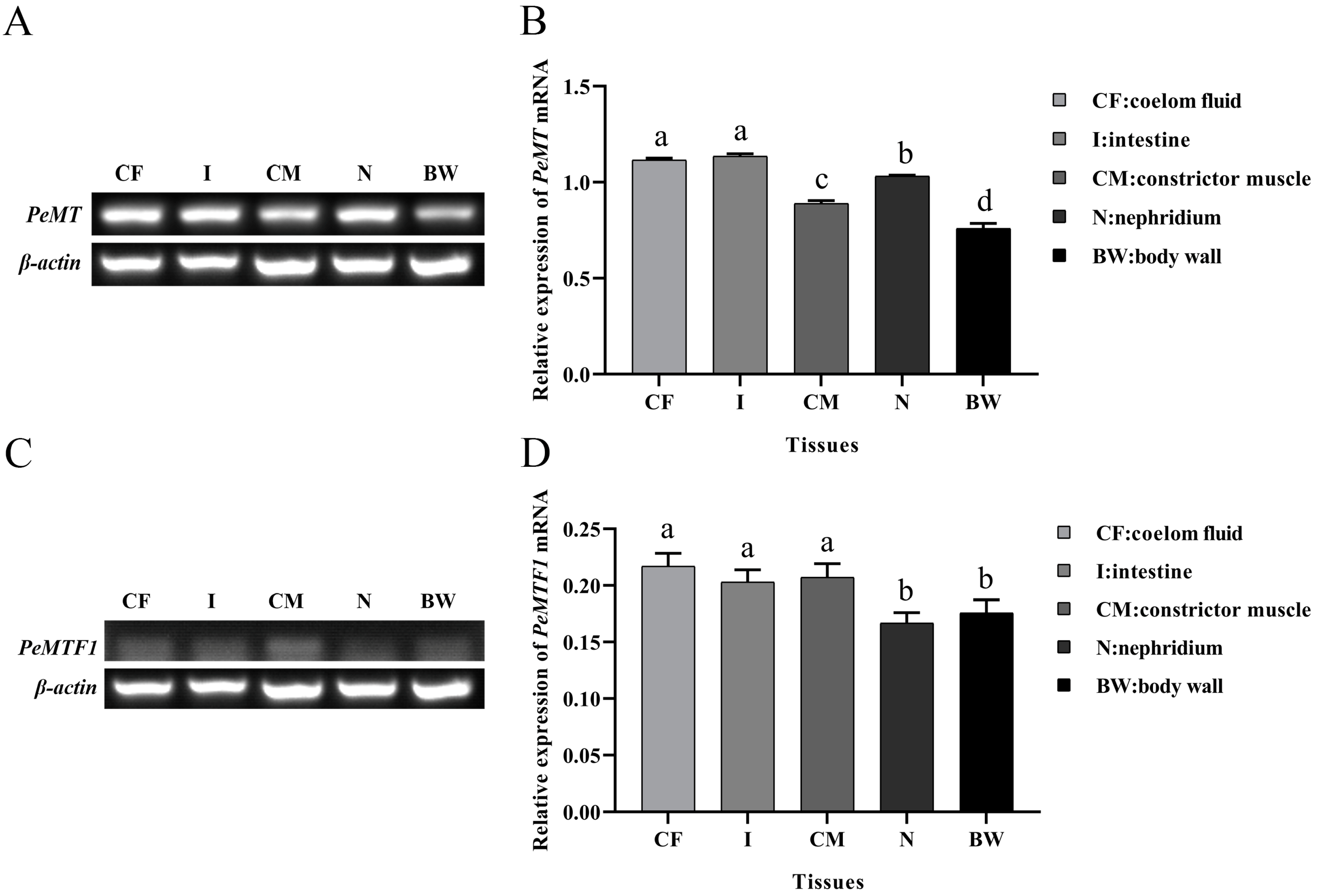
2.3. Tissue-Specific Expression of PeMT and PeMTF1 mRNA
2.4. Expression Changes of PeMT and PeMTF1 mRNA in the Intestine under Zn Stress
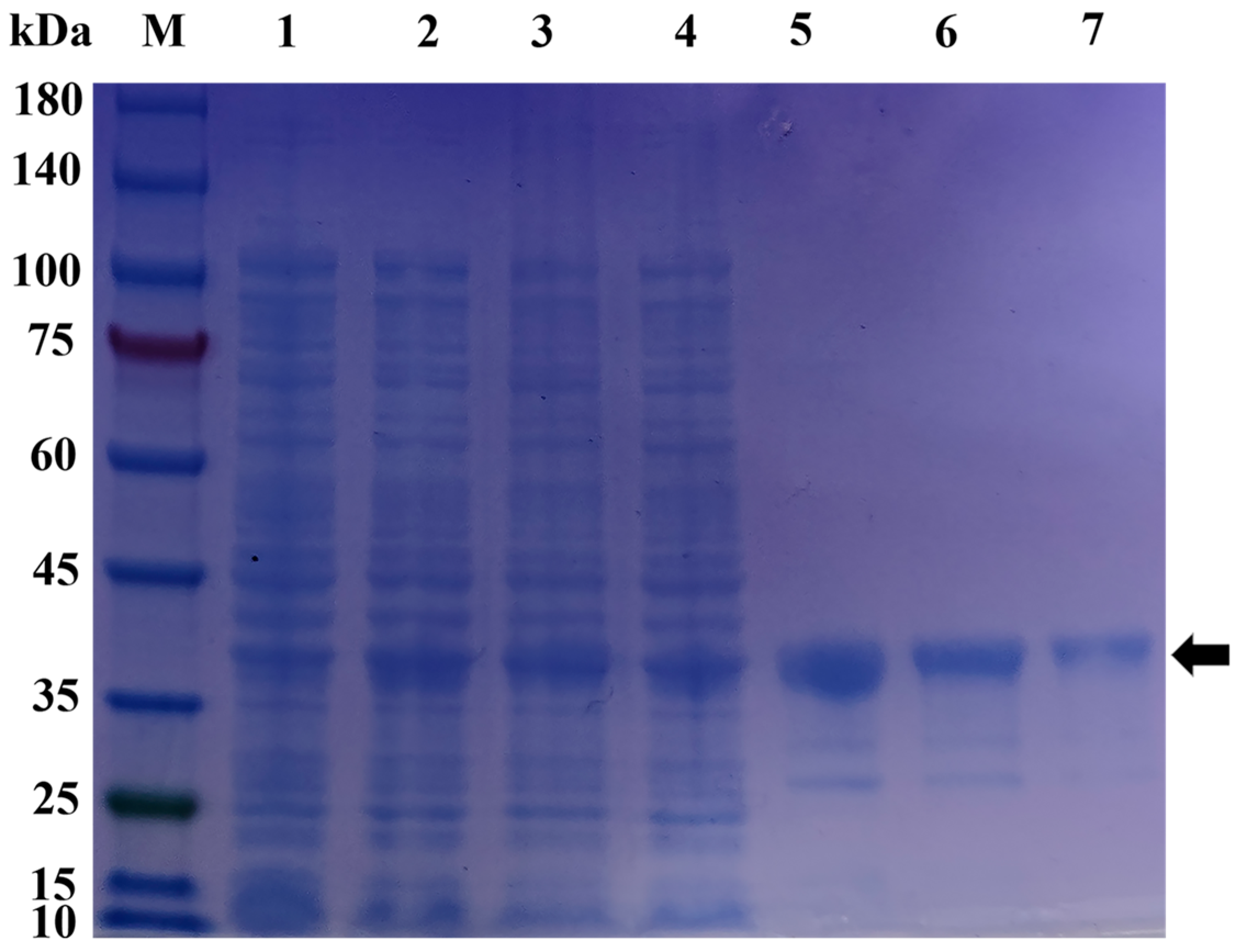
2.5. Prokaryotic Expression and Protein Purification of pGEX-6p-1-MT
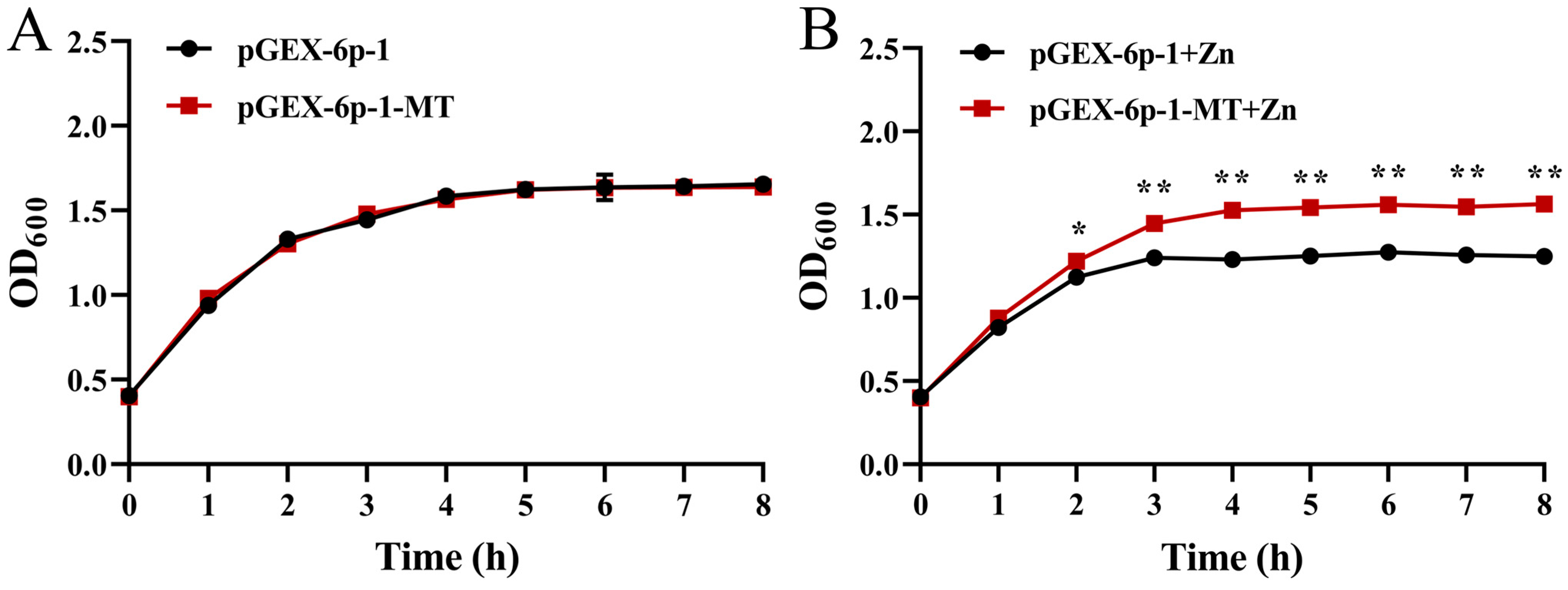
2.6. Zn Tolerance of the Recombinant E. coli
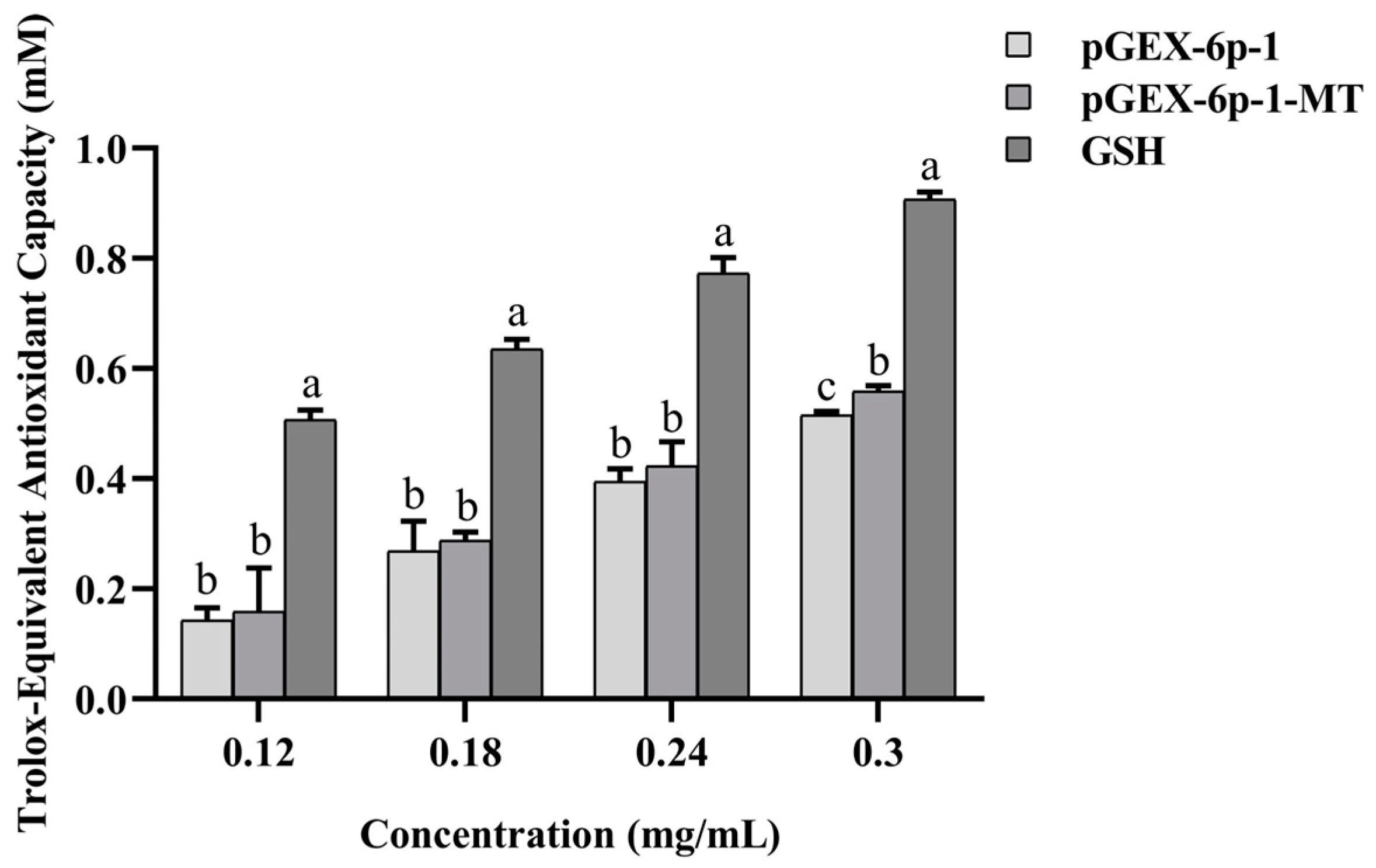
2.7. Antioxidant Capacity of the Recombinant pGEX-6p-1-MT
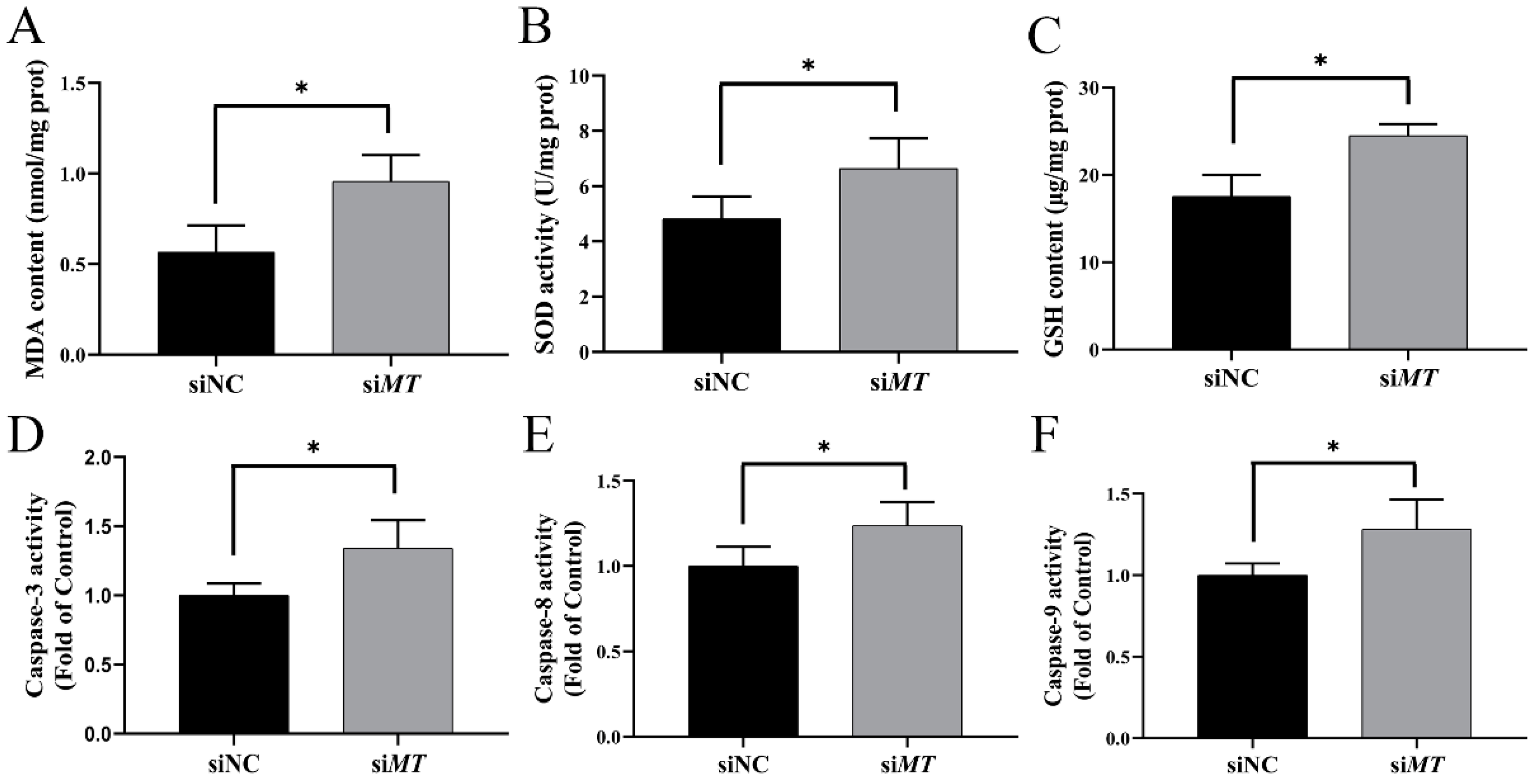
2.8. Changes in Intestinal Oxidative Stress and Apoptosis Indices after RNAi with PeMT and Zn Stress
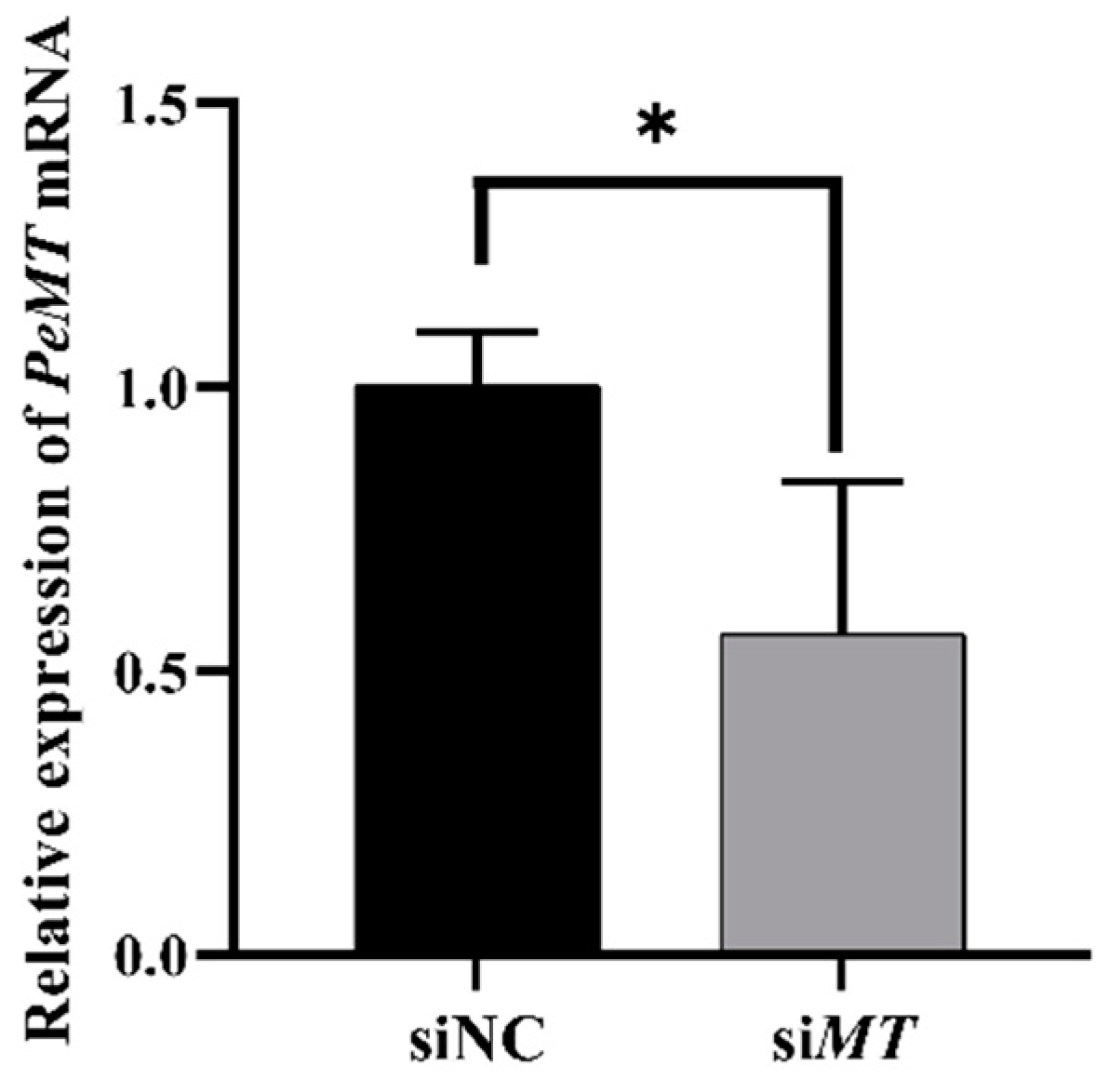
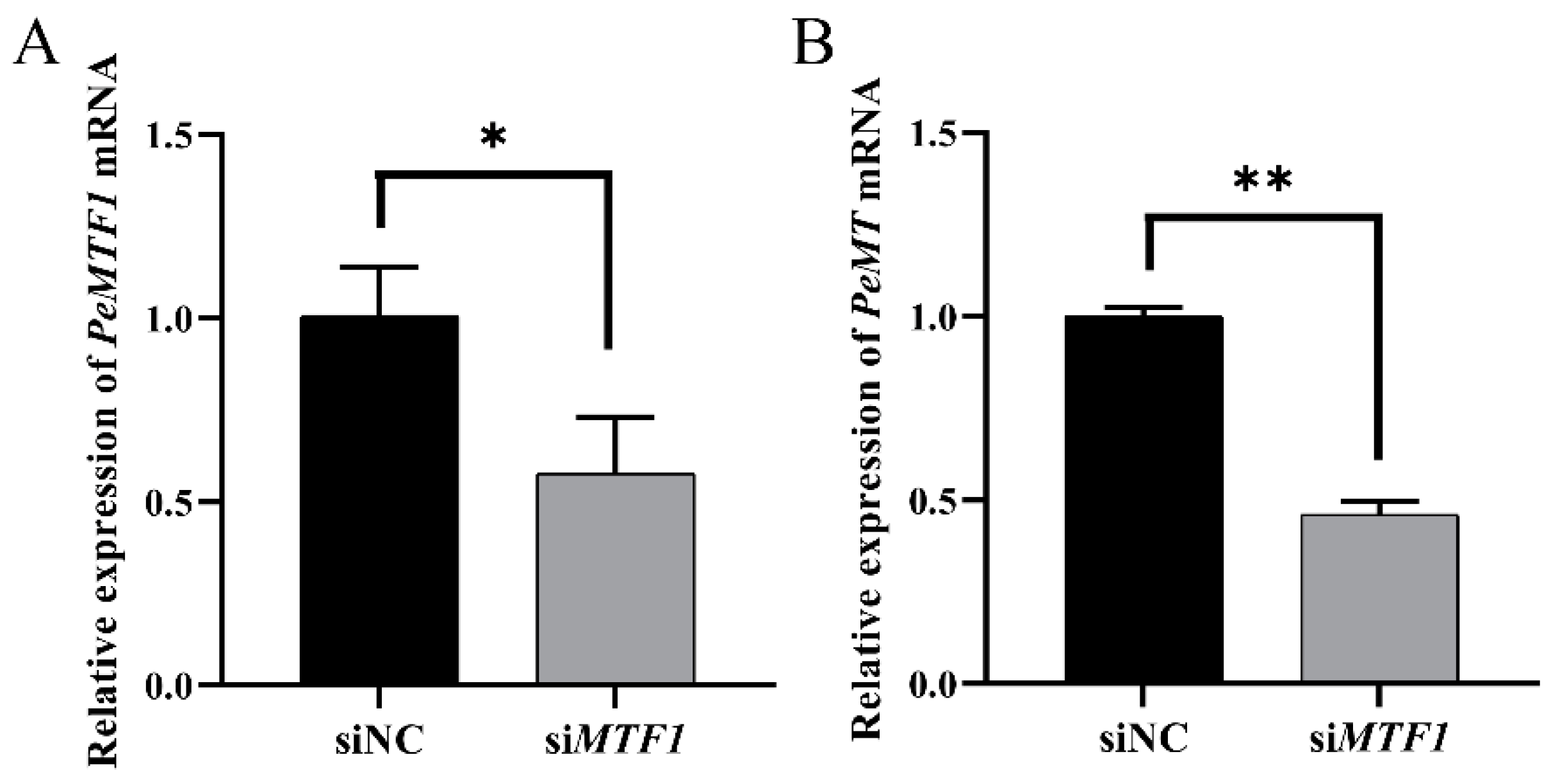
2.9. Changes in Intestinal PeMT mRNA Expression Level after RNAi with PeMTF1 and Zn Stress
3. Discussion
3.1. Sequence Features and Protein Structure of PeMT and PeMTF1
3.2. Tissue-Specific Expression of PeMT and PeMTF1 and Their Expression Changes under Zn Stress
3.3. Detoxification, Antioxidant, and Anti-Apoptosis Functions of PeMT
3.4. Regulation Effect of PeMTF1 on PeMT
4. Materials and Methods
4.1. Animals
4.2. Treatments and Sampling
4.3. RNA Extraction and cDNA Synthesis
4.4. Full-Length cDNA Cloning of PeMT and PeMTF1
4.5. Bioinformatics Analysis of PeMT and PeMTF1
4.6. Tissue-Specific Expression of PeMT and PeMTF1 mRNA and Its Expression Changes under Zn Stress
4.7. Expression and Purification of the Recombinant pGEX-6p-1-MT
4.8. Zn Tolerance of the Recombinant E. coli
4.9. Antioxidant Capacity of the Recombinant pGEX-6p-1-MT
4.10. Detection of Intestinal Oxidative Stress and Apoptosis Indices after RNAi with PeMT and Zn Stress
4.11. Detection of Intestinal PeMT mRNA Expression Level after RNAi with PeMTF1 and Zn Stress
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tchounwou, P.B.; Yedjou, C.G.; Patlolla, A.K.; Sutton, D.J. Heavy Metal Toxicity and the Environment. Exp. Suppl. 2012, 101, 133–164. [Google Scholar] [PubMed]
- Cui, Y.B.; Bai, L.; Li, C.H.; He, Z.J.; Liu, X.R. Assessment of heavy metal contamination levels and health risks in environmental media in the northeast region. Sustain. Cities Soc. 2022, 80, 103796. [Google Scholar] [CrossRef]
- Wu, J.; Lu, J.; Zhang, C.; Zhang, Y.X.; Lin, Y.C.; Xu, J. Pollution, sources, and risks of heavy metals in coastal waters of China. Hum. Ecol. Risk. Assess. 2019, 26, 2011–2026. [Google Scholar] [CrossRef]
- Wu, D.; Feng, H.; Zou, Y.; Xiao, J.; Zhang, P.F.; Ji, Y.X.; Lek, S.; Guo, Z.Q.; Fu, Q.Y. Feeding Habit-Specific Heavy Metal Bioaccumulation and Health Risk Assessment of Fish in a Tropical Reservoir in Southern China. Fishes 2023, 8, 211. [Google Scholar] [CrossRef]
- Roohani, N.; Hurrell, R.; Kelishadi, R.; Schulin, R. Zinc and its importance for human health: An integrative review. J. Res. Med. Sci. 2013, 18, 144–157. [Google Scholar] [PubMed]
- Chandra, R.K. Excessive intake of zinc impairs immune responses. JAMA 1984, 252, 1443–1446. [Google Scholar] [CrossRef] [PubMed]
- Evans, P.; Halliwell, B. Micronutrients: Oxidant/antioxidant status. Br. J. Nutr. 2007, 85, 67–84. [Google Scholar] [CrossRef]
- Ighodaro, O.M.; Akinloye, O.A. First line defence antioxidants-superoxide dismutase (SOD), catalase (CAT) and glutathione peroxidase (GPX): Their fundamental role in the entire antioxidant defence grid. Alex. J. Med. 2019, 54, 287–293. [Google Scholar] [CrossRef]
- Plum, L.M.; Rink, L.; Haase, H. The Essential Toxin: Impact of Zinc on Human Health. Int. J. Environ. Res. Public Health 2010, 7, 1342–1365. [Google Scholar] [CrossRef]
- Bakiu, R.; Pacchini, S.; Piva, E.; Schumann, S.; Tolomeo, A.M.; Ferro, D.; Irato, P.; Santovito, G. Metallothionein Expression as a Physiological Response against Metal Toxicity in the Striped Rockcod Trematomus hansoni. Int. J. Mol. Sci. 2022, 23, 12799. [Google Scholar] [CrossRef]
- Formigari, A.; Boldrin, F.; Santovito, G.; Cassidy-Hanley, D.; Clark, T.G.; Piccinni, E. Functional Characterization of the 5′-upstream Region of MTT5 Metallothionein Gene from Tetrahymena thermophila. Protist 2010, 161, 71–77. [Google Scholar] [CrossRef]
- Gautam, N.; Tiwari, M.; Kidwai, M.; Dutta, P.; Chakrabarty, D. Functional characterization of rice metallothionein OsMT-I-Id: Insights into metal binding and heavy metal tolerance mechanisms. J. Hazard. Mater. 2023, 458, 131815. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.F.; Chen, P.M.; Pan, C.S.; Huang, C.C.; Chiang, E.P.I. Association of metallothionein 2A rs10636 with low mean corpuscular volume (MCV), low mean corpuscular haemoglobin (MCH) in healthy Taiwanese. Sci. Rep. 2023, 13, 1292. [Google Scholar] [CrossRef] [PubMed]
- Monjane Mabuie, A.; Mondlane Milisse, A.; Pedro, O.; Leão Buchir, J.; Correia, D. Mercury pollution assessment and metallothionein gene expression in tilapia (Oreochromis mossambicus) a case study of Revue River in Manica, Mozambique. Rend. Lincei-Sci. Fis. 2022, 33, 513–526. [Google Scholar] [CrossRef]
- Niu, S.Y.; Wang, J.J.; Chang, X.R.; Shang, M.T.; Guo, M.H.; Sun, Z.Y.; Li, Y.J.; Xue, Y.Y. Comparative oxidative damages induced by silver nanoparticles with different sizes and coatings in Caenorhabditis elegans. Toxicol. Res. 2023, 12, 833–842. [Google Scholar] [CrossRef] [PubMed]
- Shimoda, R.; Achanzar, W.E.; Qu, W.; Nagamine, T.; Takagi, H.; Mori, M.; Waalkes, M.P. Metallothionein is a potential negative regulator of apoptosis. Toxicol. Sci. 2003, 73, 294–300. [Google Scholar] [CrossRef]
- Chen, G.H.; Lv, W.H.; Xu, Y.H.; Wei, X.L.; Xu, Y.C.; Luo, Z. Functional analysis of MTF-1 and MT promoters and their transcriptional response to zinc (Zn) and copper (Cu) in yellow catfish Pelteobagrus fulvidraco. Chemosphere 2020, 246, 125792. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.L.; Sheng, Z.; Liu, G.D.; Long, L.L.; Wang, Y.F.; Yang, W.X.; Zhu, J.Q. Cloning, characterization and cadmium inducibility of metallothionein in the testes of the mudskipper Boleophthalmus pectinirostris. Ecotoxicol. Environ. Saf. 2015, 119, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Hardivillier, Y.; Leignel, V.; Denis, F.; Uguen, G.; Cosson, R.; Laulier, M. Do organisms living around hydrothermal vent sites contain specific metallothioneins? The case of the genus Bathymodiolus (Bivalvia, Mytilidae). Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2004, 139, 111–118. [Google Scholar] [CrossRef]
- Gao, X.M.; Feng, B.B.; Tang, D.J.; Du, C.; Hou, C.C.; Jin, S.; Zhu, J.Q. Mitochondrial Features and Expressions of MFN2 and DRP1 during Spermiogenesis in Phascolosoma esculenta. Int. J. Mol. Sci. 2022, 23, 15517. [Google Scholar] [CrossRef]
- Shen, W.L.; Liu, C.; Ni, J.; Gao, X.M.; Ni, J.J.; Wang, J.P.; Jin, S.; Hou, C.C.; Wu, X.F.; Zhu, J.Q. Effects of Low Temperature Stress on the Morphology and hsp70 and hsp90 Gene Expression of Phascolosoma esculenta. J. Ocean Univ. China 2021, 20, 159–168. [Google Scholar] [CrossRef]
- Meng, J.J.; Gao, X.M.; Luo, S.Y.; Lin, C.W.; Du, C.; Hou, C.C.; Wang, J.P.; Jin, S.; Tang, D.J.; Zhang, C.D.; et al. Cloning, Functional Characterization and Response to Cadmium Stress of the Thioredoxin-like Protein 1 Gene from Phascolosoma esculenta. Int. J. Mol. Sci. 2021, 23, 332. [Google Scholar] [CrossRef] [PubMed]
- Binz, P.A.; Kägi, J.H.R. Metallothionein: Molecular evolution and classification. In Metallothionein IV; Birkhäuser: Basel, Switzerland, 1999; pp. 7–13. [Google Scholar]
- Hesketh, J. 3′-Untranslated regions are important in mRNA localization and translation. Biochem. Soc. Trans. 2004, 32, 990–993. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.Y.; Sheng, J.Q.; Hong, Y.J.; Peng, K.; Wang, J.H.; Wu, D.; Shi, J.W.; Hu, B.J. Molecular characterization and expression of metallothionein from freshwater pearl mussel, Hyriopsis schlegelii. Biosci. Biotechnol. Biochem. 2016, 80, 1327–1335. [Google Scholar] [CrossRef] [PubMed]
- Leignel, V.; Marchand, J.; Moreau, B.; Chénais, B. Metallothionein genes from hydrothermal crabs (Bythograeidae, Decapoda): Characterization, sequence analysis, gene expression and comparison with coastal crabs. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2008, 148, 6–13. [Google Scholar] [CrossRef] [PubMed]
- Sheng, Z.; Yang, W.X.; Zhu, J.Q. Metallothionein from Pseudosciaena crocea: Expression and response to cadmium-induced injury in the testes. Ecotoxicology 2015, 24, 779–794. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Shi, Z.H.; Gao, Z.H.; Wen, Y.T.; Wang, W.Q.; Liu, W.; Wang, X.P.; Zhu, F. Identification of three metallothioneins in the black soldier fly and their functions in Cd accumulation and detoxification. Environ. Pollut. 2021, 286, 117146. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.M.; Wu, H.H.; Kou, L.H.; Liu, X.J.; Zhang, J.Z.; Guo, Y.P.; Ma, E.B. Two Metallothionein Genes in Oxya chinensis: Molecular Characteristics, Expression Patterns and Roles in Heavy Metal Stress. PLoS ONE. 2014, 9, 112759. [Google Scholar] [CrossRef] [PubMed]
- Lv, X.F.; Deng, W.H.; Yu, P.; Hua, Z.Y. Metallothionein cDNA Cloning and Metallothionein Expression in Sea Cucumber (Apostichopus japonicus, Slenka) from the WeiHai Coast of China. Ocean. Sci. J. 2018, 53, 719–726. [Google Scholar] [CrossRef]
- Xiang, D.F.; Zhu, J.Q.; Jin, S.; Hu, Y.J.; Tan, F.Q.; Yang, W.X. Expression and function analysis of metallothionein in the testis of Portunus trituberculatus exposed to cadmium. Aquat. Toxicol. 2013, 140, 1–10. [Google Scholar] [CrossRef]
- Mao, H.; Tan, F.Q.; Wang, D.H.; Zhu, J.Q.; Zhou, H.; Yang, W.X. Expression and function analysis of metallothionein in the testis of stone crab Charybdis japonica exposed to cadmium. Aquat. Toxicol. 2012, 124, 11–21. [Google Scholar] [CrossRef]
- Rigby Duncan, K.E.; Kirby, C.W.; Stillman, M.J. Metal exchange in metallothioneins—A novel structurally significant Cd5 species in the alpha domain of human metallothionein 1a. FEBS J. 2008, 275, 2227–2239. [Google Scholar] [CrossRef]
- Tanguy, A.; Moraga, D. Cloning and characterization of a gene coding for a novel metallothionein in the Pacific oyster Crassostrea gigas (CgMT2): A case of adaptive response to metal-induced stress? Gene 2001, 273, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Valls, M.; Bofill, R.; Romero-Isart, N.; Gonzàlez-Duarte, R.; Abián, J.; Carrascal, M.; Gonzàlez-Duarte, P.; Capdevila, M.; Atrian, S. Drosophila MTN: A metazoan copper-thionein related to fungal forms. FEBS Lett. 2000, 467, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Jin, M.; He, S.; Kannan, R.; Ryan, S.J.; Hinton, D.R. Regulation of Metallothionein Gene Expression by Oxidative Stress and HGF. Investig. Ophth. Vis. Sci. 2003, 44, 1627. [Google Scholar]
- Ren, L.X.; Yu, Z.T.; Liu, X.J.; Zhang, J.Z.; Ma, E.B.; Guo, Y.P. Molecular characterization and expression analysis of metal-responsive transcription factor-1 gene OcMTF-1 from Oxya chinensis (Orthoptera: Acridoidea). Acta Entomol. Sin. 2014, 57, 1001–1007. (In Chinese) [Google Scholar]
- Ferencz, A.; Hermesz, E. Identification of a splice variant of the metal-responsive transcription factor MTF-1 in common carp. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2009, 150, 113–117. [Google Scholar] [CrossRef]
- Ren, F.; Jiang, H.; Sun, J.L.; He, L.; Li, W.W.; Wang, Y.; Wang, Q. Cloning, characterization, expression, and copper sensitivity of the metallothionein-1 gene in the Chinese mitten crab, Eriocheir sinensis. Mol. Biol. Rep. 2010, 38, 2383–2393. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Guo, Y.; Hu, J.J.; Bao, Z.M.; Wang, M.Q. A metallothionein gene from hard clam Meretrix meretrix: Sequence features, expression patterns, and metal tolerance activities. Dev. Comp. Immunol. 2023, 149, 105057. [Google Scholar] [CrossRef]
- Lee, S.Y.; Nam, Y.K. Transcriptional responses of metallothionein gene to different stress factors in Pacific abalone (Haliotis discus hannai). Fish Shellfish Immunol. 2016, 58, 530–541. [Google Scholar] [CrossRef]
- Felix Portillo, M.; Martinez Quintana, J.A.; Peregrino Uriarte, A.B.; Yepiz Plascencia, G. The metallothionein gene from the white shrimp Litopenaeus vannamei: Characterization and expression in response to hypoxia. Mar. Environ. Res. 2014, 101, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Hui, K.M.; Ren, Q.; Cao, J. Insights into the intestine immune of Marsupenaeus japonicus under the white spot syndrome virus challenge using RNA sequencing. Vet. Immunol. Immunopathol. 2019, 208, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.F.; Wang, Y.; Ding, X.; Xiong, D.L.; Zhang, J.S. Response of intestine microbiota, digestion, and immunity in Pacific white shrimp Litopenaeus vannamei to dietary succinate. Aquaculture 2020, 517, 734762. [Google Scholar] [CrossRef]
- Ge, D.L.; Zhang, L.; Long, Z.H.; Chi, C.F.; Liu, H.H. A novel biomarker for marine environmental pollution: A metallothionein from Mytilus coruscus. Aquacult. Rep. 2020, 17, 100364. [Google Scholar] [CrossRef]
- Sakatoku, A.; Ishikawa, M.; Yamazaki, K.; Nakamachi, T.; Kamachi, H.; Tanaka, D.; Nakamura, S. Molecular Identification, Characterization, and Expression Analysis of a Metallothionein Gene from Septifer virgatus. Mar. Biotechnol. 2020, 22, 488–497. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.H.; Meng, X.; Gan, H.T.; Liu, T.H.; Yao, H.Y.; Zhu, X.Y.; Xu, G.C.; Xu, J.T. Immune response, MT and HSP70 gene expression, and bioaccumulation induced by lead exposure of the marine crab, Charybdis japonica. Aquat. Toxicol. 2019, 210, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.R.; Liu, Y.; Yu, M.J.; Pang, Z.H.; Chen, W.B.; Xu, Z.C. Identification and functional characterization of MRE-binding transcription factor (MTF) in Crassostrea gigas and its conserved role in metal-induced response. Mol. Biol. Rep. 2013, 40, 3321–3331. [Google Scholar] [CrossRef] [PubMed]
- Siscar, R.; Koenig, S.; Torreblanca, A.; Solé, M. The role of metallothionein and selenium in metal detoxification in the liver of deep-sea fish from the NW Mediterranean Sea. Sci. Total Environ. 2014, 466–467, 898–905. [Google Scholar] [CrossRef]
- Wang, L.; Yang, H.Z.; Ma, W.L.; Chen, C.M.; Wang, L. Study on metal binding capacity of the freshwater crab Sinopotamon henanense’s recombinant copper specific binding metallothionein expressed in Escherichia coli. Ecotoxicology 2021, 31, 149–160. [Google Scholar] [CrossRef]
- Li, R.; Yang, Y.; Cao, H.P.; Peng, X.; Yu, Q.; He, L.S.; Chen, J.; Xiang, L.E.; Liu, W.H. Heterologous expression of the tobacco metallothionein gene NtMT2F confers enhanced tolerance to Cd stress in Escherichia coli and Arabidopsis thaliana. Plant Physiol. Biochem. 2023, 195, 247–255. [Google Scholar] [CrossRef]
- Amiard, J.C.; Amiard Triquet, C.; Barka, S.; Pellerin, J.; Rainbow, P.B. Metallothioneins in aquatic invertebrates: Their role in metal detoxification and their use as biomarkers. Aquat. Toxicol. 2006, 76, 160–202. [Google Scholar] [CrossRef]
- Ruttkay Nedecky, B.; Nejdl, L.; Gumulec, J.; Zitka, O.; Masarik, M.; Eckschlager, T.; Stiborova, M.; Adam, V.; Kizek, R. The Role of Metallothionein in Oxidative Stress. Int. J. Mol. Sci. 2013, 14, 6044–6066. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.Y.; Qin, J.X.; Yuan, H.L.; Guo, M.H.; Shang, M.T.; Niu, S.Y.; Li, Y.J.; Li, Q.; Xue, Y.Y. Recombinant human metallothionein-III alleviates oxidative damage induced by copper and cadmium in Caenorhabditis elegans. J. Appl. Toxicol. 2023, 43, 1242–1252. [Google Scholar] [CrossRef] [PubMed]
- Atif, F.; Kaur, M.; Yousuf, S.; Raisuddin, S. In vitro free radical scavenging activity of hepatic metallothionein induced in an Indian freshwater fish, Channa punctata Bloch. Chem. Biol. Interact. 2006, 162, 172–180. [Google Scholar] [CrossRef]
- Wang, N.; Guo, Z.Y.; Zhang, Y.L.; Zhang, P.J.; Liu, J.; Cheng, Y.; Zhang, L.; Li, Y.H. Effect on intestinal microbiota, bioaccumulation, and oxidative stress of Carassius auratus gibelio under waterborne cadmium exposure. Fish Physiol. Biochem. 2020, 46, 2299–2309. [Google Scholar] [CrossRef]
- Mruk, D.D.; Silvestrini, B.; Mo, M.Y.; Cheng, C.Y. Antioxidant superoxide dismutase—A review: Its function, regulation in the testis, and role in male fertility. Contraception 2002, 65, 305–311. [Google Scholar] [CrossRef] [PubMed]
- Xia, L.P.; Chen, S.H.; Dahms, H.U.; Ying, X.P.; Peng, X. Cadmium induced oxidative damage and apoptosis in the hepatopancreas of Meretrix meretrix. Ecotoxicology 2016, 25, 959–969. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.L.; Zhao, L.L.; Liao, L.; Tang, X.H.; Cui, C.; Liu, Q.; He, K.; Ma, J.D.; Jin, L.; Yan, T.; et al. Interactive effect of thermal and hypoxia on largemouth bass (Micropterus salmoides) gill and liver: Aggravation of oxidative stress, inhibition of immunity and promotion of cell apoptosis. Fish Shellfish Immunol. 2020, 98, 923–936. [Google Scholar] [CrossRef]
- Meng, J.; Zhang, L.L.; Li, L.; Li, C.Y.; Wang, T.; Zhang, G.F. Transcription factor CgMTF-1 regulates CgZnT1 and CgMT expression in Pacific oyster (Crassostrea gigas) under zinc stress. Aquat. Toxicol. 2015, 165, 179–188. [Google Scholar] [CrossRef]
- Troadec, M.B.; Ward, D.M.; Lo, E.; Kaplan, J.; De Domenico, I. Induction of FPN1 transcription by MTF-1 reveals a role for ferroportin in transition metal efflux. Blood 2010, 116, 4657–4664. [Google Scholar] [CrossRef]
- Atanesyan, L.; Günther, V.; Celniker, S.E.; Georgiev, O.; Schaffner, W. Characterization of MtnE, the fifth metallothionein member in Drosophila. J. Biol. Inorg. Chem. 2011, 16, 1047–1056. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gu, S.; Wang, J.; Gao, X.; Zheng, X.; Liu, Y.; Chen, Y.; Sun, L.; Zhu, J. Expression and Functional Analysis of the Metallothionein and Metal-Responsive Transcription Factor 1 in Phascolosoma esculenta under Zn Stress. Int. J. Mol. Sci. 2024, 25, 7368. https://doi.org/10.3390/ijms25137368
Gu S, Wang J, Gao X, Zheng X, Liu Y, Chen Y, Sun L, Zhu J. Expression and Functional Analysis of the Metallothionein and Metal-Responsive Transcription Factor 1 in Phascolosoma esculenta under Zn Stress. International Journal of Molecular Sciences. 2024; 25(13):7368. https://doi.org/10.3390/ijms25137368
Chicago/Turabian StyleGu, Shenwei, Jingqian Wang, Xinming Gao, Xuebin Zheng, Yang Liu, Yiner Chen, Lianlian Sun, and Junquan Zhu. 2024. "Expression and Functional Analysis of the Metallothionein and Metal-Responsive Transcription Factor 1 in Phascolosoma esculenta under Zn Stress" International Journal of Molecular Sciences 25, no. 13: 7368. https://doi.org/10.3390/ijms25137368
APA StyleGu, S., Wang, J., Gao, X., Zheng, X., Liu, Y., Chen, Y., Sun, L., & Zhu, J. (2024). Expression and Functional Analysis of the Metallothionein and Metal-Responsive Transcription Factor 1 in Phascolosoma esculenta under Zn Stress. International Journal of Molecular Sciences, 25(13), 7368. https://doi.org/10.3390/ijms25137368





