PpGATA21 Enhances the Expression of PpGA2ox7 to Regulate the Mechanism of Cerasus humilis Rootstock-Mediated Dwarf in Peach Trees
Abstract
:1. Introduction
2. Result
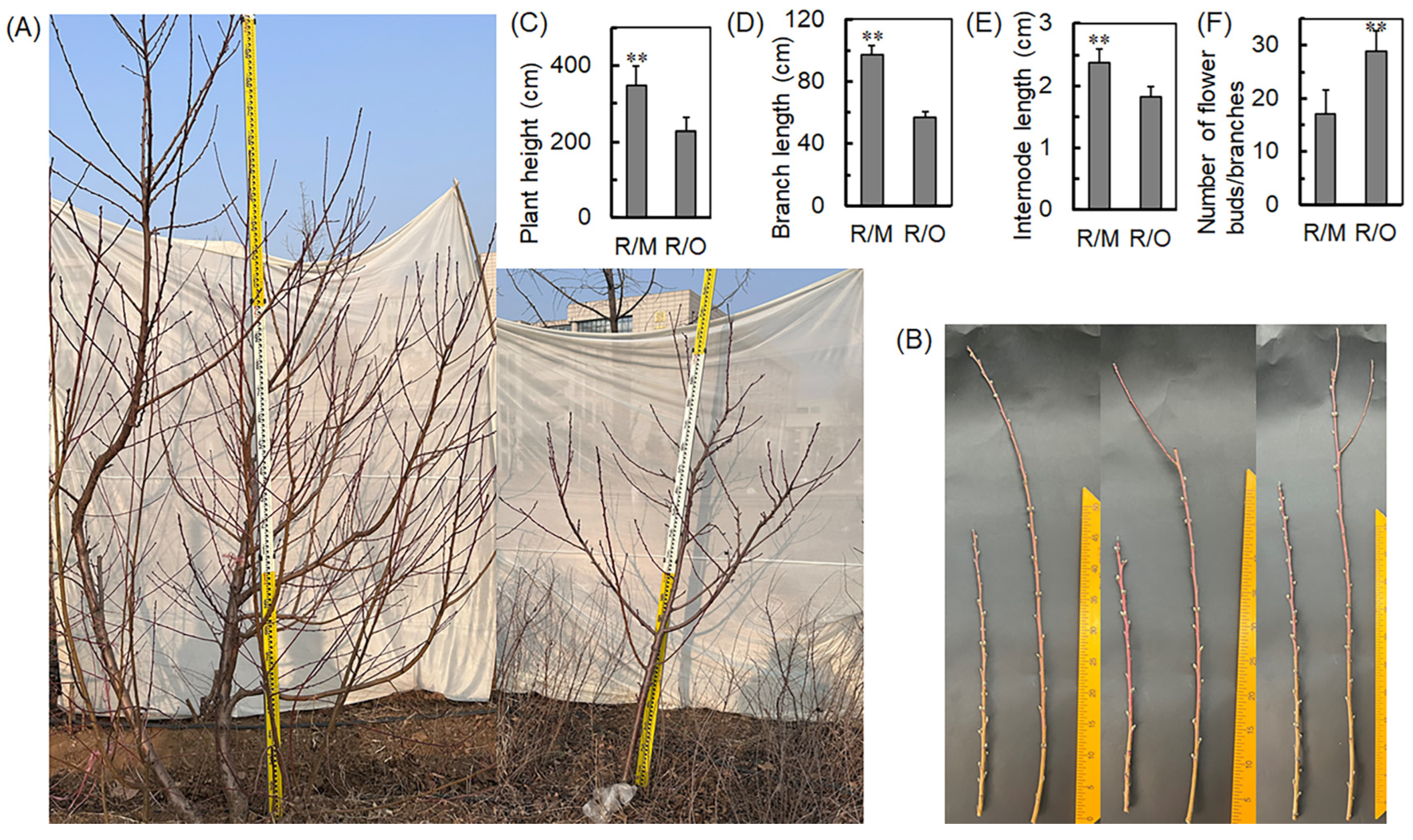
2.1. C. humilis Significantly Reduces the Height of Scions
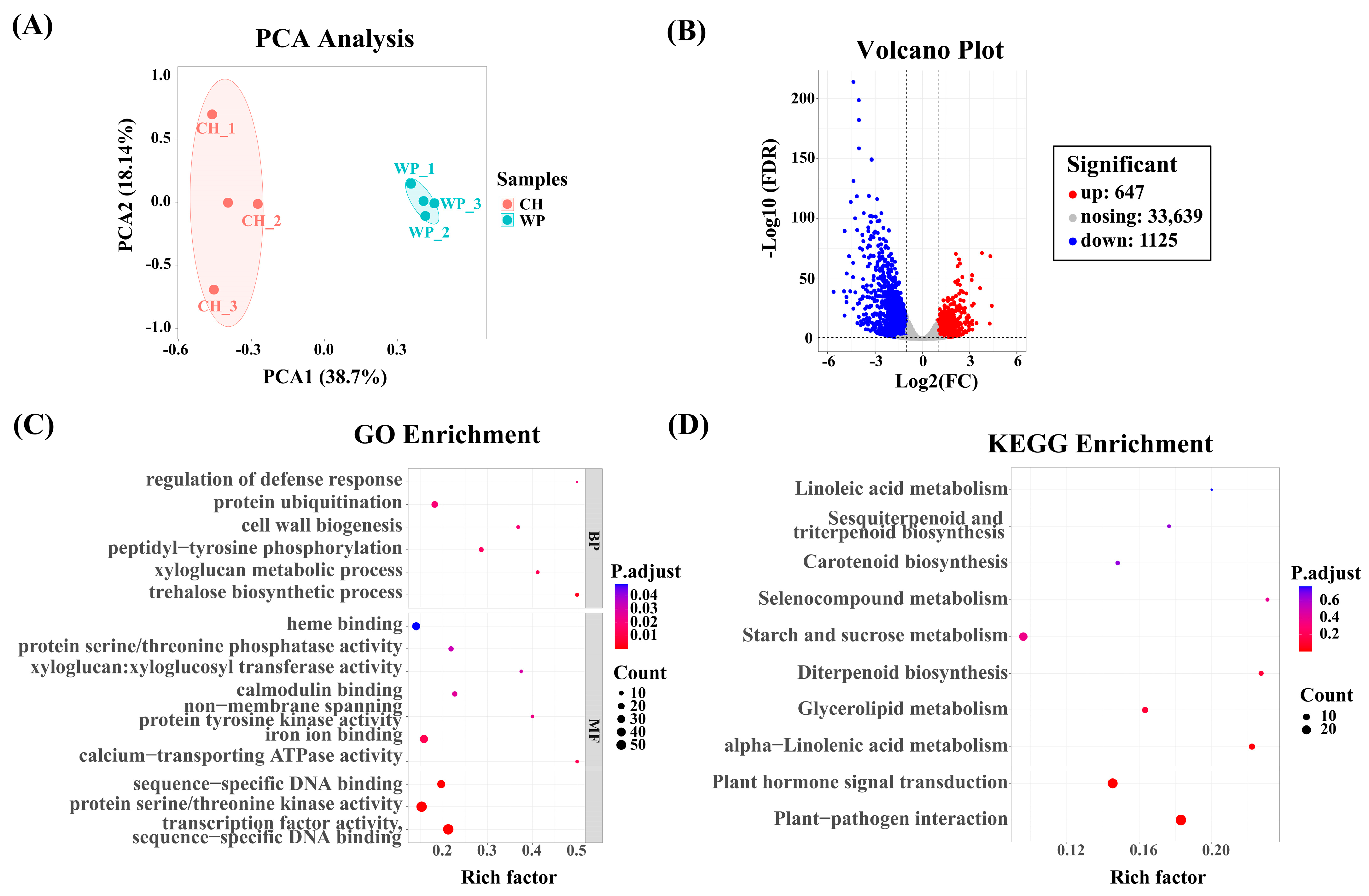
2.2. Differentially Expressed Genes (DEGs) between R/O and R/M
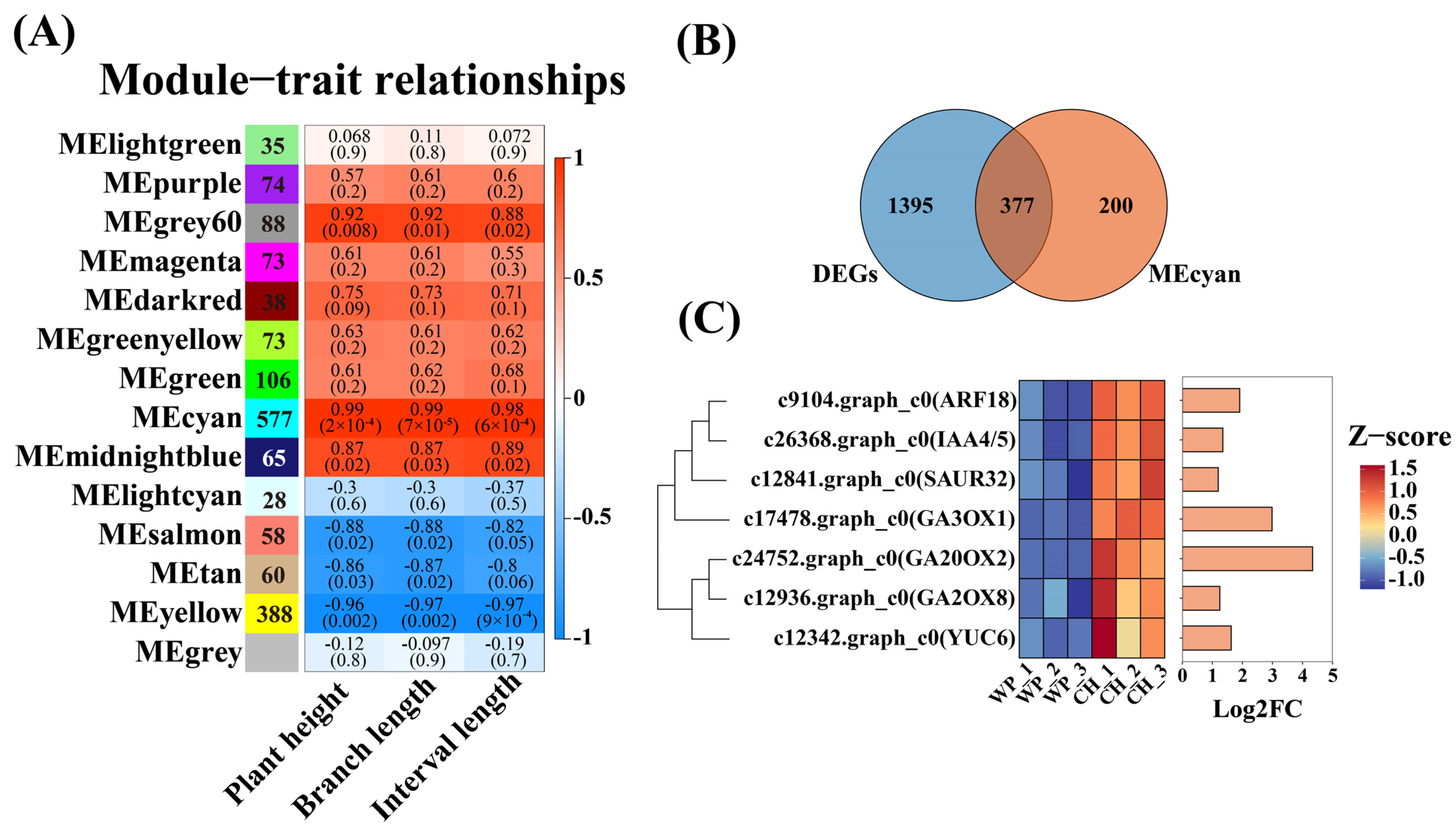
2.3. Weighted Gene Co-Expression Network Analysis (WGCNA Analysis)
2.4. PpGA2ox7 Regulates GA Levels in Peach Trees
2.5. PpGATA21 Directly Binds and Activates PpGA2ox7 Expression
2.6. PpTATA21 Is a GATA-Type Transcriptional Activator
3. Discussion
4. Materials and Methods
4.1. Fruit Materials and Treatments
4.2. Measurement of Plant Growth Parameters
4.3. RNA Extraction and RNA-Seq Analysis
4.4. RT-qPCR
4.5. Quantification of Endogenous Hormones
4.6. Quantitative GUS Assay
4.7. Yeast One-Hybrid (Y1H)
4.8. Transcriptional Activation Analysis in Yeast Cells
4.9. Subcellular Localization Analysis
4.10. Dual-Luciferase Reporter (DLR)
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, X.; Wang, Y.; Zhao, L.; Chen, S.; Yuan, Y.; Wei, T.; Geng, J. Effects of Cerasus humilis (Bge). Sok. Rootstock on Peach Growth, Development, and Expression of Growth-Related Genes. Horticulturae 2023, 9, 576. [Google Scholar] [CrossRef]
- Neri, D.; Crescenzi, S.; Massetani, F.; Manganaris, G.A.; Giorgi, V. Current trends and future perspectives towards sustainable and economically viable peach training systems. Sci. Hortic. 2022, 305, 111348. [Google Scholar] [CrossRef]
- Basile, B.; DeJong, T.M. Control of fruit tree vigor induced by dwarfing rootstocks. Hortic. Rev. 2018, 46, 39–97. [Google Scholar] [CrossRef]
- Shi, J.; Dong, Z.; Song, C.; Xie, B.; Zheng, X.; Song, S.; Jiao, J.; Wang, M.; Bai, T. Establishment of an efficient micropropagation system in enhancing rooting efficiency via stem cuttings of apple rootstock M9T337. Hortic. Sci. 2021, 48, 63–72. [Google Scholar] [CrossRef]
- Habibi, F.; Liu, T.; Folta, K.; Sarkhosh, A. Physiological, biochemical, and molecular aspects of grafting in fruit trees. Hortic. Res. 2022, 9, uhac032. [Google Scholar] [CrossRef]
- Hayat, F.; Ma, C.; Iqbal, S.; Ma, Y.; Khanum, F.; Tariq, R.; Altaf, M.A.; Khan, U.; Coulibaly, D.; Huang, X.; et al. Comprehensive transcriptome profiling and hormonal signaling reveals important mechanism related to dwarfing effect of rootstocks on scion in Japanese apricot (Prunus mume). Sci. Hortic. 2023, 321, 112267. [Google Scholar] [CrossRef]
- Hayat, F.; Li, J.; Liu, W.; Li, C.; Song, W.; Iqbal, S.; Khan, U.; Javed, H.U.; Altaf, M.A.; Tu, P.; et al. Influence of citrus rootstocks on scion growth, hormone levels, and metabolites profile of ‘Shatangju’ mandarin (Citrus reticulata Blanco). Horticulturae 2022, 8, 608. [Google Scholar] [CrossRef]
- Li, W.; Chu, C.; Li, H.; Zhang, H.; Sun, H.; Wang, S.; Wang, Z.; Li, Y.; Foster, T.M.; López-Girona, E.; et al. Near-gapless and haplotype-resolved apple genomes provide insights into the genetic basis of rootstock-induced dwarfing. Nat. Genet. 2024, 56, 505–516. [Google Scholar] [CrossRef]
- Duan, X.; Zhang, W.; Huang, J.; Hao, L.; Wang, S.; Wang, A.; Meng, D.; Zhang, Q.; Chen, Q.; Li, T. PbWoxT1 mRNA from pear (Pyrus betulaefolia) undergoes long-distance transport assisted by a polypyrimidine tract binding protein. New Phytol. 2016, 210, 511–524. [Google Scholar] [CrossRef]
- Soares, J.M.; Weber, K.C.; Qiu, W.; Stanton, D.; Mahmoud, L.M.; Wu, H.; Huyck, P.; Zale, J.; Al Jasim, K.; Grosser, J.W.; et al. The vascular targeted citrus FLOWERING LOCUS T3 gene promotes non-inductive early flowering in transgenic Carrizo rootstocks and grafted juvenile scions. Sci. Rep. 2020, 10, 21404. [Google Scholar] [CrossRef]
- Font i Forcada, C.; Reig, G.; Mestre, L.; Mignard, P.; Betrán, J.; Moreno, M. Scion × rootstock response on production, mineral composition and fruit quality under heavy-calcareous soil and hot climate. Agronomy 2020, 10, 1159. [Google Scholar] [CrossRef]
- Zhou, Y.; Underhill, S.J.R. Expression of gibberellin metabolism genes and signalling components in dwarf phenotype of breadfruit (Artocarpus altilis) plants growing on marang (Artocarpus odoratissimus) rootstocks. Plants 2020, 9, 634. [Google Scholar] [CrossRef] [PubMed]
- Ben Yahmed, J.; Ghrab, M.; Moreno, M.; Pinochet, J.; Ben Mimoun, M. Leaf mineral nutrition and tree vigor of ‘Subirana’ flat peach cultivar grafted on different Prunus rootstocks in a warm Mediterranean area. J. Plant Nutr. 2020, 43, 811–822. [Google Scholar] [CrossRef]
- Szymajda, M.; Pruski, K.; Żurawicz, E.; Sitarek, M. The nursery value of new Prunus persica seedling rootstocks for peach cultivars. Sci. Hortic. 2020, 266, 109282. [Google Scholar] [CrossRef]
- Foster, T.M.; A McAtee, P.; Waite, C.N.; Boldingh, H.L.; McGhie, T.K. Apple dwarfing rootstocks exhibit an imbalance in carbohydrate allocation and reduced cell growth and metabolism. Hortic. Res. 2017, 4, 17009. [Google Scholar] [CrossRef] [PubMed]
- Gullo, G.; Motisi, A.; Zappia, R.; Dattola, A.; Diamanti, J.; Mezzetti, B. Rootstock and fruit canopy position affect peach [Prunus persica (L.) Batsch] (cv. Rich May) plant productivity and fruit sensorial and nutritional quality. Food Chem. 2014, 153, 234–242. [Google Scholar] [CrossRef] [PubMed]
- Anthony, B.M.; Chaparro, J.M.; Sterle, D.G.; Prenni, J.E.; Minas, I.S. Metabolic signatures of the true physiological impact of canopy light environment on peach fruit quality. Environ. Exp. Bot. 2021, 191, 104630. [Google Scholar] [CrossRef]
- Han, H.; Zhang, L.; Liu, S.; Li, N.; Huo, J.; Mu, X. Temporal Changes in Flavonoid Components, Free Radical Scavenging Activities and Metabolism-Related Gene Expressions during Fruit Development in Chinese Dwarf Cherry (Prunus humilis). Horticulturae 2023, 9, 1040. [Google Scholar] [CrossRef]
- Ma, Y.; Xue, H.; Zhang, L.; Zhang, F.; Ou, C.; Wang, F.; Zhang, Z. Involvement of auxin and brassinosteroid in dwarfism of autotetraploid apple (Malus × domestica). Sci. Rep. 2016, 6, 26719. [Google Scholar] [CrossRef]
- Zheng, X.; Zhao, Y.; Shan, D.; Shi, K.; Wang, L.; Li, Q.; Wang, N.; Zhou, J.; Yao, J.; Xue, Y.; et al. MdWRKY9 overexpression confers intensive dwarfing in the M26 rootstock of apple by directly inhibiting brassinosteroid synthetase MdDWF4 expression. New Phytol. 2018, 217, 1086–1098. [Google Scholar] [CrossRef]
- Mäkilä, R.; Wybouw, B.; Smetana, O.; Vainio, L.; Solé-Gil, A.; Lyu, M.; Ye, L.; Wang, X.; Siligato, R.; Jenness, M.K.; et al. Gibberellins promote polar auxin transport to regulate stem cell fate decisions in cambium. Nat. Plants 2023, 9, 631–644. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Ye, X.; Xiong, A.; Zhu, N.; Jiang, L.; Qu, S. The regulatory role of gibberellin related genes DKGA2ox1 and MIR171f_3 in persimmon dwarfism. Plant Sci. 2021, 310, 110958. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, J.; Sabir, I.A.; Sun, W.; Wang, L.; Xu, Y.; Zhang, N.; Liu, H.; Jiu, S.; Liu, L.; et al. PavGA2ox-2L inhibits the plant growth and development interacting with PavDWARF in sweet cherry (Prunus avium L.). Plant Physiol. Biochem. 2022, 186, 299–309. [Google Scholar] [CrossRef]
- Li, Y.; Shan, X.; Jiang, Z.; Zhao, L.; Jin, F. Genome-wide identification and expression analysis of the GA2ox gene family in maize (Zea mays L.) under various abiotic stress conditions. Plant Physiol. Biochem. 2021, 166, 621–633. [Google Scholar] [CrossRef] [PubMed]
- Korkmaz, K.; Bolat, I.; Uzun, A.; Sahin, M.; Kaya, O. Selection and Molecular Characterization of Promising Plum Rootstocks (Prunus cerasifera L.) among Seedling-Origin Trees. Life 2023, 13, 1476. [Google Scholar] [CrossRef] [PubMed]
- Cirilli, M.; Geuna, F.; Babini, A.R.; Bozhkova, V.; Catalano, L.; Cavagna, B.; Dallot, S.; Decroocq, V.; Dondini, L.; Foschi, S.; et al. Fighting sharka in peach: Current limitations and future perspectives. Front. Plant Sci. 2016, 7, 1290. [Google Scholar] [CrossRef] [PubMed]
- Bakır, A.G.; Bolat, I.; Korkmaz, K.; Hasan, M.; Kaya, O. Exogenous nitric oxide and silicon applications alleviate water stress in apricots. Life 2022, 12, 1454. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Mu, X.; Gao, Y.G.; Zhang, J.; Du, J. Successful induction and the systematic characterization of tetraploids in cerasus humilis for subsequent breeding. Sci. Hortic. 2020, 265, 109216. [Google Scholar] [CrossRef]
- Li, Q.; Gao, Y.; Wang, K.; Feng, J.; Sun, S.; Lu, X.; Liu, Z.; Zhao, D.; Li, L.; Wang, D. Transcriptome analysis of the effects of grafting interstocks on apple rootstocks and scions. Int. J. Mol. Sci. 2023, 24, 807. [Google Scholar] [CrossRef]
- Gu, Q.; Wei, Q.; Hu, Y.; Chen, M.; Chen, Z.; Zheng, S.; Ma, Q.; Luo, Z. Physiological and Full-length transcriptome analyses reveal the dwarfing regulation in trifoliate orange (Poncirus trifoliata L.). Plants 2023, 12, 271. [Google Scholar] [CrossRef]
- Zhao, H.; Wan, S.; Huang, Y.; Li, X.; Jiao, T.; Zhang, Z.; Ma, B.; Zhu, L.; Ma, F.; Li, M. The transcription factor MdBPC2 alters apple growth and promotes dwarfing by regulating auxin biosynthesis. Plant Cell 2024, 36, 585–604. [Google Scholar] [CrossRef]
- Shani, E.; Hedden, P.; Sun, T.-P. Highlights in gibberellin research: A tale of the dwarf and the slender. Plant Physiol. 2024, 195, 111–134. [Google Scholar] [CrossRef]
- Liu, J.-L.; Zhang, C.-X.; Li, T.-T.; Liang, C.-L.; Yang, Y.-J.; Li, D.-L.; Cui, Z.-H.; Wang, R.; Song, J.-K. Phenotype and mechanism analysis of plant dwarfing in pear regulated by abscisic acid. J. Integr. Agric. 2022, 21, 1346–1356. [Google Scholar] [CrossRef]
- Wang, J.; Xue, L.; Zhang, X.; Hou, Y.; Zheng, K.; Fu, D.; Dong, W. A new function of MbIAA19 identified to modulate Malus plants dwarfing growth. Plants 2023, 12, 3097. [Google Scholar] [CrossRef]
- Hu, F.; Chen, Z.; Zhao, J.; Wang, X.; Su, W.; Qin, Y.; Hu, G. Differential gene expression between the vigorous and dwarf litchi cultivars based on RNA-Seq transcriptome analysis. PLoS ONE 2018, 13, e0208771. [Google Scholar] [CrossRef]
- Shu, K.; Chen, Q.; Wu, Y.; Liu, R.; Zhang, H.; Wang, P.; Li, Y.; Wang, S.; Tang, S.; Liu, C.; et al. ABI4 mediates antagonistic effects of abscisic acid and gibberellins at transcript and protein levels. Plant J. 2016, 85, 348–361. [Google Scholar] [CrossRef]
- Meraj, T.A.; Fu, J.; Raza, M.A.; Zhu, C.; Shen, Q.; Xu, D.; Wang, Q. Transcriptional factors regulate plant stress responses through mediating secondary metabolism. Genes 2020, 11, 346. [Google Scholar] [CrossRef]
- Waseem, M.; Nkurikiyimfura, O.; Niyitanga, S.; Jakada, B.H.; Shaheen, I.; Aslam, M.M. GRAS transcription factors emerging regulator in plants growth, development, and multiple stresses. Mol. Biol. Rep. 2022, 49, 9673–9685. [Google Scholar] [CrossRef]
- Hao, Y.; Zong, X.; Ren, P.; Qian, Y.; Fu, A. Basic helix-loop-helix (bHLH) transcription factors regulate a wide range of functions in Arabidopsis. Int. J. Mol. Sci. 2021, 22, 7152. [Google Scholar] [CrossRef]
- Shahnejat-Bushehri, S.; Tarkowska, D.; Sakuraba, Y.; Balazadeh, S. Arabidopsis NAC transcription factor JUB1 regulates GA/BR metabolism and signalling. Nat. Plants 2016, 2, 16013. [Google Scholar] [CrossRef]
- Odgerel, K.; Jose, J.; Karsai-Rektenwald, F.; Ficzek, G.; Simon, G.; Végvári, G.; Bánfalvi, Z. Effects of the repression of GIGANTEA gene StGI.04 on the potato leaf transcriptome and the anthocyanin content of tuber skin. BMC Plant Biol. 2022, 22, 249. [Google Scholar] [CrossRef] [PubMed]
- Niu, L.; Chu, H.D.; Tran, C.D.; Nguyen, K.H.; Pham, H.X.; Le, D.T.; Li, W.; Wang, W.; Le, T.D.; Tran, L.-S.P. The GATA gene family in chickpea: Structure analysis and transcriptional responses to abscisic acid and dehydration treatments revealed potential genes involved in drought adaptation. J. Plant Growth Regul. 2020, 39, 1647–1660. [Google Scholar] [CrossRef]
- Liang, J.; Wu, Z.; Zheng, J.; Koskela, E.A.; Fan, L.; Fan, G.; Gao, D.; Dong, Z.; Hou, S.; Feng, Z.; et al. The GATA factor HANABA TARANU promotes runner formation by regulating axillary bud initiation and outgrowth in cultivated strawberry. Plant J. 2022, 110, 1237–1254. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Huang, L.; Zhang, Y.; Yan, Z.; Wang, N. Overexpression of PdeGATA3 results in a dwarf phenotype in poplar by promoting the expression of PdeSTM and altering the content of gibberellins. Tree Physiol. 2022, 42, 2614–2626. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An Integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Tong, Z.; Gao, Z.; Wang, F.; Zhou, J.; Zhang, Z. Selection of reliable reference genes for gene expression studies in peach using real-time PCR. BMC Mol. Biol. 2009, 10, 71. [Google Scholar] [CrossRef]
- Shao, Y.; Zhou, H.-Z.; Wu, Y.; Zhang, H.; Lin, J.; Jiang, X.; He, Q.; Zhu, J.; Li, Y.; Yu, H.; et al. OsSPL3, an SBP-domain protein, regulates crown root development in rice. Plant Cell 2019, 31, 1257–1275. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Wang, R.; Wang, Y.; Li, X.; Shi, Q.; Yu, Y. PpGATA21 Enhances the Expression of PpGA2ox7 to Regulate the Mechanism of Cerasus humilis Rootstock-Mediated Dwarf in Peach Trees. Int. J. Mol. Sci. 2024, 25, 7402. https://doi.org/10.3390/ijms25137402
Li X, Wang R, Wang Y, Li X, Shi Q, Yu Y. PpGATA21 Enhances the Expression of PpGA2ox7 to Regulate the Mechanism of Cerasus humilis Rootstock-Mediated Dwarf in Peach Trees. International Journal of Molecular Sciences. 2024; 25(13):7402. https://doi.org/10.3390/ijms25137402
Chicago/Turabian StyleLi, Xiuzhen, Ruxin Wang, Yuman Wang, Xueqiang Li, Qiaofang Shi, and Yihe Yu. 2024. "PpGATA21 Enhances the Expression of PpGA2ox7 to Regulate the Mechanism of Cerasus humilis Rootstock-Mediated Dwarf in Peach Trees" International Journal of Molecular Sciences 25, no. 13: 7402. https://doi.org/10.3390/ijms25137402




