Potential of Polar Lipids Isolated from the Marine Sponge Haliclona (Halichoclona) vansoesti against Melanoma
Abstract
1. Introduction
2. Results
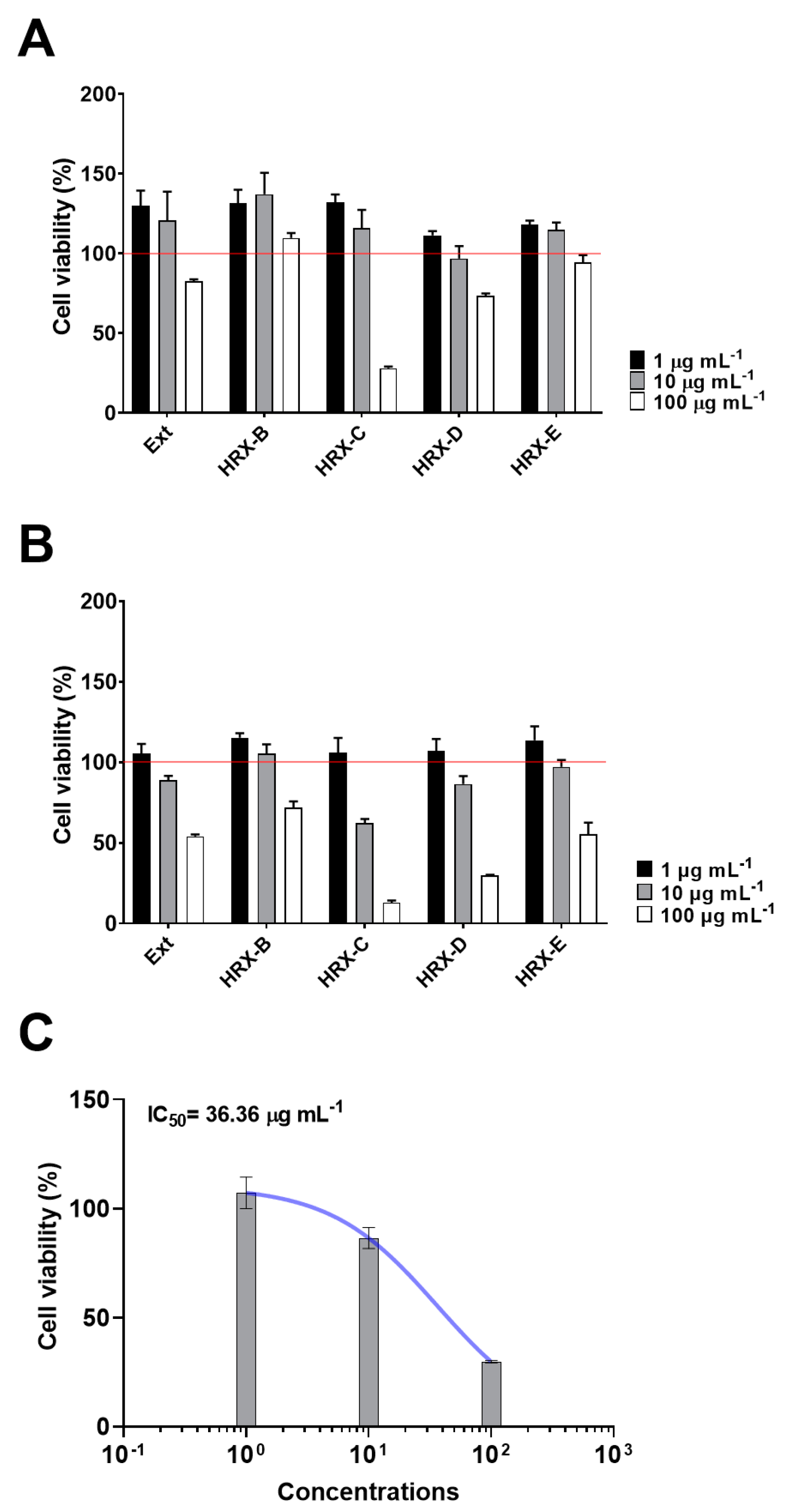
2.1. Bioassay-Guided Fractionation and Chemical Analysis of the Active Fractions
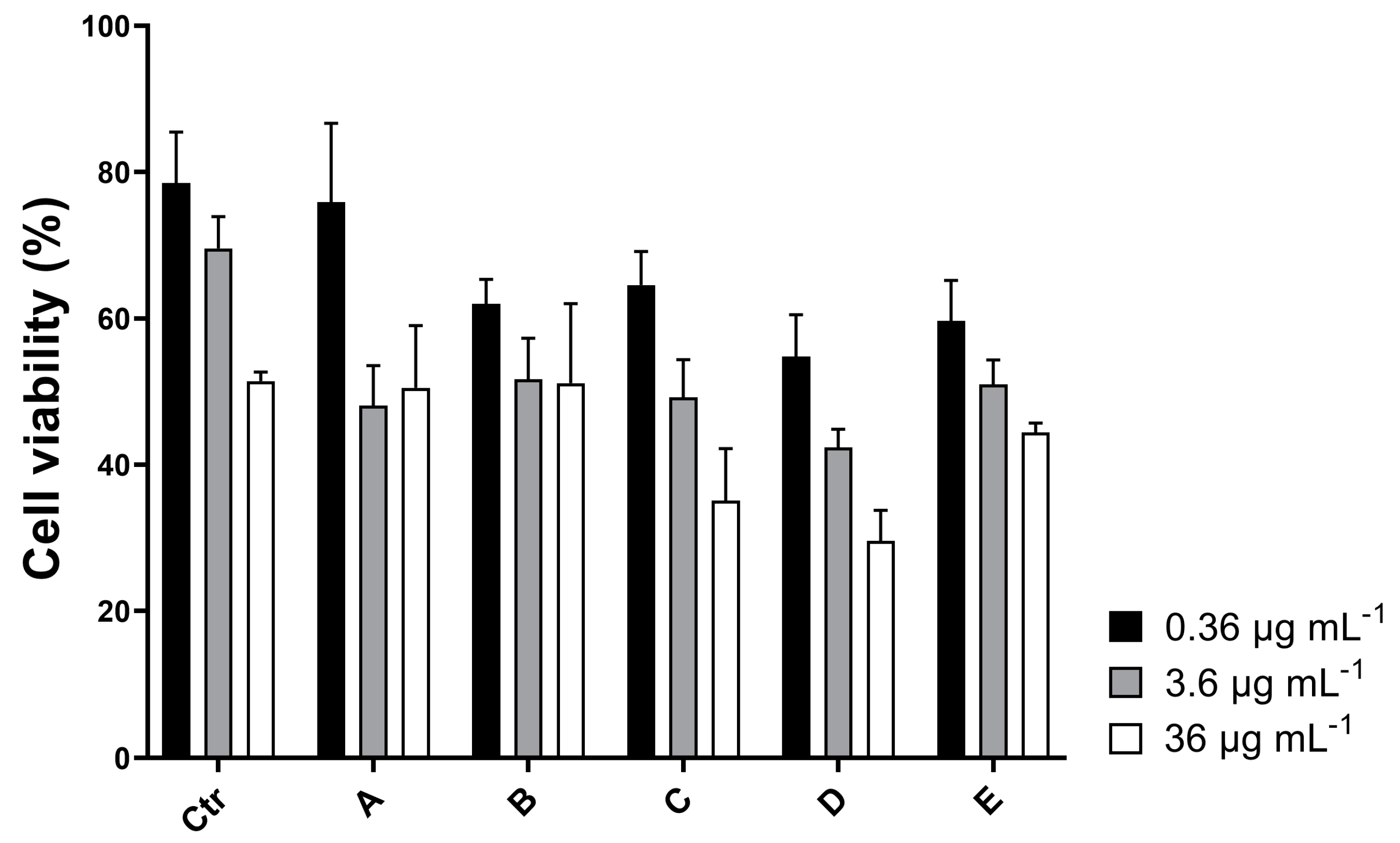
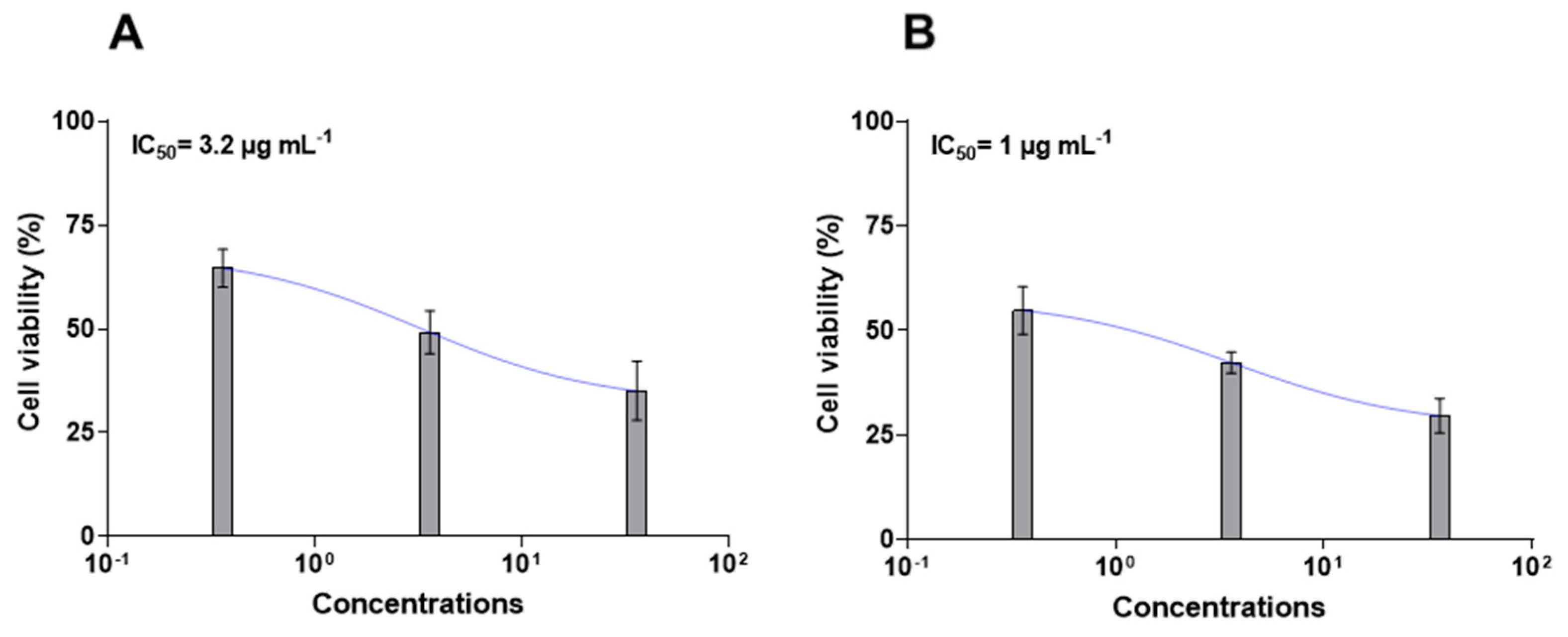
2.2. Chemical Purification and Characterization of HILIC Fraction C
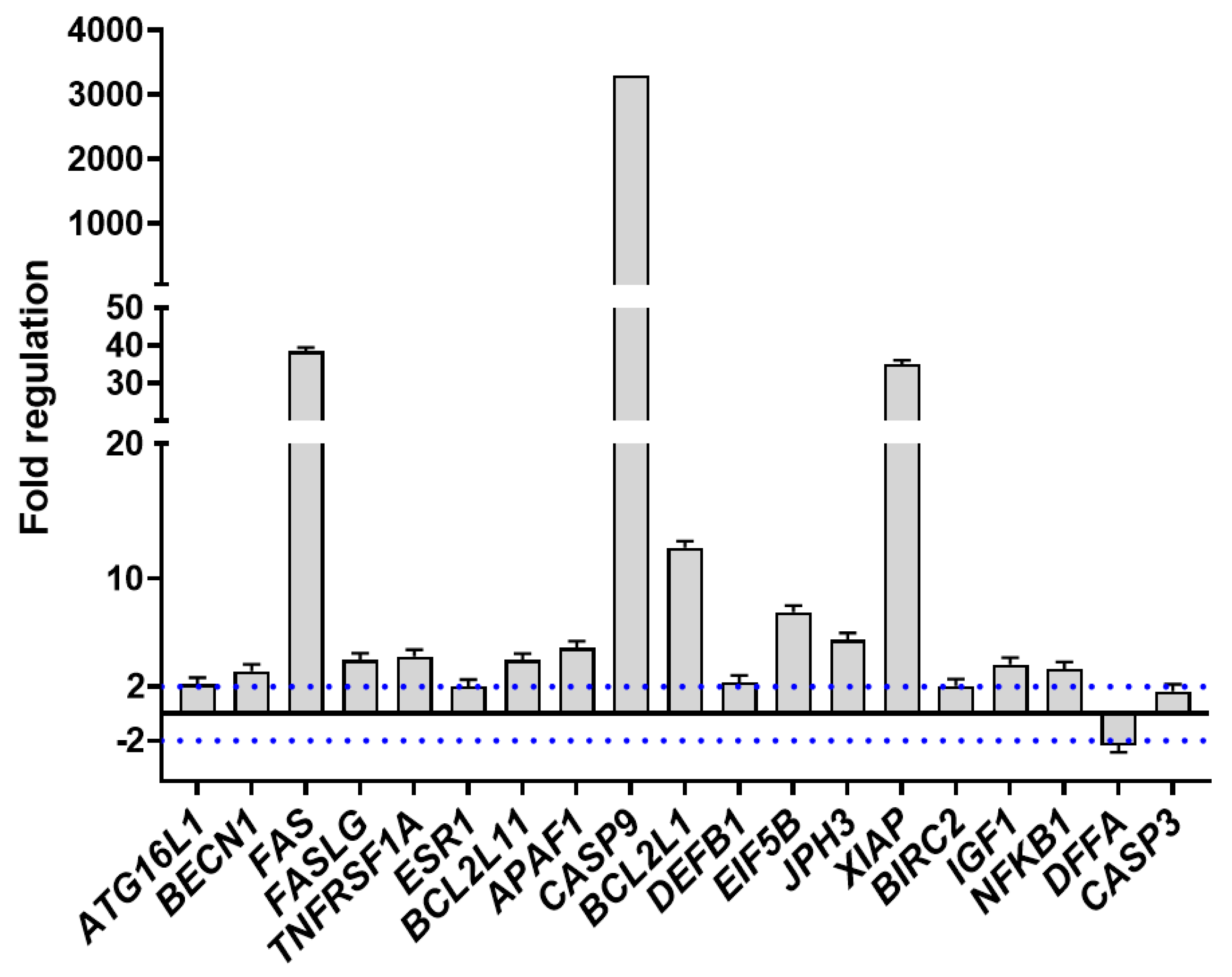
2.3. PCR Array
2.4. ImmunoArray
3. Discussion
4. Materials and Methods
4.1. Chemical Extraction and Fractionation
4.2. Cell Maintenance and Treatments
4.3. Cytotoxicity Assay
4.4. RNA Extraction, cDNA Synthesis, and RT2 Profiler PCR-Array
4.5. Protein Extraction and ImmunoArray
4.6. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Esposito, R.; Federico, S.; Glaviano, F.; Somma, E.; Zupo, V.; Costantini, M. Bioactive compounds from marine sponges and algae: Effects on cancer cell metabolome and chemical structures. Int. J. Mol. Sci. 2022, 23, 10680. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.; Sorolla, M.A.; Krishnan, P.D.G.; Sorolla, A. From seabed to bedside: A review on promising marine anticancer compounds. Biomolecules 2020, 10, 248. [Google Scholar] [CrossRef] [PubMed]
- Nuzzo, G.; Manzo, E.; Gallo, C.; D’Ippolito, G.; Fontana, A. Fractionation protocol of marine metabolites. Methods Mol. Biol. 2022, 2498, 307–313. [Google Scholar] [PubMed]
- Nadar, V.M.; Manivannan, S.; Chinnaiyan, R.; Govarthanan, M.; Ponnuchamy, K. Review on marine sponge alkaloid, aaptamine: A potential antibacterial and anticancer drug. Chem. Biol. Drug Des. 2022, 99, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Thakur, N.; Choudhary, P.; Kaushik, N.; Sharma, A. A review on pharmacological and pharmaceutical properties of Genus Stelletta from marine sponges. Mater. Today Proc. 2023, 81, 1034–1039. [Google Scholar] [CrossRef]
- Hassane, C.S.; Fouillaud, M.; Le Goff, G.; Skilrou, A.D.; Boyer, J.B.; Trougakos, I.P.; Jerabek, M.; Bignon, J.; de Voogd, N.J.; Ouazzani, J.; et al. Microorganisms associated with the marine sponge Scopalina hapalia: A reservoir of bioactive molecules to slow down the aging process. Microorganisms 2020, 8, 1262. [Google Scholar] [CrossRef] [PubMed]
- Bergmann, W.; Feeney, R.J. Contributions to the study of marine products. XXXII. The nucleosides of sponges. I. J. Org. Chem. 1951, 16, 981–987. [Google Scholar] [CrossRef]
- Hudayah, T.; Gul-E-Saba, T.M.; Ismail, N.; Muhammad, T.S.T. Methanol extracts of four selected marine sponges induce apoptosis in human breast cancer cell line, MCF-7. Int. J. Res. Pharm. Sci. 2017, 8, 667–675. [Google Scholar]
- Panigrahi, A.R.; Guntuku, G.; Kumar, M.K. Screening and identification of marine sponge associated fungus producing novel bioactive molecules. Int. J. Res. Pharm. Chem. 2017, 7, 92–106. [Google Scholar]
- Shady, N.H.; El-Hossary, E.M.; Fouad, M.A.; Gulder, T.A.M.; Kamel, M.S.; Abdelmohsen, U.R. Bioactive natural products of marine sponges from the genus Hyrtios. Molecules 2017, 22, 781. [Google Scholar] [CrossRef]
- Bashari, M.H.; Huda, F.; Tartila, T.S.; Shabrina, S.; Putri, T.; Qomarilla, N.; Atmaja, H.; Subhan, B.; Sudji, I.R.; Meiyanto, E. Bioactive compounds in the ethanol extract of marine sponge Stylissa carteri demonstrates potential anti-cancer activity in breast cancer cells. Asian Pac. J. Cancer Prev. 2019, 20, 1199–1206. [Google Scholar] [CrossRef]
- Pagliara, P.; Barca, A.; Verri, T.; Caroppo, C. The marine sponge Petrosia ficiformis harbors different cyanobacteria strains with potential biotechnological application. J. Mar. Sci. Eng. 2020, 8, 638. [Google Scholar] [CrossRef]
- Santos, J.D.; Vitorino, I.; de la Cruz, M.; Díaz, C.; Cautain, B.; Annang, F.; Pérez-Moreno, G.; Gonzalez, I.; Tormo, J.R.; Martin, J.; et al. Diketopiperazines and other bioactive compounds from bacterial symbionts of marine sponges. Antonie Leeuwenhoek 2020, 113, 875–887. [Google Scholar] [CrossRef] [PubMed]
- Santos, J.D.; Vitorino, I.; De La Cruz, M.; Díaz, C.; Cautain, B.; Annang, F.; Pérez-Moreno, G.; Gonzalez, I.; Tormo, J.R.; Martin, J.; et al. Bioactivities and extract dereplication of Actinomycetales isolated from marine sponges. Front. Microbiol. 2019, 10, 727. [Google Scholar] [CrossRef]
- Nikolaou, V.; Stratigos, A.J. Emerging trends in the epidemiology of melanoma. Br. J. Dermatol. 2014, 170, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Domingues, B.; Lopes, J.; Soares, P.; Populo, H. Melanoma treatment in review. Immunotargets Ther. 2018, 7, 35–49. [Google Scholar] [CrossRef]
- Jenkins, R.W.; Fisher, D.E. Treatment of advanced melanoma in 2020 and beyond. J. Investig. Dermatol. 2021, 141, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Vanmeerbeek, I.; Sprooten, J.; De Ruysscher, D.; Tejpar, S.; Vandenberghe, P.; Fucikova, J.; Spisek, R.; Zitvogel, L.; Kroemer, G.; Galluzzi, L.; et al. Trial watch: Chemotherapy-induced immunogenic cell death in immuno-oncology. Oncoimmunology 2020, 9, 1703449. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Wang, G.; Chen, Y.; Wang, H.; Hua, Y.; Cai, Z. Immunogenic cell death in cancer therapy: Present and emerging inducers. J. Cell. Mol. Med. 2019, 23, 4854–4865. [Google Scholar] [CrossRef]
- Nuzzo, G.; Gallo, C.; Crocetta, F.; Romano, L.; Barra, G.; Senese, G.; dell’Isola, M.; Carbone, D.; Tanduo, V.; Albiani, F.; et al. Identification of the marine alkaloid Lepadin A as potential inducer of immunogenic cell death. Biomolecules 2022, 12, 246. [Google Scholar] [CrossRef]
- Careaga, V.P.; Maier, M.S. Cerebrosides from marine organisms. Stud. Nat. Prod. Chem. 2014, 42, 59–81. [Google Scholar]
- Biegelmeyer, R.; Schröder, R.; Rambo, D.F.; Dresch, R.R.; Carraro, J.L.F.; Mothes, B.; Moreira, J.C.F.; da Frota, M.L.C.; Henriques, A.T. Sphingosines derived from marine sponge as potential multi-target drug related to disorders in cancer development. Mar. Drugs 2015, 13, 5552–5563. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Cao, M.; Lam, S.M.; Shui, G. Embracing lipidomics at single-cell resolution: Promises and pitfalls. Trends Anal. Chem. 2023, 160, 1169. [Google Scholar] [CrossRef]
- Aida, K.; Takakuwa, N.; Kinoshita, M.; Sugawara, T.; Imai, H.; Ono, J.; Ohnishi, M. Properties and physiological effects of plant cerebroside species as functional lipids. In Advanced Research on Plant Lipids; Springer: Dordrecht, The Netherlands, 2003; pp. 233–236. [Google Scholar] [CrossRef]
- Lee, M.; Lee, S.Y.; Bae, Y.S. Functional roles of sphingolipids in immunity and their implication in disease. Exp. Mol. Med. 2023, 55, 1110–1130. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, F.J.; McDonald, F.J. Isolation and identification of cerebrosides from the marine sponge Chondrilla nucula. J. Lipid Res. 1974, 15, 158–164. [Google Scholar] [CrossRef] [PubMed]
- Morita, M.; Sawa, E.; Yamaji, K.; Sakai, T.; Natori, T.; Koezuka, Y.; Fukushima, H.; Akimoto, K. Practical total synthesis of (2S,3S,4R)-1-O-(α-D-Galactopyranosyl)-N-hexacosanoyl-2-amino-1,3,4-octadecanetriol, the antitumorial and immunostimulatory α-Galactosylcer-amide, KRN7000. Biosci. Biotechnol. Biochem. 1996, 60, 288–292. [Google Scholar] [CrossRef] [PubMed]
- Crul, M.; Mathôt, R.A.; Giaccone, G.; Punt, C.A.; Rosing, H.; Hillebrand, M.X.; Ando, Y.; Nishi, N.; Tanaka, H.; Schellens, J.M.; et al. Population pharmacokinetics of the novel anticancer agent KRN7000. Cancer Chemother. Pharmacol. 2002, 49, 287–293. [Google Scholar] [CrossRef] [PubMed]
- Santalova, E.A.; Denisenko, V.A.; Dmitrenok, P.S. Structural analysis of oxidized Cerebrosides from the extract of deep-sea sponge Aulosaccus sp.: Occurrence of amide-linked allylically oxygenated fatty acids. Molecules 2020, 25, 6047. [Google Scholar] [CrossRef] [PubMed]
- Santalova, E.A.; Denisenko, V.A.; Dmitrenok, P.S.; Drozdov, A.L.; Stonik, V.A. Cerebrosides from a far-Eastern glass sponge Aulosaccus sp. Lipids 2015, 50, 57–69. [Google Scholar] [CrossRef]
- Cutignano, A.; Nuzzo, G.; Ianora, A.; Luongo, E.; Romano, G.; Gallo, C.; Sansone, C.; Aprea, S.; Mancini, F.; D’Oro, U.; et al. Development and application of a novel SPE-method for bioassay-guided fractionation of marine extracts. Mar. Drugs 2015, 13, 5736–5749. [Google Scholar] [CrossRef]
- Bertolino, M.; Costa, G.; Ruocco, N.; Esposito, R.; De Matteo, S.; Zagami, G.; Costantini, M. First certain record of Demospongiae class (Porifera) alien species from the Mediterranean Sea. Mar. Genom. 2022, 63, 100951. [Google Scholar] [CrossRef] [PubMed]
- Richelle-Maurer, E.; Braekman, J.C.; De Kluijver, M.J.; Gomez, R.; De Vyver, G.; Soest, R.W.M.; Devijver, C. Cellular location of (2R, 3R, 7Z)-2-aminotetradec-7-ene-1, 3-diol, a potent antimicrobial metabolite produced by the Caribbean sponge Haliclona vansoesti. Cell Tissue Res. 2001, 306, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Devijver, C.; Salmoun, M.; Daloze, D.; Braekman, J.C.; De Weerdt, W.H.; De Kluijver, M.J.; Gomez, R. (2R,3R,7Z)-2-Aminotetradec-7-ene-1,3-diol, a new amino alcohol from the Caribbean sponge Haliclona vansoesti. J. Nat. Prod. 2000, 63, 978–980. [Google Scholar] [CrossRef] [PubMed]
- Bodennec, J.; Koul, O.; Aguado, I.; Brighon, G.; Zwingelstein, G.; Portoukalian, J. A procedure for fractionation of sphingolipid classes by solid-phase extraction on aminopropyl cartridges. J. Lipid Res. 2000, 41, 1524–1531. [Google Scholar] [CrossRef]
- Cheng, S.Y.; Wen, Z.H.; Chiou, S.F.; Tsai, C.W.; Wang, S.K.; Hsu, C.H.; Dai, C.F.; Chiang, M.Y.; Wang, W.H.; Duh, C.Y. Ceramide and cerebrosidea from octocoral Sarcophyton ehrenbergi. J. Nat. Prod. 2009, 72, 465–468. [Google Scholar] [CrossRef] [PubMed]
- Jatai Batista, P.; Nuzzo, G.; Gallo, C.; Carbone, D.; dell’Isola, M.; Affuso, M.; Barra, G.; Albiani, F.; Crocetta, F.; Virgili, R.; et al. Chemical and pharmacological prospection of the ascidian Cystodytes dellechiajei. Mar. Drugs 2024, 22, 75. [Google Scholar] [CrossRef] [PubMed]
- Jung, S.H.; Lee, C.O.; Kim, Y.C.; Kan, S.S. New bioactive cerebrosides from Arisaema amurense. J. Nat. Prod. 1996, 59, 319–322. [Google Scholar] [CrossRef] [PubMed]
- Mansoor, T.A.; Park, T.; Luo, X.; Hong, J.; Lee, C.O.; Jung, J.H. A new Sphingosine from a marine sponge Haliclona (Reniera) sp. Chem. Pharm. Bull. 2007, 13, 247–250. [Google Scholar]
- Brinkmann, C.M.; Marker, A.; Kurtböke, D.I. An overview on marine sponge-symbiotic bacteria as unexhausted sources for natural product discovery. Diversity 2017, 9, 40. [Google Scholar] [CrossRef]
- Esposito, R.; Ruocco, N.; Viel, T.; Federico, S.; Zupo, V.; Costantini, M. Sponges and their symbionts as a source of valuable compounds in cosmeceutical field. Mar. Drugs 2021, 19, 444. [Google Scholar] [CrossRef]
- Ramanjooloo, A.; Andersen, R.J.; Bhaw-Luximon, A. Marine sponge-derived/inspired drugs and their applications in drug delivery systems. Future Med. Chem. 2021, 13, 487–504. [Google Scholar] [CrossRef]
- Varijakzhan, D.; Loh, J.Y.; Yap, W.S.; Tusoff, K.; Seboussi, R.; Lim, S.H.E.; Lai, K.S.; Chong, C.M. Bioactive compounds from marine sponges: Ufndamentals and applications. Mar. Drugs 2021, 19, 246. [Google Scholar] [CrossRef]
- Marra, M.V.; Bertolino, M.; Pansini, M.; Giacobbe, S.; Manconi, R.; Pronzato, R. Long-term turnover of the sponge fauna in Faro Lake (North-East Sicily, Mediterranean Sea). Ital. J. Zool. 2016, 83, 579–588. [Google Scholar] [CrossRef]
- De Weerdt, W.H. A monograph of the shallow-water Chalinidae (Porifera, Haplosclerida) of the Caribbean. Beaufortia 2000, 50, 1–67. [Google Scholar]
- Muricy, G.; Esteves, E.L.; Monteiro, L.C.; Rodrigues, B.R.; Albano, R.M. A new species of Haliclona (Demospongiae: Haplosclerida: Chalinidae) from Southeastern Brazil and the first record of Haliclona vansoesti from the Brazilian coast. Zootaxa 2015, 3925, 536–550. [Google Scholar] [CrossRef]
- Servello, G.; Andaloro, F.; Azzurro, E.; Castriota, L.; Catra, M.; Chiarore, A.; Crocetta, F.; D’Alessandro, M.; Denitto, F.; Froglia, C.; et al. Marine alien species in Italy: A contribution to the implementation of descriptor D2 of the marine strategy framework directive. Mediterr. Mar. Sci. 2019, 20, 1–48. [Google Scholar] [CrossRef]
- Molinski, T.F.; Biegelmeyer, R.; Stout, E.P.; Wang, X.; Frota, M.L.C., Jr.; Henriques, A.T. Halisphingosines A and B, modified Sphingoid bases from Haliclona tubifera. Assignment of configuration by circular dichroism and van’t Hoff’s principle of optical superposition. J. Nat. Prod. 2013, 76, 374–381. [Google Scholar] [CrossRef]
- Ling, L.U.; Tan, K.B.; Lin, H.; Chiu, G.N.C. The role of reactive oxygen species and autophagy in safingol-induced cell death. Cell Death Dis. 2011, 2, e129. [Google Scholar] [CrossRef] [PubMed]
- Čonková, M.; Martinková, M.; Gonda, J.; Jacková, D.; Pilátová, M.B.; Kupka, D.; Jáger, D. Stereoselective synthesis and antiproliferative activity of the isomeric sphinganine analogues. Carbohydr. Res. 2019, 472, 76–85. [Google Scholar] [CrossRef]
- Glick, D.; Barth, S.; Macleod, K.F. Autophagy: Cellular and molecular mechanisms. J. Pathol. 2010, 221, 3–12. [Google Scholar] [CrossRef]
- Denton, D.; Kumar, S. Autophagy-dependent cell death. Cell Death Differ. 2019, 26, 605–616. [Google Scholar] [CrossRef] [PubMed]
- Levine, B.; Kroemer, G. Biological functions of autophagy genes: A disease perspective. Cell 2019, 176, 11–42. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Klionsky, D.J. Autophagy and disease: Unanswered questions. Cell Death Differ. 2020, 27, 858–871. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.C.S.; Hahne, M.; Schroeter, M.; Frei, K.; Fontana, A.; Villunger, A.; Newton, K.; Tschopp, J.; Strasser, A. Activation of Fas by FasL induces apoptosis by a mechanism that cannot be blocked by Bcl-2 or Bcl-xL. Proc. Natl. Acad. Sci. USA 1999, 96, 14871–14876. [Google Scholar] [CrossRef] [PubMed]
- Campbell, K.J.; Tait, S.W.G. Targeting BCL-2 regulated apoptosis in cancer. Open Biol. 2018, 8, 180002. [Google Scholar] [CrossRef] [PubMed]
- Brentnall, M.; Rodriguez-Menocal, L.; De Guevara, R.; Cepero, E.; Boise, L. Caspase-9, caspase-3 and caspase-7 have distinct roles during intrinsic apoptosis. BMC Cell Biol. 2013, 14, 32. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Zhou, L.; Zhao, T.; Liu, X.; Zhang, P.; Liu, Y.; Zheng, X.; Li, Q. Caspase-9: Structure, mechanisms and clinical application. Oncotarget 2017, 8, 23996–24008. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Zhang, Y.; Zhao, S.; Chen, J.; Yang, J.; Wang, T.; Zou, H.; Wang, Y.; Gu, J.; Liu, X.; et al. Cadmium-induced apoptosis in neuronal cells is mediated by Fas/FasL-mediated mitochondrial apoptotic signaling pathway. Sci. Rep. 2018, 8, 8837. [Google Scholar] [CrossRef] [PubMed]
- Brindle, N.P.J.; Saharinen, P.; Alitalo, K. Signaling and functions of angiopoietin-1 in vascular protection. Circ. Res. 2006, 98, 1014–1023. [Google Scholar] [CrossRef]
- Natori, T.; Sata, M.; Washida, M.; Hirata, Y.; Nagai, R.; Makuuchi, M. G-CSF stimulates angiogenesis and promotes tumor growth: Potential contribution of bone marrow-derived endothelial progenitor cells. Biochem. Biophys. Res. Commun. 2002, 297, 1058–1061. [Google Scholar] [CrossRef]
- Bernardini, G.; Spinetti, G.; Ribatti, D.; Camarda, G.; Morbidelli, L.; Ziche, M.; Santoni, A.; Capogrossi, M.C.; Napolitano, M. I-309 binds to and activates endothelial cell functions and acts as an angiogenic molecule in vivo. Blood 2000, 96, 4039–4045. [Google Scholar] [CrossRef] [PubMed]
- Deryugina, E.I.; Quigley, J.P. Tumor angiogenesis: MMP-mediated induction of intravasation- and metastasis-sustaining neovasculature. Matrix Biol. 2015, 44–46, 94–112. [Google Scholar] [CrossRef] [PubMed]
- Quintero-Fabián, S.; Arreola, R.; Becerril-Villanueva, E.; Torres-Romero, J.C.; Arana-Argáez, V.; Lara-Riegos, J.; Ramírez-Camacho, M.A.; Alvarez-Sánchez, M.E. Role of matrix metalloproteinases in angiogenesis and cancer. Front. Oncol. 2019, 9, 1370. [Google Scholar] [CrossRef] [PubMed]
- Kohno, T.; Mizukami, H.; Suzuki, M.; Saga, Y.; Takei, Y.; Shimpo, M.; Matsushita, T.; Okada, T.; Hanazono, Y.; Kume, A.; et al. Interleukin-10-mediated inhibition of angiogenesis and tumor growth in mice bearing VEGF-producing ovarian cancer. Cancer Res. 2003, 63, 5091–5094. [Google Scholar] [PubMed]
- Dell’Eva, R.; Pfeffer, U.; Indraccolo, S.; Albini, A.; Noonan, D. Inhibition of tumor angiogenesis by angiostatin: From recombinant protein to gene therapy. Endothelium 2002, 9, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Berríos, R.R.; Ríos-Delgado, A.M.; Perdomo-Lizardo, A.P.; Cardona-Rivera, A.E.; Vidal-Rosado, Á.G.; Narváez-Lozano, G.A.; Nieves-Quiñones, I.A.; Rodríguez-Vargas, J.A.; Álamo-Diverse, K.Y.; Lebrón-Acosta, N.; et al. Extraction, isolation, characterization, and bioactivity of polypropionates and related polyketide metabolites from the Caribbean Region. Antibiotics 2023, 12, 1087. [Google Scholar] [CrossRef] [PubMed]
- Aiello, A.; Fattorusso, E.; Menna, M.; Pansini, M. Further bioactive acetylenic compounds from the Caribbean sponge Cribrochalina vasculum. J. Nat. Prod. 1992, 55, 1275–1280. [Google Scholar] [CrossRef]
- Grube, A.; Assmann, M.; Lichte, E.; Sasse, F.; Pawlik, J.R.; Köck, M. Bioactive metabolites from the Caribbean sponge Aka coralliphagum. J. Nat. Prod. 2007, 70, 504–509. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]


| Rt (min) | m/z Ion Peak [M-H]−/[M+Cl]− | m/z Fragment [M-H-Glc]− | Molecular Formula | Exact Mass |
|---|---|---|---|---|
| 19.20 | 732.56/768.54 | 570.52 | C40H79NO10 | 733.5704 |
| 20.97 | 712.54/748.53 | 550.49 | C40H75NO9 | 713.5442 |
| 22.49 | 726.56/762.54 | 564.50 | C41H77NO9 | 727.5962 |
| 23.02 | 760.60/796.59 | 598.54 | C42H83NO10 | 761.6017 |
| 23.79 | 740.57/776.56 | 578.52 | C42H79NO9 | 741.5755 |
| 26.64 | 816.66/852.65 | 654.61 | C46H91NO10 | 817.6643 |
| 27.29 | 830.67/866.65 | 668.62 | C46H89NO11 | 831.6436 |
| 1a, 1d | 1b, 1c, 1e | |||
|---|---|---|---|---|
| Position | δH (m, J in Hz) | δC, Type | δH (m, J in Hz) | δC, Type |
| 1 | 4.08 (dd, 10.5, 6.0) 3.83 (dd, 10.5, 3.8) | 69.7, CH2 | 4.14 (overlap.) 3.75 (dd, 10.5, 3.8) | 69.5, CH2 |
| 2 | 4.28 (m) | 51.5, CH | 4.02 (m) | 54.4, CH |
| 3 | 3.64 (t, 6.0) | 75.2, CH | 4.17 (overlap.) | 72.4, CH |
| 4 | 3.55 (m) | 72.9, CH | 5.51 (dd, 15.0, 7.0) | 130.9, CH |
| 5 | 1.44 (m) | 32.6, CH | 5.77 (dt, 15.0, 7.0) | 134.3, CH |
| 6 | 2.10 (m) | 33.3, CH2 | ||
| 7 | 2.11 (m) | 33.4, CH2 | ||
| 8, 9 | 5.45 (m) | 131.0, CH | ||
| 10 | 2.00 (m) | 34.8, CH2 | ||
| 11 | 1.37 (m) | 23.9, CH2 | ||
| 1′ | 176.5, C | 176.5, C | ||
| 2′ | 4.05 (dd, 8.0, 4.2) | 72.7, CH | 4.05 (dd, 8.0, 4.2) | 72.7, CH |
| 3′ | 1.64 (m) | 35.2, CH2 | 1.64 (m) | 35.2, CH2 |
| 4′ | 1.30 (m) | 30.0, CH2 | 1.30 (m) | 30.0, CH2 |
| 1″ | 4.31 (d, 7.7) | 104.1, CH | 4.31 (d, 7.7) | 104.1, CH |
| 2″ | 3.21 (m) | 74.8, CH | 3.21 (m) | 74.8, CH |
| 3″ | 3.38 (m) | 77.5, CH | 3.38 (m) | 77.5, CH |
| 4″ | 3.30 (m) | 71.7, CH | 3.30 (m) | 71.7, CH |
| 5″ | 3.31 (m) | 77.3, CH | 3.31 (m) | 77.3, CH |
| 6″ | 3.70 (dd, 12.0, 2.0) 3.90 (dd, 12.0, 3.0) | 62.5, CH2 | 3.70 (dd, 12.0, 2.0) 3.90 (dd, 12.0, 3.0) | 62.5, CH2 |
| Position | δH (m, J in Hz) | δC, Type |
|---|---|---|
| 1 | 3.74 (dd, 11.0, 3.7) 3.62 (m) | 61.8, CH2 |
| 2 | 2.96 (m) | 58.4, CH |
| 3 | 3.66 (m) | 69.7, CH |
| 4 | 1.54–1.40 (m) | 35.0, CH2 |
| 5 | 1.56–1.40 (m) | 38.0, CH2 |
| 6 | 4.39 (m) | 67.8, CH |
| 7 | 5.35 (dd, 10.0, 9.0) | 134.2, CH |
| 8 | 5.44 (overlap.) | 133.0, CH |
| 9 | 2.08 (m) | 28.6, CH2 |
| 10–17 | 1.40–1.25 | |
| 18 | 0.90 (t, 7.0) | 14.0, CH3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruocco, N.; Nuzzo, G.; Federico, S.; Esposito, R.; Gallo, C.; Ziaco, M.; Manzo, E.; Fontana, A.; Bertolino, M.; Zagami, G.; et al. Potential of Polar Lipids Isolated from the Marine Sponge Haliclona (Halichoclona) vansoesti against Melanoma. Int. J. Mol. Sci. 2024, 25, 7418. https://doi.org/10.3390/ijms25137418
Ruocco N, Nuzzo G, Federico S, Esposito R, Gallo C, Ziaco M, Manzo E, Fontana A, Bertolino M, Zagami G, et al. Potential of Polar Lipids Isolated from the Marine Sponge Haliclona (Halichoclona) vansoesti against Melanoma. International Journal of Molecular Sciences. 2024; 25(13):7418. https://doi.org/10.3390/ijms25137418
Chicago/Turabian StyleRuocco, Nadia, Genoveffa Nuzzo, Serena Federico, Roberta Esposito, Carmela Gallo, Marcello Ziaco, Emiliano Manzo, Angelo Fontana, Marco Bertolino, Giacomo Zagami, and et al. 2024. "Potential of Polar Lipids Isolated from the Marine Sponge Haliclona (Halichoclona) vansoesti against Melanoma" International Journal of Molecular Sciences 25, no. 13: 7418. https://doi.org/10.3390/ijms25137418
APA StyleRuocco, N., Nuzzo, G., Federico, S., Esposito, R., Gallo, C., Ziaco, M., Manzo, E., Fontana, A., Bertolino, M., Zagami, G., Zupo, V., Sansone, C., & Costantini, M. (2024). Potential of Polar Lipids Isolated from the Marine Sponge Haliclona (Halichoclona) vansoesti against Melanoma. International Journal of Molecular Sciences, 25(13), 7418. https://doi.org/10.3390/ijms25137418














